Key Points
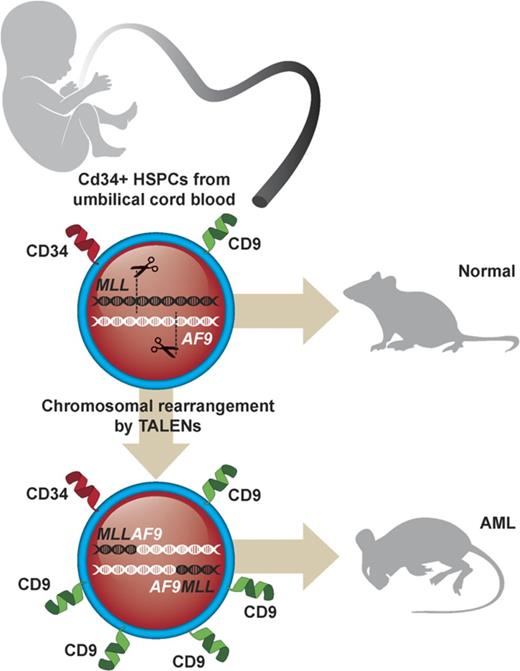
Genome editing induces t(9;11) chromosomal translocations and transforms primary CD34+ human cord blood cells leading to acute leukemia.
CD9 is upregulated in primary t(9;11) cells and is a useful marker for enrichment of genome-edited MLL-rearranged cells in vitro.
Abstract
Genome editing provides a potential approach to model de novo leukemogenesis in primary human hematopoietic stem and progenitor cells (HSPCs) through induction of chromosomal translocations by targeted DNA double-strand breaks. However, very low efficiency of translocations and lack of markers for translocated cells serve as barriers to their characterization and model development. Here, we used transcription activator-like effector nucleases to generate t(9;11) chromosomal translocations encoding MLL-AF9 and reciprocal AF9-MLL fusion products in CD34+ human cord blood cells. Selected cytokine combinations enabled monoclonal outgrowth and immortalization of initially rare translocated cells, which were distinguished by elevated MLL target gene expression, high surface CD9 expression, and increased colony-forming ability. Subsequent transplantation into immune-compromised mice induced myeloid leukemias within 48 weeks, whose pathologic and molecular features extensively overlap with de novo patient MLL-rearranged leukemias. No secondary pathogenic mutations were revealed by targeted exome sequencing and whole genome RNA-sequencing analyses, suggesting the genetic sufficiency of t(9;11) translocation for leukemia development from human HSPCs. Thus, genome editing enables modeling of human acute MLL-rearranged leukemia in vivo, reflecting the genetic simplicity of this disease, and provides an experimental platform for biological and disease-modeling applications.
Introduction
Leukemias constitute a heterogeneous group of diseases defined by their diverse molecular abnormalities, which dictate disease pathogenesis, treatment response, and clinical prognosis.1 The MLL/KMT2A proto-oncogene is a frequent target for chromosomal translocations in a subset of acute leukemia that is generally associated with a poor prognosis.2 The roles of MLL fusion proteins have been investigated in various model systems, each of which has specific technical advantages as well as limitations with uncertain implications for human MLL leukemia pathogenesis.3-8 Recently, we used custom nucleases for genome editing to activate MLL oncogenes in primary human hematopoietic stem and progenitor cells (HSPCs) that generated leukemia after transplantation in mice.9 Although this knock-in approach recapitulated many features of the clinical disease presented in patients, it did not reconstitute all genetic aspects of the disease context (eg, lacking the reciprocal product of the translocation).9 However, induction of reciprocal MLL-AF4 and MLL-AF9 translocations in vitro resulted in translocated cell frequencies that were extremely low, and the cells died out during long-term culture and failed to induce leukemia in mice.10 These findings are supported by other studies showing that the generation of translocations using human primary cells is not robust, with detectable translocation frequencies between 1 × 10−3 and 1 × 10−5 independent of the type of nucleases employed for genome editing.11-14 Despite the increasing feasibility of using novel genome editing tools to generate alterations in primary human HSPCs mimicking the nature of patient disease, it remains difficult to achieve sufficient numbers due to both low efficiency as well as the lack of marker proteins that allow for selective recognition and analysis of the translocated cells.
In this report, we employed non–virally expressed transcription activator-like effector nucleases (TALENs) to specifically engineer reciprocal chromosomal translocations of the MLL and AF9 genes in primary human HSPCs that induced myeloid leukemias in transplanted mice and shared pathologic and molecular features with patient MLL-rearranged (MLLr) acute myeloid leukemia (AML). Our results demonstrate that optimized culture conditions and surface markers expressed preferentially on the translocated cells can overcome the existing limitations in genome editing techniques to induce chromosomal translocations in primary human HSPCs and develop reliable in vivo models of human MLLr leukemia.
Methods
TALENs
Cell culture and nucleofection
CD34+ HSPCs were isolated from fresh human umbilical cord blood (huCB) obtained from the maternity ward of Stanford Hospital (under an institutional review board–approved research protocol) and maintained as previously described,9 with the following changes: granulocyte colony-stimulating factor (G-CSF) (50 ng/mL, PeproTech) and UM729 (0.75 μM, STEMCELL Technologies) were added into the culture media. A total of 300 000 CD34+ cells were nucleofected in one reaction, and the viability (30% to 50%) and nucleofection efficiency (40% to 70% GFP positive) were measured by flow cytometry 2 days after nucleofection. Nucleofected cells were incubated at 37°C, 5% CO2 in serum-free media (StemSpan II) plus cytokines, 20 μM Z-Vad-FMK (Enzo Life Sciences), and Rho-associated kinase pathway inhibitor/thiazovivin (STEMCELL Technologies), and 20% filtered fetal bovine serum was added 48 hours later. Viable cell counts were assessed 2 to 3 times per week, and cultures were split into fresh media to maintain a cell density of 7.5 × 105 cells/mL.
Humanized MLL-AF9 leukemia model
Monoclonal MLL-AF9 translocated cells were transplanted into immune-compromised NOD.Cg-PrkdcscidIL2rgtm1Wjl/SzJ (NSG) mice as previously described.9 Primary recipient female NSG mice were sublethally irradiated (200 cGy), and xenotransplants were performed either by IV injection with 2 × 106 or intrafemoral injection with 1 × 106MLL-AF9–rearranged cells. Human cell engraftment in the bone marrow was determined by the detection of human CD45 by flow cytometry. All experiments on mice were performed with the approval of and in accordance with the Stanford University Administrative Panel on Laboratory Animal Care.
PCR for translocation detection and sequencing
For each polymerase chain reaction (PCR), 150 ng genomic DNA was used with the translocation-specific primers listed in supplemental Table 1. The following PCR parameters were used: denature at 95°C for 5 min (denature at 95°C for 30 s and anneal at 60°C for 45 s, with extension at 72°C for 1 min) for 35 cycles, and final extension at 72°C for 5 min. PCR products were visualized on a 1% agarose gel with ethidium bromide. PCR products were extracted from the gel (QIAGEN), cloned into the pCRII-Blunt-TOPO vector (Invitrogen), and transformed into competent cells. Plasmid DNA of the transformed competent cells was subjected to Sanger sequencing using the M13 forward and M13 reverse primers.
RT-PCR and qPCR
RNA was isolated using the RNeasy Mini Kit (QIAGEN) and used to generate complementary DNA using the SuperScript III First-Strand Synthesis System (Invitrogen). PCR was subsequently performed for MLL-AF9 and AF9-MLL fusion transcripts with the reverse transcription polymerase chain reaction (RT-PCR) primers listed in supplemental Table 1. PCR products were visualized, gel extracted, and subjected to Sanger sequencing. Quantitative PCR (qPCR) was performed for detection of target genes MEIS1 (HS00180020_m1), HOXA6 (HS00430615_m1), HOXA9 (HS00266821_m1), and HOXA10 (HS00365956_m1) by qPCR TaqMan Gene Expression Assays (Life Technologies).15 qPCR was performed in triplicate followed by melting curve analysis in the Bio-Rad CFX384 C1000 Real-Time System. Cq values of undetectable transcripts were artificially set to the maximum amplification cycle numbers. Results were normalized to the housekeeping gene ACTB and then compared with the value of Mono Mac-6.
Flow cytometry and FACS
Analyses were performed using an LSR II flow cytometer (BD Biosciences). Fluorescence-activated cell sorting (FACS) was performed by using a FACS Aria (BD Biosciences) and FACS DIVA software (BD Biosciences). Data were analyzed using FlowJo volume 10 (Tree Star). For analysis, the following fluorochrome-conjugated monoclonal antibodies were used: CD34-allophycocyanin (clone 4H11, eBioscience), CD38-phycoerythrin /Cy7 (clone HIT2, BioLegend), CD33-PerCP/Cy5.5 (clone WM53, BioLegend), CD14-Alexa Fluor 700 (clone HCD14, BioLegend), CD64-allophycocyanin/Cy7 (clone 10.1, BioLegend), CD117-brilliant violet 421 (clone104D2, BioLegend), CD15-PerCP/Cy5.5 (clone W6D3, BioLegend), and CD9-phycoerythrin (clone eBioSN4/SN4 C3-3A2, eBioscience)
Results
TALEN-mediated genome editing of primary human HSPCs induces t(9;11) chromosomal translocations and MLL fusion gene expression
To induce t(9;11) translocations in HSPCs, CD34+ cells isolated from huCB were nucleofected with 4 plasmids expressing 2 sets of TALENs targeting MLL or AF9, respectively (supplemental Figure 1). Plasmids encoding GFP or TALENs targeting only MLL were used as controls. The TALENs were designed to cleave intron 11 of the MLL gene and intron 5 of the AF9 gene, respectively, corresponding to the genomic regions of frequent chromosomal translocation breakpoints in patients with MLL-AF9 leukemias.3,9-10 After nucleofection, the unselected cell population was maintained in vitro in liquid culture (∼0.75 × 106/mL) supplemented with cytokines (stem cell factor [SCF], thrombopoietin, FLT3L, interleukin-6 [IL-6], IL-3, and G-CSF) and aryl hydrocarbon receptor antagonists (SR1 and UM729) optimized for growth of normal human HSPCs.16-17
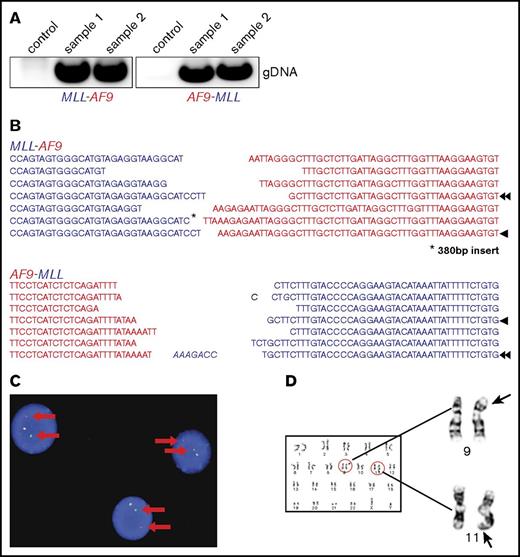
MLL-AF9 and AF9-MLL fusion junctions were detected by PCR in 4 out of 8 TALEN-treated samples, 2 of which showed long-term outgrowth (Figure 1A) during extended in vitro culture. Sanger sequencing demonstrated that the PCR products in early cultures (days 7-14) constituted a heterogeneous mixture of several fusion sequences (Figure 1B). To confirm the chromosomal rearrangements and quantify the percentage of cells with translocations, fluorescence in situ hybridization (FISH) and karyotype analyses were performed. On day 54 in liquid culture, an MLL break-apart probe detected MLL translocations in 96% and 78% of the genome-edited cells, respectively, in 2 independent cultures (Figure 1C), which increased to 100% by day 76. G-banding analyses demonstrated the presence of both derivative chromosomes 9 and 11 resulting from reciprocal t(9;11) translocation (Figure 1D).
Molecular and cytogenetic features of MLL-AF9–rearranged cells induced by genome editing. (A) Representative PCR to detect MLL-AF9 (left) and AF9-MLL (right) translocation breakpoints in gene-edited cells at day 27 of culture (samples 1 and 2). (B) Data shown are a composite alignment of PCR products from multiple experiments (days 7-14 of culture) showing a variety of distinct translocations. Arrowheads indicate the sequence of long-term–survived clones. (C) FISH analysis using an MLL break-apart probe was performed on genome-edited cells maintained in culture for over 50 days. A representative image is shown. Arrows indicate the split signals of the break-apart probe indicating MLL translocation. (D) Representative metaphase chromosomes from karyotype analysis of genome-edited cells shows a balanced t(9;11) chromosomal translocation.
Molecular and cytogenetic features of MLL-AF9–rearranged cells induced by genome editing. (A) Representative PCR to detect MLL-AF9 (left) and AF9-MLL (right) translocation breakpoints in gene-edited cells at day 27 of culture (samples 1 and 2). (B) Data shown are a composite alignment of PCR products from multiple experiments (days 7-14 of culture) showing a variety of distinct translocations. Arrowheads indicate the sequence of long-term–survived clones. (C) FISH analysis using an MLL break-apart probe was performed on genome-edited cells maintained in culture for over 50 days. A representative image is shown. Arrows indicate the split signals of the break-apart probe indicating MLL translocation. (D) Representative metaphase chromosomes from karyotype analysis of genome-edited cells shows a balanced t(9;11) chromosomal translocation.
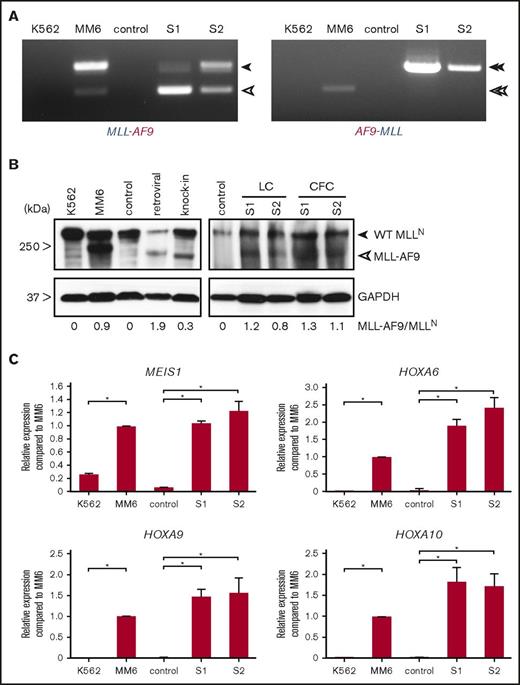
RT-PCR analysis revealed the expression of MLL-AF9 fusion transcripts as in the MLL-AF9 human leukemia cell line Mono Mac 6 vs non-MLLr control cells (Figure 2A). Two alternatively transcribed fusion transcripts were detected as previously reported in MLL-AF9 cell lines and patient samples.18-19 Western blot analysis demonstrated the presence of MLL-AF9 fusion proteins at comparable levels to the MLL-AF9 cell line (Figure 2B). Similarly, the genome-edited MLL-AF9 cells demonstrated increased expression levels of common MLL target genes (MEIS1, HOXA6, HOXA9, and HOXA10) compared with control cells (Figure 2C). These results indicate that TALEN-mediated genome editing targeting MLL and AF9 creates t(9;11) chromosomal translocations, which result in MLL-AF9 gene fusions, which in turn induce MLL target gene expression at levels comparable to human leukemia cell lines harboring similar chromosomal translocations.
Transcriptional features of genome-edited MLL-AF9 translocated cells. (A) RT-PCR was performed on complementary DNA of genome-edited MLLr cells from 2 independent cultures to detect fusion transcripts. Closed arrowheads indicate MLL-AF9 and AF9-MLL fusion transcripts. Open arrowheads indicate the alternatively processed fusion transcripts, which lack MLL exon 11 and MLL exon 12 from MLL-AF9 and AF9-MLL, respectively. (B) Representative western blot analysis of MLL proteins in controls (nucleofected with GFP alone) and MLL-AF9–rearranged cells cultured either in liquid or semisolid medium. Retroviral and knock-in indicate transformed human cord blood cells by retroviral MLL-AF9 or MLL-AF9 knock-in. CFC, colony-formed cells; GAPDH, loading control; LC, liquid culture. Numbers below indicate relative MLL-AF9 band intensities compared with WT MLLN. (C) Representative qPCR analyses show expression levels of MLL target genes compared with control (nucleofected with GFP) at day 65 of culture and human leukemia cell lines. *P < .02. Error bars indicate standard deviation of triplicate analyses. MM6, Mono Mac 6; S1, sample 1; S2, sample 2; WT, wild-type.
Transcriptional features of genome-edited MLL-AF9 translocated cells. (A) RT-PCR was performed on complementary DNA of genome-edited MLLr cells from 2 independent cultures to detect fusion transcripts. Closed arrowheads indicate MLL-AF9 and AF9-MLL fusion transcripts. Open arrowheads indicate the alternatively processed fusion transcripts, which lack MLL exon 11 and MLL exon 12 from MLL-AF9 and AF9-MLL, respectively. (B) Representative western blot analysis of MLL proteins in controls (nucleofected with GFP alone) and MLL-AF9–rearranged cells cultured either in liquid or semisolid medium. Retroviral and knock-in indicate transformed human cord blood cells by retroviral MLL-AF9 or MLL-AF9 knock-in. CFC, colony-formed cells; GAPDH, loading control; LC, liquid culture. Numbers below indicate relative MLL-AF9 band intensities compared with WT MLLN. (C) Representative qPCR analyses show expression levels of MLL target genes compared with control (nucleofected with GFP) at day 65 of culture and human leukemia cell lines. *P < .02. Error bars indicate standard deviation of triplicate analyses. MM6, Mono Mac 6; S1, sample 1; S2, sample 2; WT, wild-type.
Genome-edited MLL-AF9-rearranged cells display distinct cytokine requirements for proliferation and maturation arrest
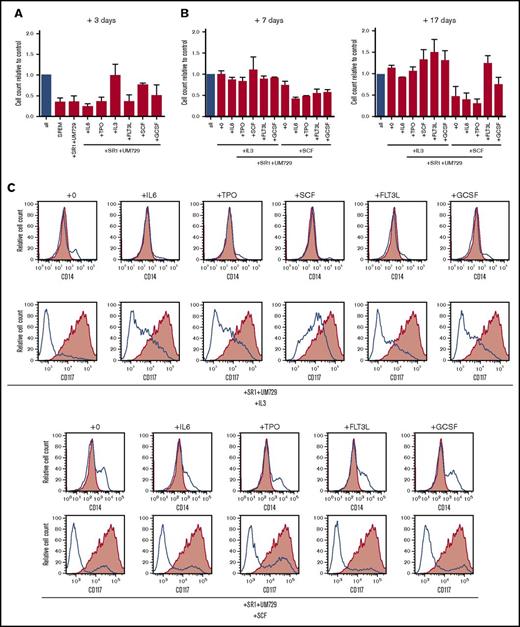
To assess their cytokine requirements, proliferation of the translocated cells was measured after three days culture in the presence of aryl hydrocarbon receptor inhibitors SR-1 and UM729 with single cytokines. Under these conditions, only IL-3 or SCF alone supported robust proliferation (Figure 3A). To further assess the necessary combination of cytokines, cells were cultured for an additional 7 and 17 days (total of 27 days) in cytokine milieus containing IL-3 or SCF and additionally one of the remaining cytokines of the standard culture conditions (Figure 3B). Whereas IL-3 alone was sufficient to sustain proliferation, it was further increased by the addition of SCF, FLT3L, or G-CSF. Conversely, SCF alone was not sufficient to support proliferation in long-term culture but was partially rescued by the addition of G-CSF and completely rescued by the addition of FLT3L, indicating important roles for both cytokines. Interestingly, cells cultured with SCF alone showed not only a decrease of cell proliferation but also an increase in cell differentiation after 27 days of culture, indicated by the loss of immature marker CD117 and gain of maturation marker CD14 (Figure 3C). Whereas IL-3 was the major driving factor for proliferation, the inclusion of other cytokines present in our standard culture conditions was necessary to prevent maturation. These results demonstrate that MLL-AF9 translocated cells remain cytokine dependent for in vitro proliferation and maturation arrest.
MLL-AF9 translocated cells display specific cytokine dependence profiles. (A) MLL-AF9–rearranged cells cultured in the presence of SR-1 and UM729 plus the indicated single cytokines were quantified by trypan blue dye exclusion after 3 days and compared with control (all cytokines, blue bar). (B) Cells cultured with IL-3 or SCF were additionally cultured for 7 and 17 days in the presence of SR-1, UM729, the respective cytokines IL-3 or SCF, and the indicated single cytokine to evaluate their sensitivity compared with the control (all cytokines, blue bar). Error bars indicate SEM for 2 analyses. (C) Flow cytometry profiles show representative phenotypes of cells cultured in the indicated cytokine milieus after 27 days (blue lines) vs cells culture in the complete cytokine cocktail (red shading). TPO, thrombopoietin.
MLL-AF9 translocated cells display specific cytokine dependence profiles. (A) MLL-AF9–rearranged cells cultured in the presence of SR-1 and UM729 plus the indicated single cytokines were quantified by trypan blue dye exclusion after 3 days and compared with control (all cytokines, blue bar). (B) Cells cultured with IL-3 or SCF were additionally cultured for 7 and 17 days in the presence of SR-1, UM729, the respective cytokines IL-3 or SCF, and the indicated single cytokine to evaluate their sensitivity compared with the control (all cytokines, blue bar). Error bars indicate SEM for 2 analyses. (C) Flow cytometry profiles show representative phenotypes of cells cultured in the indicated cytokine milieus after 27 days (blue lines) vs cells culture in the complete cytokine cocktail (red shading). TPO, thrombopoietin.
TALEN-induced chromosomal translocations in primary CD34+ cells confer enhanced long-term proliferation and survival in vitro
In long-term culture, the numbers of gene-edited MLL-AF9 cells progressively increased while control cell proliferation peaked at around day 40 and decreased afterward (Figure 4A). Similarly, viability of control cells declined after day 40, whereas MLL-AF9 cells maintained their viability (Figure 4B). PCR analysis of DNA from the cultures of gene-edited cells showed an extremely low level of MLL-AF9 and AF9-MLL fusion junctions in early cultures (day 7) with progressive increase as culture time proceeded (Figure 4C). Sanger sequencing of PCR bands at day 40 revealed that long-term cultures typically contained one distinct translocation product for the MLL-AF9 fusion and the reciprocal AF9-MLL fusion, respectively (Figure 1B, arrowheads), indicating that a single clone of cells with the MLL-AF9 translocation outgrew in each of the cultures.
Genome-edited primary CD34+cells display survival advantage and clonal expansion in vitro. (A) Representative growth curves chart the differences in proliferative capacity of genome-edited CD34+ cells compared with controls (GFP or MLL TALENs alone). Arrows indicate time point of FISH analyses and karyotyping. (B) Graph shows cell viability in liquid culture monitored over time by flow cytometry and further confirmed by trypan blue dye exclusion. (C) PCR was performed for MLL-AF9 and reciprocal AF9-MLL breakpoints on genomic DNA over progressive time of culture. (D) Representative morphologies are shown for control and translocated cells on day 76 by May-Grünwald-Giemsa staining. Arrowheads indicate differentiating cells. Scale bar, 20 μm. (E) Colony-forming assays were performed on day 60 of liquid culture. Bars represent the mean number of colony-forming units (CFUs) per 104 seeded cells. (F) Plot indicates cell numbers after each replating. Experiments were performed in triplicate, and data from 2 independent experiments are shown. (G) Images show representative morphologies of colonies after each replating (R). Scale bar, 200 μm.
Genome-edited primary CD34+cells display survival advantage and clonal expansion in vitro. (A) Representative growth curves chart the differences in proliferative capacity of genome-edited CD34+ cells compared with controls (GFP or MLL TALENs alone). Arrows indicate time point of FISH analyses and karyotyping. (B) Graph shows cell viability in liquid culture monitored over time by flow cytometry and further confirmed by trypan blue dye exclusion. (C) PCR was performed for MLL-AF9 and reciprocal AF9-MLL breakpoints on genomic DNA over progressive time of culture. (D) Representative morphologies are shown for control and translocated cells on day 76 by May-Grünwald-Giemsa staining. Arrowheads indicate differentiating cells. Scale bar, 20 μm. (E) Colony-forming assays were performed on day 60 of liquid culture. Bars represent the mean number of colony-forming units (CFUs) per 104 seeded cells. (F) Plot indicates cell numbers after each replating. Experiments were performed in triplicate, and data from 2 independent experiments are shown. (G) Images show representative morphologies of colonies after each replating (R). Scale bar, 200 μm.
Consistent with this, MLL-AF9–rearranged cells displayed immature morphologies in contrast to the differentiated control cells (Figure 4D). Following transfer at day 60 from liquid to semisolid medium supplemented with the same cytokines as the liquid culture, MLL-AF9 translocated cells formed predominantly compact colonies that increased in size and numbers as well as cell counts after each round of plating, whereas control cells were unable to form compact colonies and failed to replate (Figure 4E-G). These studies demonstrate that generation of MLL-AF9 chromosomal translocations by genome editing stimulates the long-term growth and survival of primary human HSPCs.
MLL-AF9 translocated cells display immature phenotypes and preferentially express CD9
The clonal cells bearing t(9;11) chromosomal translocations displayed immature myelomonocytic phenotypes with expression of CD34 and CD117 with virtually no expression of the mature marker CD14, whereas control cells lacked immature markers and displayed high expression of CD14, indicating that in long-term liquid culture conditions, genome-edited CD34+ cells had markedly impaired terminal myeloid differentiation (Figure 5A).
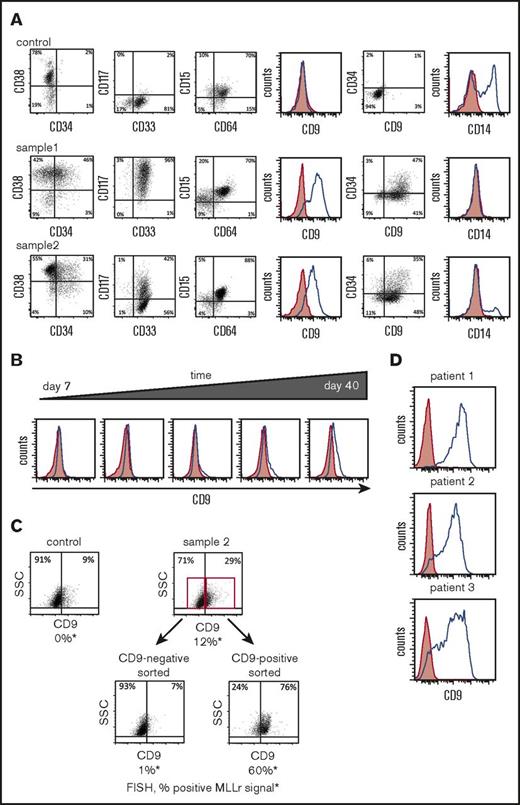
MLL-AF9 translocated cells preferentially express CD9. (A) Representative flow cytometry analyses are shown for cell-surface protein expression of genome-edited MLLr cells compared with control cells (CD34+ cells nucleofected with GFP alone) at day 65. (B) Representative flow cytometry plots of CD9 expression over time in culture for genome-edited MLLr cells. (C) Representative flow cytometry plots before and after FACS sorting of genome-edited MLLr cells based on CD9 expression at day 37 of in vitro culture followed by FISH analysis for MLL translocation. (D) Representative flow cytometry plots of CD9 expression on MLLr leukemic patient samples. Red shading indicates control (fluorescence minus one); blue line denotes expression of indicated marker. SSC, side scatter.
MLL-AF9 translocated cells preferentially express CD9. (A) Representative flow cytometry analyses are shown for cell-surface protein expression of genome-edited MLLr cells compared with control cells (CD34+ cells nucleofected with GFP alone) at day 65. (B) Representative flow cytometry plots of CD9 expression over time in culture for genome-edited MLLr cells. (C) Representative flow cytometry plots before and after FACS sorting of genome-edited MLLr cells based on CD9 expression at day 37 of in vitro culture followed by FISH analysis for MLL translocation. (D) Representative flow cytometry plots of CD9 expression on MLLr leukemic patient samples. Red shading indicates control (fluorescence minus one); blue line denotes expression of indicated marker. SSC, side scatter.
Similar to the previous reports that CD9 is preferentially expressed on MLLr acute lymphoblastic leukemia (ALL) cells, but not normal cells from the same patient,20 most of the MLL-AF9 translocated cells expressed CD9 irrespective of CD34 status, in contrast to control cells, which lacked CD9 expression (Figure 5A). CD9 expression progressively increased over time in culture comparable to PCR band intensity of MLL-AF9 and reciprocal AF9-MLL fusions (compare Figure 4C and Figure 5B), suggesting a positive correlation between CD9 expression and translocation.
To assess the relationship between t(9;11) chromosomal rearrangement and CD9 expression, CD9-positive and CD9-negative cells on day 36 of culture were sorted by FACS and subjected to FISH analysis. Unsorted cells showed a FISH signal in 12% of cells, whereas ∼60% of CD9-positive sorted cells were FISH positive compared with 1% of CD9-negative sorted cells (Figure 5C). Colony-forming assays also showed a substantial enrichment of clonogenic cells in the CD9-positive versus negative MLLr populations (supplemental Figure S2A), indicating a positive correlation between self-renewal and CD9 expression of MLL-AF9 translocated cells.
CD9 is highly expressed on primary MLLr patient AML cells (Figure 5D) and was increased in MLLr AMLs compared with non-MLLr AMLs in previously published datasets (supplemental Figure 2C).21-22 In contrast, only a small minority of mononuclear cells from bone marrow and cord blood of healthy donors expressed CD9, which was predominantly restricted to the CD34− population (supplemental Figure 2B). Thus, acute induction of MLL-AF9 chromosomal translocation antagonizes differentiation of CD34+ cells in prolonged in vitro culture and presents CD9 as a surface marker for their identification and enrichment
MLL-AF9 translocated cells created by genome editing induce AML without evidence of secondary genetic mutations
Immune-compromised NSG mice were transplanted with genome-edited cells harvested from cultures containing 100% t(9;11) cells by FISH analysis (Figures 4A and 6A). The cells were transplanted IV (2 × 106) or intrafemorally (1 × 106) directly from liquid media.9 Cells engrafted into recipient mice (tail vein 72.7%, n = 11; femur 37.5%, n = 8; Table 1) and developed leukemia with a mean latency of ∼48 weeks (∼37-60 weeks, n = 10; Figure 6B; Tables 1 and 2).
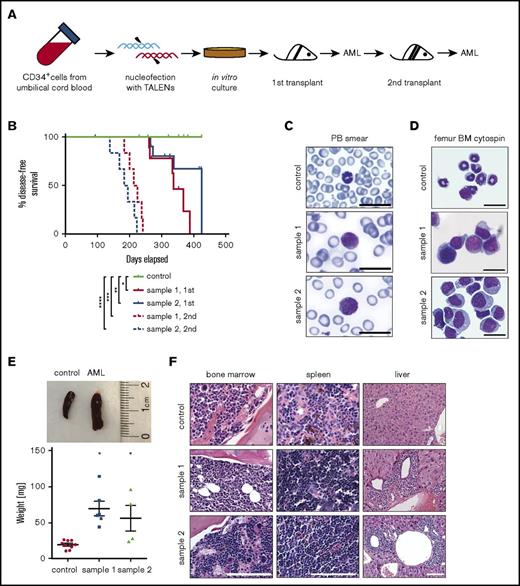
Genome-edited cells with t(9;11) chromosomal translocations induce AML. (A) Experimental scheme for induction of AML by genome-edited cells following transplantation in sublethally irradiated NSG recipient mice after 98 days of in vitro culture. (B) Disease-free survival of transplanted mice; *P < .009 (control vs sample 1, 1st), **P < .07 (control vs sample 2, 1st), ***P < .0001 (control vs sample 1, 2nd), and ****P < .0001 (control vs sample 2, 2nd) by log-rank test. (C-D) May-Grünwald-Giemsa staining shows blast cells in peripheral blood (PB) smear (C) and monomorphic bone marrow (BM) cells (D). Scale bar, 20 μm. (E) Increased spleen size (top) and weight (bottom) of leukemic mice compared with control; *P < .01. (F) Hematoxylin and eosin–stained sections of bone marrow, spleen, and liver. Scale bar, 100 μm.
Genome-edited cells with t(9;11) chromosomal translocations induce AML. (A) Experimental scheme for induction of AML by genome-edited cells following transplantation in sublethally irradiated NSG recipient mice after 98 days of in vitro culture. (B) Disease-free survival of transplanted mice; *P < .009 (control vs sample 1, 1st), **P < .07 (control vs sample 2, 1st), ***P < .0001 (control vs sample 1, 2nd), and ****P < .0001 (control vs sample 2, 2nd) by log-rank test. (C-D) May-Grünwald-Giemsa staining shows blast cells in peripheral blood (PB) smear (C) and monomorphic bone marrow (BM) cells (D). Scale bar, 20 μm. (E) Increased spleen size (top) and weight (bottom) of leukemic mice compared with control; *P < .01. (F) Hematoxylin and eosin–stained sections of bone marrow, spleen, and liver. Scale bar, 100 μm.
Recipient mice transplanted with MLL-AF9 translocated cells
| Injection method (no. of cells) . | Injected cells . | No. engrafted/no. of mice . | AML/no. engrafted . |
|---|---|---|---|
| IV (2 × 106) | Control | 0/5 | 0 |
| Sample 1 | 4/6 | 4/4 | |
| Sample 2 | 4/5 | 4/4 | |
| Intrafemural (1 × 106) | Control | 0/4 | 0 |
| Sample 1 | 2/3 | 2/2 | |
| Sample 2 | 1/5 | 0/1 |
| Injection method (no. of cells) . | Injected cells . | No. engrafted/no. of mice . | AML/no. engrafted . |
|---|---|---|---|
| IV (2 × 106) | Control | 0/5 | 0 |
| Sample 1 | 4/6 | 4/4 | |
| Sample 2 | 4/5 | 4/4 | |
| Intrafemural (1 × 106) | Control | 0/4 | 0 |
| Sample 1 | 2/3 | 2/2 | |
| Sample 2 | 1/5 | 0/1 |
Pathological features of leukemias arising in mice transplanted with rearranged cells
| Mouse number . | Injected cells . | Latency, d . | BM hCD45, % . | BM hCD34, % . | SP hCD45, % . | LV hCD45, % . | PB hCD45, % . | SP wt, mg . | LV wt, mg . | Secondary transplant . |
|---|---|---|---|---|---|---|---|---|---|---|
| S1-AML1 | S1 | 259 | 92.4 | 53.8 | ND | ND | ND | 44.0 | ND | ND |
| S1-AML2 | S1 | 335 | 82.5 | 30.2 | 9.5 | ND | ND | 61.6 | 865.0 | 1/2 |
| S1-AML3 | S1 | 386 | 93.3 | 51.6 | 14.27 | 15.9 | ND | 55.6 | 1331.8 | ND |
| S1-AML4 | S1 | 367 | 98.7 | 19.3 | 22.79 | 17.8 | 8.88 | 114.3 | 1380.9 | 2/2 |
| S1-AML5 | S1 | 331 | 94.72 | 18.1 | 52.2 | 7.9 | 18.6 | 80.8 | 1035.2 | 2/2 |
| S1-AML6 | S1 | 261 | 84.24 | 10.9 | ND | ∼1.0 | ND | 60 | 3460 | ND |
| S2-AML1 | S2 | 265 | 94.6 | 34.7 | 10.2 | ND | ND | 96.0 | 1140 | 3/3 |
| S2-AML2 | S2 | 272 | 87.4 | 43.7 | 3.0 | 0.4 | ND | 23.6 | 581.6 | 1/2 |
| S2-AML3 | S2 | 336 | 66.5 | 15.6 | 1.9 | 0.5 | ND | 75.8 | 2367 | 2/2 |
| S2-AML4 | S2 | 423 | 54.9 | 34.9 | ND | ND | ND | 27.7 | ND | ND |
| Mouse number . | Injected cells . | Latency, d . | BM hCD45, % . | BM hCD34, % . | SP hCD45, % . | LV hCD45, % . | PB hCD45, % . | SP wt, mg . | LV wt, mg . | Secondary transplant . |
|---|---|---|---|---|---|---|---|---|---|---|
| S1-AML1 | S1 | 259 | 92.4 | 53.8 | ND | ND | ND | 44.0 | ND | ND |
| S1-AML2 | S1 | 335 | 82.5 | 30.2 | 9.5 | ND | ND | 61.6 | 865.0 | 1/2 |
| S1-AML3 | S1 | 386 | 93.3 | 51.6 | 14.27 | 15.9 | ND | 55.6 | 1331.8 | ND |
| S1-AML4 | S1 | 367 | 98.7 | 19.3 | 22.79 | 17.8 | 8.88 | 114.3 | 1380.9 | 2/2 |
| S1-AML5 | S1 | 331 | 94.72 | 18.1 | 52.2 | 7.9 | 18.6 | 80.8 | 1035.2 | 2/2 |
| S1-AML6 | S1 | 261 | 84.24 | 10.9 | ND | ∼1.0 | ND | 60 | 3460 | ND |
| S2-AML1 | S2 | 265 | 94.6 | 34.7 | 10.2 | ND | ND | 96.0 | 1140 | 3/3 |
| S2-AML2 | S2 | 272 | 87.4 | 43.7 | 3.0 | 0.4 | ND | 23.6 | 581.6 | 1/2 |
| S2-AML3 | S2 | 336 | 66.5 | 15.6 | 1.9 | 0.5 | ND | 75.8 | 2367 | 2/2 |
| S2-AML4 | S2 | 423 | 54.9 | 34.9 | ND | ND | ND | 27.7 | ND | ND |
h, human; LV, liver; ND, not determined; SP, spleen; wt, weight.
Transplanted mice with disease symptoms were euthanized for analysis. Leukemic mice shared similar disease profiles characterized by blasts in peripheral blood (Figure 6C), hypercellular bone marrow with human blast cells, myelomonocytic differentiation and occasional megakaryocytic hyperplasia (Figure 6D,F), splenomegaly (Figure 6E), and infiltration of peripheral tissues (Figure 6F). Human cells were only detected in the peripheral blood at a very late stage of disease progression, and the blood panel was not dramatically changed (data not shown), indicating the disease is primarily localized to the bone marrow and spleen. Leukemic bone marrow cells displayed high expression of CD34, CD117, and CD64 and lack of the differentiation marker CD14 and were CD9 positive at levels similar to injected cells (supplemental Figure 3; compare with Figure 5A). PCR of genomic DNA followed by Sanger sequencing showed monoclonal breakpoint sequences identical to the injected MLLr cells (supplemental Figure 4A,C; compare with Figure 1B, arrowheads), and RT-PCR confirmed expression of MLL-AF9 and AF9-MLL fusion transcripts (supplemental Figure 4B). Injection of AML bone marrow cells into secondary recipient mice induced AML with a shorter latency (median 28 weeks, n = 12; Figure 6B; supplemental Figure 5). These results demonstrate that genome editing to create balanced t(9;11) chromosomal translocations transforms primary CD34+ human cord blood cells, leading to acute myelomonocytic leukemia.
The latency observed for AML development in primary transplant recipients, and its acceleration following secondary transplantation, suggested the possibility that collaborating mutations may arise during progression of genome-edited cells in vitro or in vivo. To test this, targeted exome sequencing was performed to assess 53 genes, including RAS pathway genes and FLT3-ITD (TruSight Myeloid Sequencing Panel, Illumina) that are often mutated in hematological malignancies, including a subset of MLLr AML (accession number GSE103811).23-25 Analysis of mouse leukemia bone marrow cells (n = 2) compared with the original cord blood cells (n = 1), and in vitro–cultured translocated cells at different time points (n = 6), revealed no pathogenic mutations in either leukemic samples or the in vitro culture cells at any time point (data not shown). This prompted a whole-genome approach based on RNA sequencing to identify potential pathogenic expressed single-nucleotide polymorphisms or indels not detected by the targeted exome approach (GSE103811). No recurrent pathogenic secondary mutations were detected that distinguished the original in vitro genome-edited cells used for transplant (n = 2) from their leukemic counterparts (n = 8 primary leukemia; n = 2 secondary leukemia; supplemental Table 2). Although we cannot exclude that critical additional mutations were not detected in our approach, these results support the possibility that t(9;11) chromosomal translocation is genetically sufficient to induce AML in a human xenograft model.
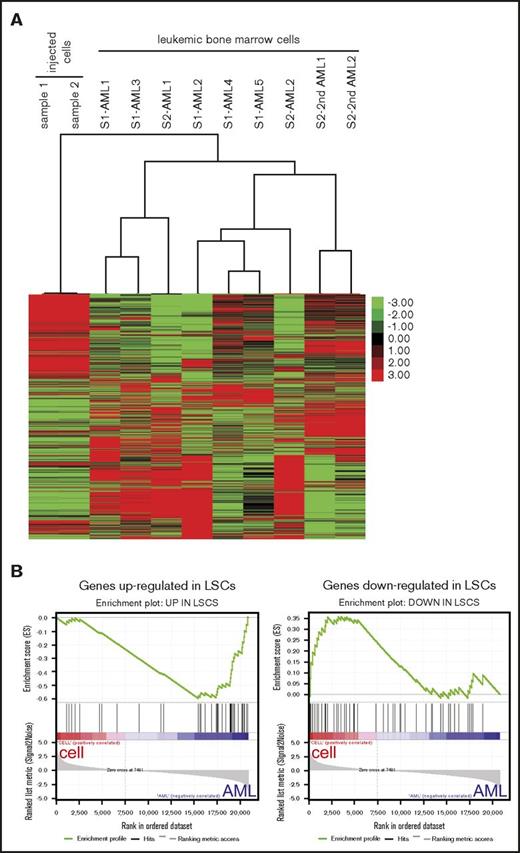
Unsupervised hierarchical cluster analysis of the RNA-sequencing data demonstrated distinctive gene expression patterns of the various AMLs that clustered separately from the genome-edited cell lines from which they were respectively derived (Figure 7A). Analysis of differentially expressed genes revealed that MLLr leukemia signature gene changes, such as upregulation of HOXA cluster and downregulation of HOXB cluster,23,26-28 were more prominent in leukemic bone marrow cells than injected cells (supplemental Figure 6A-B). Furthermore, gene set enrichment analysis29-30 revealed that genes upregulated in human leukemia stem cells (LSCs) were more enriched in the leukemic bone marrow cells compared with the cultured cells, whereas genes downregulated in human LSCs were enriched in the cultured cells used for injection (Figure 7B).31 These data suggested that LSCs may be more abundant in AML in vivo than in cultured genome-edited cells, consistent with much shorter latencies for disease emergence in secondary transplanted mice (Figure 6B). Thus, although t(9;11) may be sufficient for development of AML from genome-edited primary human cells, environmental or nongenetic factors may also influence disease progression.
AML signature gene expression changes in the MLL-AF9 AMLs. (A) Heatmap displays gene expression profiles of MLL-AF9 rearranged cultured cells and their derived leukemic bone marrow cells. Unbiased hierarchical cluster analysis is shown for 7 primary (AML) and 2 secondary (2nd AML) leukemias compared with 2 cultured cell lines used for injection (S1, sample 1; S2, sample 2). (B) Gene set enrichment analysis plots show that genes upregulated in human LSCs were enriched in the AMLs (left, normalized enrichment score = −1.84, false discovery rate q value = 0) whereas genes downregulated in LSCs were enriched in the injected cells (right, normalized enrichment score = 1.33, false discovery rate q value <0.074).
AML signature gene expression changes in the MLL-AF9 AMLs. (A) Heatmap displays gene expression profiles of MLL-AF9 rearranged cultured cells and their derived leukemic bone marrow cells. Unbiased hierarchical cluster analysis is shown for 7 primary (AML) and 2 secondary (2nd AML) leukemias compared with 2 cultured cell lines used for injection (S1, sample 1; S2, sample 2). (B) Gene set enrichment analysis plots show that genes upregulated in human LSCs were enriched in the AMLs (left, normalized enrichment score = −1.84, false discovery rate q value = 0) whereas genes downregulated in LSCs were enriched in the injected cells (right, normalized enrichment score = 1.33, false discovery rate q value <0.074).
Discussion
Using genome editing techniques, we generated t(9;11) chromosomal translocations encoding the MLL-AF9 and reciprocal AF9-MLL fusion products in primary human HSPCs to model the consequences of endogenous oncogene activation in human leukemia. Our studies demonstrate that genome editing is a feasible approach to modify CD34+ huCB cells to induce balanced translocations in a minor fraction of cells, stimulating their long-term clonal outgrowth in supportive cytokine conditions in vitro and promoting leukemia in vivo. Human cells bearing productive translocations driving MLL-AF9 oncogene expression under control of the endogenous MLL promoter, as verified by multiple techniques, induced leukemias in transplanted mice without apparent secondary pathogenic mutations.
Our data are consistent with previous reports showing that genome editing in human cell lines or primary cells can lead to detectable translocations in vitro11-13 but significantly extend prior studies by demonstrating the leukemic potential of cells with engineered translocations. These advances spurred by the availability of novel genetic tools may provide more representative models to recapitulate the early events in leukemogenesis. However, despite the utility of these techniques, their application remains challenging because of low translocation frequencies, although recent refinements in TALEN32 and clustered regularly interspaced short palindromic repeats/Cas933 methods that are less toxic to transfected primary human cells may allow for increased translocation frequencies. Nevertheless, the genome editing approach employed in our studies addressed these limitations in part by using a cytokine milieu based on the requirements of rearranged cells that lead to their monoclonal outgrowth and replacement of normal hematopoietic elements in vitro comparable to the progressive in vivo development of patient leukemia. The monoclonal outgrowths arose from initial oligoclonal populations of genome-edited cells with diverse MLL-AF9 translocation breakpoints, suggesting a competitive advantage but nevertheless not associated with detectable secondary mutations in vitro or in vivo. Because the CD34+ HSPCs used for our studies consisted of a mixed hematopoietic cell population, the monoclonal outgrowths may have resulted from translocations occurring in a “primed” stem or precursor population. Further experiments using different subgroups of primary cells are needed to determine which may be most susceptible to initiation by an MLL rearrangement.
The frequency of cells with chromosomal translocations generated by genome editing is initially exceedingly low (1 in 300 000 nucleofected cells),10 and the inability to specifically mark translocated cells complicates efforts for their isolation and study. Expression of CD9 at increased levels by MLL-AF9 cells may assist in circumventing this limitation. CD9 is a known cell-surface glycoprotein that belongs to the tetraspanin family and is expressed by various hematologic malignancies and solid tumors.34-38 Recently, Aoki et al described CD9 expression on the surface of MLLr ALL cells from patients, whereas normal HSPCs from the same patients did not express CD9.20 Our studies and analysis of public databases (www.oncomine.org) showed relatively high expression of CD9 in MLLr AMLs.21-22 Thus, although CD9 is not exclusively expressed by MLLr leukemia cells, our data suggest that it may serve as a surrogate marker for the detection and possible enrichment of genome-edited HSPCs with t(9;11) from among the much more abundant normal HSPCs in early cultures.
The need for complementing mutations in MLL leukemias has been less clear. Genomic analyses of primary patient leukemias show that most MLL-associated AMLs do not harbor recurrent secondary mutations, consistent with our results,26 whereas other studies claim that a majority of MLLr AML patients have mutations, especially in Ras pathways.24,39 In this study, we did not detect recurrent pathogenic mutations, even at hot spots in the RAS genes. Although, we cannot exclude possible secondary mutations beyond our detection limit and need more samples for better conclusion, our data suggest that MLL rearrangement is sufficient for AML development in cord blood HSPCs. A recent study showed a dynamic relationship between developmental stage and leukemogenic response to MLL fusion, where fetal and neonatal hematopoietic progenitors are more sensitive to MLL-ENL–driven leukemogenesis than adult cells.40 It would be interesting to test the effect of MLL-AF9 translocation in adult hematopoietic progenitors compared with cord blood cells. Nevertheless, our studies clearly demonstrate selection for monoclonal outgrowth of translocated cells in vitro and enrichment of LSCs in vivo. This may reflect the contributions of cell of origin, environmental factors, and/or epigenetic effects as recently discussed for mouse models of MLLr leukemia.41 Our studies provide an experimental platform for further investigation focused on these issues.
Human HSPCs bearing t(9;11) chromosome translocations induced by genome editing developed leukemia, but with relatively long latency. These results differ from those obtained using a genome editing knock-in approach wherein human MLL-AF9/ENL leukemias developed within 2 months9 compared with a median latency of ∼11 months in our current study. This may result in part from the longer duration of in vitro culture to obtain sufficient numbers of translocated cells for xenotransplantation. In previous studies, culture of knock-in cells affected leukemia lineage, inducing AML, ALL, and mixed-phenotype acute leukemia when transduced huCB cells were transplanted without antecedent in vitro culture, whereas only AML was observed after 3 weeks of culture prior to transplantation.9 Restrictive effects of progressive pretransplant culture were also observed following retroviral transduction of MLL-AF9 or MLL-ENL in huCB cells, which initially promoted different human graft phenotypes but eventually extinguished engraftment in immune-deficient mice,5 whereas induction of leukemia in mice by transformed huCB with the same technique after long-term culture was observed in another report.42 Our data suggest that despite the competitive growth advantage of genome-edited t(9;11) cells in long-term culture, the frequency of leukemia-initiating cells or their “stemness” properties may be relatively low, possibly due to cytokine addiction of the cells and the lack of those cytokines in NSG mice. Human HSPCs with engineered MLL chromosomal translocations appear unable to override the antagonistic effects of progressive in vitro culture on leukemogenesis. This further emphasizes the need for earlier detection and potential enrichment of rare translocated cells for improved MLLr leukemia modeling.
In summary, our studies highlight the feasibility of engineering chromosomal translocations at their endogenous loci in primary human cells to induce acute leukemia from initially rare cell populations to model human disease. The genome editing approach demonstrates the ontogeny of MLL rearrangements and overcomes the limitations of creating endogenous MLL translocations in human HSPCs in vitro, providing the basis for further in vivo studies to prospectively study leukemia etiology and pathogenesis.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank the members of Cleary laboratory for constructive discussions, Carmencita E. Nicolas and Jason H. Kurzer for technical assistance, and Norm Cyr for graphical assistance. The authors also thank Carol D. Jones and Linda Gojenola (Stanford Healthcare) and Jung-Wook Park (University of California, Los Angeles) for help with bioinformatics analysis, Dan Voytas (University of Minnesota) for generously providing the TALEN Golden Gate library, and Daniel E. Vega Salazar (Stanford Hospital) for his efforts and assistance in the collection of huCB.
This work was supported in part by grants from the National Institutes of Health, National Cancer Institute (CA116606) (M.L.C.), Alex’s Lemonade Stand Foundation (M.L.C.), Hyundai Hope on Wheels (M.P.), the Dr. Mildred Scheel Stiftung (C.S. and D.S.), the St. Baldrick’s Foundation (E.H.B.), the German Research Foundation (J.D.-A.), and the Lucile Packard Foundation for Children's Health (M.L.C.).
Authorship
Contribution: C.S. and J.J. designed and performed the research, analyzed data, and wrote the manuscript; D.S., I.-S.K., E.H.B., J.D.-A, S.H.K.W., and M.I. performed research and analyzed data; J.L.Z. and M.P. provided fruitful discussions; M.L.C. provided overall guidance; and all authors edited the manuscript for content.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael L. Cleary, Department of Pathology, Lokey Stem Cell Research Building, Room G2034, 1291 Welch Rd, Stanford, CA 94305; e-mail: mcleary@stanford.edu.
References
Author notes
C.S. and J.J. contributed equally to this study.