Key Points
RTEL1 variants associate with AA, idiopathic cytopenias, and hypocellular myelodysplastic syndromes.
Detailed clinical/family history, functional assays, and in silico tools are critical for interpreting the pathogenicity of RTEL1 variants.
Abstract
Biallelic germline mutations in RTEL1 (regulator of telomere elongation helicase 1) result in pathologic telomere erosion and cause dyskeratosis congenita. However, the role of RTEL1 mutations in other bone marrow failure (BMF) syndromes and myeloid neoplasms, and the contribution of monoallelic RTEL1 mutations to disease development are not well defined. We screened 516 patients for germline mutations in telomere-associated genes by next-generation sequencing in 2 independent cohorts; one constituting unselected patients with idiopathic BMF, unexplained cytopenia, or myeloid neoplasms (n = 457) and a second cohort comprising selected patients on the basis of the suspicion of constitutional/familial BMF (n = 59). Twenty-three RTEL1 variants were identified in 27 unrelated patients from both cohorts: 7 variants were likely pathogenic, 13 were of uncertain significance, and 3 were likely benign. Likely pathogenic RTEL1 variants were identified in 9 unrelated patients (7 heterozygous and 2 biallelic). Most patients were suspected to have constitutional BMF, which included aplastic anemia (AA), unexplained cytopenia, hypoplastic myelodysplastic syndrome, and macrocytosis with hypocellular bone marrow. In the other 18 patients, RTEL1 variants were likely benign or of uncertain significance. Telomeres were short in 21 patients (78%), and 3′ telomeric overhangs were significantly eroded in 4. In summary, heterozygous RTEL1 variants were associated with marrow failure, and telomere length measurement alone may not identify patients with telomere dysfunction carrying RTEL1 variants. Pathogenicity assessment of heterozygous RTEL1 variants relied on a combination of clinical, computational, and functional data required to avoid misinterpretation of common variants.
Introduction
The telomeropathies, or telomere diseases, are a group of disorders caused by germline mutations in telomere-associated genes, resulting in excessive telomere attrition, reducing the stem cell pool and regenerative potential, and clinically affecting the bone marrow (BM), lung, liver, and skin, among other tissues.1,2 To date, mutations in DKC1, TERC, TERT, USB1, CTC1, NHP2, NOP10, WRAP53, TINF2, RTEL1, PARN, and ACD, all involved in telomere biology, have been identified in patients with a spectrum of clinical manifestations, from bone marrow failure (BMF) and myelodysplastic syndromes (MDS) to pulmonary fibrosis and cirrhosis.1,3
RTEL1 is a helicase critical to genome integrity, DNA repair, and telomere maintenance: it disassembles the t-loops and G4 quadruplexes (DNA secondary structures) at telomeres required for replication.4,5 Biallelic mutations in RTEL1, either in homozygosity or compound heterozygosity, provoke extreme telomere erosion and severe phenotype, clinically manifested as dyskeratosis congenita (DC) and Hoyeraal-Hreidarsson (HH) syndrome in early childhood.6-10 TERT or TERC haploinsufficiency due to heterozygous mutations also results in telomere shortening and is identified in up to 10% of patients with aplastic anemia (AA) or myeloid malignancies.11-13 Heterozygous RTEL1 mutations have been reported in idiopathic pulmonary fibrosis (IPF)14,15 but are not common in hematologic disorders.
We screened 2 independent cohorts for germline RTEL1 mutations: the first cohort comprised 457 unselected patients diagnosed with idiopathic BMF, MDS, or acute myeloid leukemia (AML) from King’s College Hospital (KCH); a second cohort comprised 59 patients selected on the suspicion of constitutional BMF from the National Institutes of Health (NIH). Germline RTEL1 variants were found in 27 (5%) patients, and only 2 patients were compound heterozygotes. Coinheritance of a mutation in another telomere-associated gene was present in 6 instances.
Methods
Patients
Two independent cohorts were enrolled in this study and screened for mutations in telomere-associated genes using an inherited BMF targeting next-generation sequencing panel. The KCH cohort (n = 457) consisted of 285 patients with idiopathic AA, BMF, or unexplained cytopenias (idiopathic cytopenia of uncertain significance [ICUS]) and 172 patients diagnosed with either MDS or AML randomly selected from a 600-sample bank (supplemental Figure 1). The NIH cohort comprised 59 patients selected for the study on the basis of a personal or familial medical history suggestive of constitutional BMF (features included early graying of hair, liver cirrhosis, pulmonary fibrosis, chronic cytopenias, clonal evolution to MDS, and family history of BMF or hematopoietic neoplasms). The diagnosis of AA and hypoplastic MDS (hypoMDS) was defined according to standard criteria.16-18 Full hematologic data, including subtypes of AA/BMF and MDS on the 2 cohorts, are shown in supplemental Table 1 and supplemental Figure 1. Peripheral blood (PB) or BM samples were used for analysis and were obtained from all patients after written informed consent, following the Declaration of Helsinki and under protocols approved by the institutional review board of the National Heart, Lung, and Blood Institute and KCH local research ethics committee and institutional review board.
Targeting sequencing
Two targeting sequencing approaches were used to identify variants in the KCH and NIH cohorts. For the KCH cohort, patients were screened for mutations in 12 telomere biology genes using an Illumina Nextera-amplicon sequencing on MiSeq platform (supplemental Table 4). For the NIH cohort, patients were screened with the Inherited Bone Marrow Failure Sequencing panel (49 candidate genes) performed by the University of Chicago Genetics Services Laboratory. Patients with isolated cytopenias were previously screened for other germline mutations (see supplemental data), and all were wild-type.
Variant annotation
The Variant Effect Predictor (https://useast.ensembl.org) was used to annotate all variants with in silico impact prediction by SIFT and Polyphen2 software, analysis of 3-dimensional (3D) structure, amino acid conservation, and the Combined Annotation Dependent Depletion (CADD; http://cadd.gs.washington.edu/home). Population-variant allele frequencies were determined from the Exome Aggregation Consortium (ExAC; http://exac.broadinstitute.org), and 1000 Genomes (http://www.internationalgenome.org/) (supplemental data). The RTEL1 variants identified in our study were annotated using the isoform 3 (1300 aa; NM_001283009.1; Ensembl: ENST00000360203.9). A variant was considered rare if it was novel or had a minor allele frequency of less than 0.1% in ExAC or 1000 Genomes. RTEL1 variants were classified as pathogenic, likely pathogenic (LP), of uncertain significance (US), likely benign (LB), and benign according to the established American College of Medical Genetics and Genomics and Association for Molecular Pathology (ACMG) consensus criteria (supplemental Table 3).19
Functional assays
To investigate RTEL1 variants’ impacts on telomere functions, we measured patients’ overall telomere length (TL) in nucleated PB or BM cells in both KCH and NIH cohorts, as has been described previously20,21 (supplemental Figure 2). The correlation of TL measured in PB and BM mononuclear cells is shown in supplemental Figure 3. A further functional analysis was performed only in the NIH cohort because of the lack of KCH patients’ samples. Telomeric single-stranded 3′ overhang was measured, as previously described.22,23 RNA and DNA were extracted from NIH patients’ PB for gene expression analysis and telomere circle (TC) assay, respectively, as described with minor modifications.5,10,24 Signaling pathways related to DNA damage, apoptosis, and senescence or telomere and telomerase maintenance were evaluated in patient samples by polymerase chain reaction (PCR) array.
In vitro, 293T cells were infected with a pLV[Exp]-Puro-CMV>RTEL1[NM_001283009.1]-FLAG vector carrying RTEL1 wild-type (WT) or one of the following RTEL1 variants: P82L, M652T, D719N, G951S, or F1262L. The control used was a 293T infected with an empty vector. Stable 293T cell lines expressing RTEL1-FLAG were established by puromycin selection (bulk cells). FLAG and TRF2 expression in 293T bulk cells was assessed by western blot. To validate our results, we assessed both FLAG and TRF2 expression in an isolated clone of 293T-RTEL1-FLAG obtained by single-cell dilution in a 10-cm2 petri dish. The RTEL1 binding to TRF2 was evaluated by FLAG immunoprecipitation of cell lysates extracted from synchronized WT and 293T clones carrying RTEL1 variants. Both FLAG and TRF2 were quantified in the immunoprecipitants by western blot. Detailed protocols are described in the supplemental data.
Results
Assessment of variant pathogenicity
We screened the KCH and NIH cohorts (n = 516) for inherited mutations in telomere-associated genes. A total of 23 rare RTEL1 variants were identified in 27 unrelated patients from both cohorts (Figure 1A). According to ACMG criteria, 7 variants were classified as pathogenic or likely pathogenic (c.301+1 G>A, M652T, D719N, G951S, F1262L, R974X, and R986X; Table 1 and Figure 2B), including 2 stop codons previously reported in patients with HH syndrome or IPF.6,7,9 Pathogenic/likely pathogenic variants were identified in 3 (5%) of 59 patients from the NIH cohort and in 6 (1.3%) of 457 from the KCH cohort (Figure 2A). Twelve variants were of uncertain significance, because data were insufficient to fulfill ACMG criteria. These variants were found in 3 NIH patients (5%) and 10 KCH patients (2.2%). There was strong evidence of pathogenicity for 7 variants grouped as uncertain significance; the V178A, D220N, Q397E, A549T, A902G, and D942N variants were well conserved, absent in controls, and predicted to be damaging, but functional or pedigree data were not available (Table 1; supplemental Figure 6; supplemental Table S2). Four variants classified as likely benign were identified in 4 unrelated patients and in the compound heterozygous NIH-1, which also carried a likely pathogenic variant (Table 1; Figure 2A).
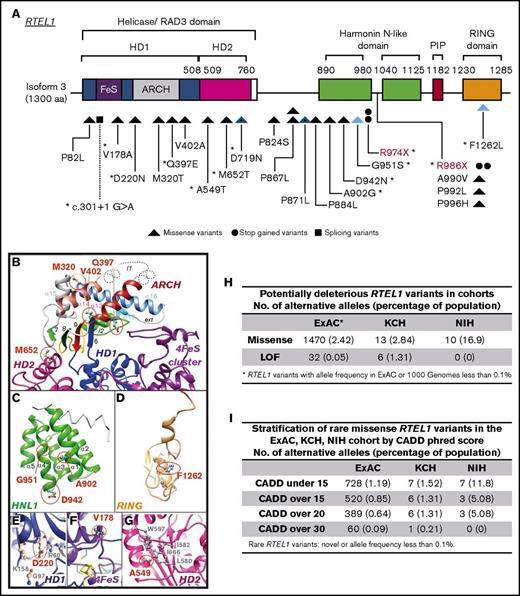
Heterozygous RTEL1 variants. (A) Linear representation of RTEL1 isoform 3 (1300 amino acids; NM_001283009.1). Colored boxes indicate the conserved RTEL1 domains, and lines indicate the positions of individual variants in the gene. Twenty-one novel (black) and 2 previously described (red) heterozygous variants were identified in 8 patients from the NIH cohort (n = 59) and in 19 patients from the KCH cohort (n = 457). Variants are represented by triangles, circles, and squares, which also represent the type of the variant (missense variants, stop-gained variants, or splicing variants, respectively). Blue triangles represent 2 compound heterozygous patients. Splicing variants are indicated by dashed lines. *Eleven variants had a CADD phred score of >15 (this score was selected to predicted RTEL1 deleteriousness; supplemental Table 2). (B-G) Ribbon representation of the 3D structure models of the RTEL1 domains. Amino acid changes observed in patients are colored in red and displayed in an atomic representation in the figure within their close neighborhood. Panel B focuses on variants located on the ARCH and HD2 domains and panel C on the first harmonin-N-like domain. Panels D-G focus on variants located on the C4C4-RING domain, helicase domain 1, the FeS cluster, and helicase domain 2, respectively. The RTEL1 3D structure information obtained by comparative modeling predicted 14 of 23 RTEL1 variants as substitutions that may destabilize the domain 3D structure in which they are located or affect the protein partner interactions of RTEL1 (supplemental data). (H) The frequency of potential deleterious RTEL1 missense and LoF in the ExAC database (n = 60 706), KCH (n = 457), and NIH (n = 59) cohorts. Missense variants are enriched in the NIH cohort but not in the KCH cohort when compared with the ExAC database. However, the KCH cohort had a very significant enrichment in LoF variants when compared with ExAC. (I) Frequency of missense RTEL1 variants stratified by CADD phred score in the ExAC database, and the KCH and NIH cohorts. The CADD predicts deleteriousness of a given variant. All variants with CADD over 15 from KCH and NIH had CADD over 20 as well. The CADD of 15 was selected as a cutoff to predict a variant as deleterious on the basis of the frequency identified in the ExAC database. HD1, helicase domain 1; HD2, helicase domain 2; HNL-1, first harmonin-N-like domain.
Heterozygous RTEL1 variants. (A) Linear representation of RTEL1 isoform 3 (1300 amino acids; NM_001283009.1). Colored boxes indicate the conserved RTEL1 domains, and lines indicate the positions of individual variants in the gene. Twenty-one novel (black) and 2 previously described (red) heterozygous variants were identified in 8 patients from the NIH cohort (n = 59) and in 19 patients from the KCH cohort (n = 457). Variants are represented by triangles, circles, and squares, which also represent the type of the variant (missense variants, stop-gained variants, or splicing variants, respectively). Blue triangles represent 2 compound heterozygous patients. Splicing variants are indicated by dashed lines. *Eleven variants had a CADD phred score of >15 (this score was selected to predicted RTEL1 deleteriousness; supplemental Table 2). (B-G) Ribbon representation of the 3D structure models of the RTEL1 domains. Amino acid changes observed in patients are colored in red and displayed in an atomic representation in the figure within their close neighborhood. Panel B focuses on variants located on the ARCH and HD2 domains and panel C on the first harmonin-N-like domain. Panels D-G focus on variants located on the C4C4-RING domain, helicase domain 1, the FeS cluster, and helicase domain 2, respectively. The RTEL1 3D structure information obtained by comparative modeling predicted 14 of 23 RTEL1 variants as substitutions that may destabilize the domain 3D structure in which they are located or affect the protein partner interactions of RTEL1 (supplemental data). (H) The frequency of potential deleterious RTEL1 missense and LoF in the ExAC database (n = 60 706), KCH (n = 457), and NIH (n = 59) cohorts. Missense variants are enriched in the NIH cohort but not in the KCH cohort when compared with the ExAC database. However, the KCH cohort had a very significant enrichment in LoF variants when compared with ExAC. (I) Frequency of missense RTEL1 variants stratified by CADD phred score in the ExAC database, and the KCH and NIH cohorts. The CADD predicts deleteriousness of a given variant. All variants with CADD over 15 from KCH and NIH had CADD over 20 as well. The CADD of 15 was selected as a cutoff to predict a variant as deleterious on the basis of the frequency identified in the ExAC database. HD1, helicase domain 1; HD2, helicase domain 2; HNL-1, first harmonin-N-like domain.
Description of 27 patients with RTEL1 variants
| Patient ID . | Age/sex . | TL* . | RTEL1 variants . | Other germline variants† . | RTEL1 ACGM‡ . | Hematologic diagnosis . | Somatic anomalies and other clinical features . | Family history . | Outcome . |
|---|---|---|---|---|---|---|---|---|---|
| Heterozygous pathogenic or likely pathogenic variants | |||||||||
| KCH-1 | 36/M | <1st | R974X, c.2920 C>T§ | TERT, R756C c.2266 C>T | Pathogenic | Familial hypoMDS | Abnormal lung function: TLCOc 72% | Brother has hypoMDS | Responded to cyclosporine |
| KCH-3 | 25/M | <1st | R974X, c.2920 C>T§ | None | Pathogenic | HypoMDS | Suspected to have inherited disease | FH of cancer | No treatment |
| KCH-8 | 29/F | <1st | c.301+1G>A | None | LP | ICUS: neutropenia | Bicuspid aortic valve, high arched palate | Pedigree (Figure 3) | Treated with G-CSF |
| KCH-10 | 31/F | <1st | R986X, c.2956 C>T§ | None | Pathogenic | ICUS: neutropenia | Dystrophic nails, neutropenia for 16 y | None | Treated with G-CSF |
| KCH-11 | 54/F | <10th | R986X, c.2956 C>T§ | Not tested | Pathogenic | Macrocytosis, hypocellular BM | Dystrophic nail | Pedigree (Figure 3) | No treatment |
| KCH-15 | 30/F | <1st | c.301+1G>A | None | LP | AA/PNH | None | None | Treated with IST, LTFU |
| NIH-4 | 31/F | Normal | M652T, c.1955 T>C | SLX4, T750M c.2249 C>T | LP | ICUS: neutropenia | Normal BM cellularity | None | No treatment |
| Biallelic pathogenic or likely pathogenic variants | |||||||||
| NIH-1 | 32/M | <1st | D719N, c.2155 G>A P871L, c.2612 C>T | None | LP | MAA | Liver cirrhosis, pulmonary fibrosis | Pedigree (Figure 3) | Responded to androgen (danazol) |
| LB | |||||||||
| NIH-2 | 6/F | <1st | G951, c.2851 G>A F1262L, c.3786 C>G | None | LP | SAA | Prolonged thrombocytopenia | Pedigree (Figure 3) | Awaiting HSCT |
| Pathogenic | |||||||||
| Variants of uncertain significance | |||||||||
| KCH-2 | 71/M | <10th | V178A, c.533 T>C | None | US | MDS (RAEB1) | Laryngeal squamous cell cancer, age of 60 y; small cell lung cancer, age of 66 y | FH of cancer | Treated with 5-azacitidine, progressed to AML, died |
| KCH-4 | 37/M | <1st | A549T, c.1645 G>A | None | US | HypoMDS | Reticulate skin pigmentation, skin warts, liver fibrosis, portal hypertension, esophageal varices | FH of cancer | No treatment |
| KCH-5 | 33/F | <10th | Q397E, c.1189 C>G | None | US | ICUS | None | None | Evolved to hypoMDS |
| KCH-9 | 19/F | <1st | D220N, c.658 G>A | TERT, A1062T c.3184 G>A | US | MAA | Short stature, reticulate skin pigmentation, dystrophic nails, Lichen planus, epiphora | Pedigree (Figure 3) | No treatment |
| KCH-12 | 33/M | <10th | A990V, c.2969 C>T | None | US | BMFS, ICUS | Learning difficulties, epilepsy, urogenital anomalies, hyperostosis cranium, occult spina bifida | None | G-CSF, died lymphoma |
| KCH-13 | 73/M | <1st | P992L, c.2975 C>T | None | US | CMML1 | None | FH of cancer | No treatment |
| KCH-14 | 53/F | <1st | Q397E, c.1189 C>G | None | US | Thrombocytopenia/MAA | Short stature | FH of cancer, brother has short TL | Partial response to IST, relapsed |
| KCH-16 | 29/F | <1st | P824S, c.2470 C>T | None | US | MAA | Presented in pregnancy | None | No treatment |
| KCH-17 | 38/M | <1st | V402A, c.1205 T>C | None | US | HypoMDS | None | None | No treatment |
| KCH-19 | 63/M | <1st | D942N, c.2824 G>A | None | US | AML | None | None | Chemotherapy, MUD HSCT, died GVHD |
| NIH-3 | 17/M | Normal | P884L, c.2651 C>T | None | US | SAA | None | FH of cancer | Treated with IST/EPAG and evolved to −7/MDS |
| NIH-5 | 63/F | Normal | P82L, c.245 C>T | None | US | MAA | Mitral valve prolapse | Pedigree (Figure 3) | Progressed to SAA on EPAG and evolved to MDS/AML; died following HSCT of relapsed AML |
| NIH-7 | 32/F | <1st | A902G, c.2705 C>G | TERC, r.287 C>G§ | US | MAA | Frequent miscarriages, early graying of hair | Maternal early graying of hair, MDS in maternal uncle, BMF in paternal grandmother | No treatment |
| Likely benign variants | |||||||||
| KCH-6 | 52/M | <1st | M320T, c.959 T>C | TERT, A279T c.835 G>A | LB | HypoMDS | Celiac disease, hepatosplenomegaly, abnormal lung function | None | MUD HSCT; complete response |
| KCH-7 | 38/F | <10th | P867L, c.2600 C>T | None | LB | Macrocytosis | Short stature, normal BM cellularity | None | No treatment |
| KCH-18 | 73/M | Normal | M320T, c.959 T>C | None | LB | MDS (RAEB2) | None | None | Best supportive care; died |
| NIH-6 | 23/F | Normal | P867L, c.2672 C>T | TERT, R537C c.1609 C>T | LB | MAA | Eczema | Father and brother with early graying of hair, mother with macrocytic anemia | No treatment |
| NIH-8 | 47/M | Normal | P996H, c.2987 C>A | None | LB | hypoMDS | None | Sister with SLE, paternal grandfather with polycythemia vera | Responded to EPAG |
| Patient ID . | Age/sex . | TL* . | RTEL1 variants . | Other germline variants† . | RTEL1 ACGM‡ . | Hematologic diagnosis . | Somatic anomalies and other clinical features . | Family history . | Outcome . |
|---|---|---|---|---|---|---|---|---|---|
| Heterozygous pathogenic or likely pathogenic variants | |||||||||
| KCH-1 | 36/M | <1st | R974X, c.2920 C>T§ | TERT, R756C c.2266 C>T | Pathogenic | Familial hypoMDS | Abnormal lung function: TLCOc 72% | Brother has hypoMDS | Responded to cyclosporine |
| KCH-3 | 25/M | <1st | R974X, c.2920 C>T§ | None | Pathogenic | HypoMDS | Suspected to have inherited disease | FH of cancer | No treatment |
| KCH-8 | 29/F | <1st | c.301+1G>A | None | LP | ICUS: neutropenia | Bicuspid aortic valve, high arched palate | Pedigree (Figure 3) | Treated with G-CSF |
| KCH-10 | 31/F | <1st | R986X, c.2956 C>T§ | None | Pathogenic | ICUS: neutropenia | Dystrophic nails, neutropenia for 16 y | None | Treated with G-CSF |
| KCH-11 | 54/F | <10th | R986X, c.2956 C>T§ | Not tested | Pathogenic | Macrocytosis, hypocellular BM | Dystrophic nail | Pedigree (Figure 3) | No treatment |
| KCH-15 | 30/F | <1st | c.301+1G>A | None | LP | AA/PNH | None | None | Treated with IST, LTFU |
| NIH-4 | 31/F | Normal | M652T, c.1955 T>C | SLX4, T750M c.2249 C>T | LP | ICUS: neutropenia | Normal BM cellularity | None | No treatment |
| Biallelic pathogenic or likely pathogenic variants | |||||||||
| NIH-1 | 32/M | <1st | D719N, c.2155 G>A P871L, c.2612 C>T | None | LP | MAA | Liver cirrhosis, pulmonary fibrosis | Pedigree (Figure 3) | Responded to androgen (danazol) |
| LB | |||||||||
| NIH-2 | 6/F | <1st | G951, c.2851 G>A F1262L, c.3786 C>G | None | LP | SAA | Prolonged thrombocytopenia | Pedigree (Figure 3) | Awaiting HSCT |
| Pathogenic | |||||||||
| Variants of uncertain significance | |||||||||
| KCH-2 | 71/M | <10th | V178A, c.533 T>C | None | US | MDS (RAEB1) | Laryngeal squamous cell cancer, age of 60 y; small cell lung cancer, age of 66 y | FH of cancer | Treated with 5-azacitidine, progressed to AML, died |
| KCH-4 | 37/M | <1st | A549T, c.1645 G>A | None | US | HypoMDS | Reticulate skin pigmentation, skin warts, liver fibrosis, portal hypertension, esophageal varices | FH of cancer | No treatment |
| KCH-5 | 33/F | <10th | Q397E, c.1189 C>G | None | US | ICUS | None | None | Evolved to hypoMDS |
| KCH-9 | 19/F | <1st | D220N, c.658 G>A | TERT, A1062T c.3184 G>A | US | MAA | Short stature, reticulate skin pigmentation, dystrophic nails, Lichen planus, epiphora | Pedigree (Figure 3) | No treatment |
| KCH-12 | 33/M | <10th | A990V, c.2969 C>T | None | US | BMFS, ICUS | Learning difficulties, epilepsy, urogenital anomalies, hyperostosis cranium, occult spina bifida | None | G-CSF, died lymphoma |
| KCH-13 | 73/M | <1st | P992L, c.2975 C>T | None | US | CMML1 | None | FH of cancer | No treatment |
| KCH-14 | 53/F | <1st | Q397E, c.1189 C>G | None | US | Thrombocytopenia/MAA | Short stature | FH of cancer, brother has short TL | Partial response to IST, relapsed |
| KCH-16 | 29/F | <1st | P824S, c.2470 C>T | None | US | MAA | Presented in pregnancy | None | No treatment |
| KCH-17 | 38/M | <1st | V402A, c.1205 T>C | None | US | HypoMDS | None | None | No treatment |
| KCH-19 | 63/M | <1st | D942N, c.2824 G>A | None | US | AML | None | None | Chemotherapy, MUD HSCT, died GVHD |
| NIH-3 | 17/M | Normal | P884L, c.2651 C>T | None | US | SAA | None | FH of cancer | Treated with IST/EPAG and evolved to −7/MDS |
| NIH-5 | 63/F | Normal | P82L, c.245 C>T | None | US | MAA | Mitral valve prolapse | Pedigree (Figure 3) | Progressed to SAA on EPAG and evolved to MDS/AML; died following HSCT of relapsed AML |
| NIH-7 | 32/F | <1st | A902G, c.2705 C>G | TERC, r.287 C>G§ | US | MAA | Frequent miscarriages, early graying of hair | Maternal early graying of hair, MDS in maternal uncle, BMF in paternal grandmother | No treatment |
| Likely benign variants | |||||||||
| KCH-6 | 52/M | <1st | M320T, c.959 T>C | TERT, A279T c.835 G>A | LB | HypoMDS | Celiac disease, hepatosplenomegaly, abnormal lung function | None | MUD HSCT; complete response |
| KCH-7 | 38/F | <10th | P867L, c.2600 C>T | None | LB | Macrocytosis | Short stature, normal BM cellularity | None | No treatment |
| KCH-18 | 73/M | Normal | M320T, c.959 T>C | None | LB | MDS (RAEB2) | None | None | Best supportive care; died |
| NIH-6 | 23/F | Normal | P867L, c.2672 C>T | TERT, R537C c.1609 C>T | LB | MAA | Eczema | Father and brother with early graying of hair, mother with macrocytic anemia | No treatment |
| NIH-8 | 47/M | Normal | P996H, c.2987 C>A | None | LB | hypoMDS | None | Sister with SLE, paternal grandfather with polycythemia vera | Responded to EPAG |
The RTEL1 variants identified in our study were annotated using the isoform 3 (1300 aa; NM_001283009.1).
CMML, chronic myelomonocytic leukemia; EPAG, eltrombopag; F, female; FH, family history; GVHD, graft-versus-host disease; HSCT, hematopoietic stem cell transplantation; ID, identification; IST, immunosuppressive therapy with antithymocyte globulin and cyclosporine; LTFU, loss to follow-up; M, male; MAA, moderate aplastic anemia; MUD, matched unrelated donor; PNH, paroxysmal nocturnal hemoglobinuria; RAEB, MDS with refractory anemia with excess blasts; SAA, severe AA; SLE, systemic lupus erythematosus; TLCO, transfer factor of the lung for carbon monoxide.
<1st, TL below the first percentile of age matched controls (very short telomeres); <10th, TL below tenth percentile (short telomeres).
For the KCH cohort, the targeting next-generation sequencing panel had 12 candidate genes. For the NIH cohort, the targeting next-generation sequencing panel had 49 candidate genes.
ACMG consensus criteria.
Variants previously reported as pathogenic.
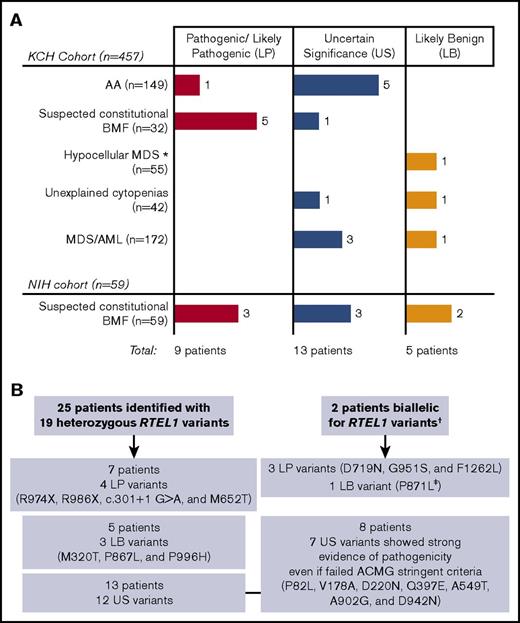
Schematic diagram representing RTEL1 variants identified in this study. (A) We screened 516 patients from 2 independent cohorts (the KCH and NIH), identifying 23 RTEL1 variants in 27 unrelated patients. The number of patients with pathogenic/likely pathogenic, uncertain significance, or likely benign variants are presented by colored bars according to their clinical diagnosis. (B) Twenty-five patients carried heterozygous RTEL1 variants, whereas 2 patients were biallelic. The number of patients with LP, US, and LB variants are summarized in the figure. Heterozygous LP variants were enriched in the group of patients suspected to have inherited BMF (see supplemental Figure 1 for cohort’s data). *Patients with hypoMDS and not suspected to have constitutional BMF. †Four RTEL1 variants identified in patients NIH-1 and NIH-2 were associated with an autosomal recessive form of AA because they were compound heterozygous. ‡Despite being classified as LB, the P871L variant was seen together with the D719N variant in compound heterozygosity.
Schematic diagram representing RTEL1 variants identified in this study. (A) We screened 516 patients from 2 independent cohorts (the KCH and NIH), identifying 23 RTEL1 variants in 27 unrelated patients. The number of patients with pathogenic/likely pathogenic, uncertain significance, or likely benign variants are presented by colored bars according to their clinical diagnosis. (B) Twenty-five patients carried heterozygous RTEL1 variants, whereas 2 patients were biallelic. The number of patients with LP, US, and LB variants are summarized in the figure. Heterozygous LP variants were enriched in the group of patients suspected to have inherited BMF (see supplemental Figure 1 for cohort’s data). *Patients with hypoMDS and not suspected to have constitutional BMF. †Four RTEL1 variants identified in patients NIH-1 and NIH-2 were associated with an autosomal recessive form of AA because they were compound heterozygous. ‡Despite being classified as LB, the P871L variant was seen together with the D719N variant in compound heterozygosity.
Comparison of RTEL1 rare variants to population database
To ascertain whether likely pathogenic RTEL1 variants were enriched in inherited BMF syndromes, we compared frequencies of these variants from our cohorts to the ExAC database. As we identified both loss of function (LoF) and missense RTEL1 variants in our patients, ExAC data were also stratified for comparison purposes.
In the KCH cohort, both LoF and missense RTEL1 variants were identified, but all likely pathogenic variants led to RTEL1 LoF. These variants were enriched in this cohort in comparison with ExAC (1.31% vs 0.05%; odds ratio [OR], 25.2; 95% confidence interval [CI], 11.27-58.67) (Figure 1H). No difference was observed between frequency of rare missense RTEL1 variants in the KCH cohort (2.8%) and ExAC population (2.4%).
Missense RTEL1 variants (but not LoF) were identified in the NIH cohort. Frequency of rare RTEL1 missense variants was increased in comparison with ExAC (16.9% vs 2%; OR, 7.90; 95% CI, 3.845-15.55). However, not all missense variants were predicted to be deleterious. Because it is not feasible to assess the pathogenicity of RTEL1 variants in ExAC on the basis of ACMG criteria, we stratified RTEL1 variants from this database on the basis of their CADD phred scores. Missense variants with CADD scores higher than 20, but not greater than 30, were still enriched in the NIH cohort in comparison with ExAC (Figure 1I). In both cohorts, the enrichment of missense RTEL1 variants, even when stratified by CADD scores, appeared to be dependent on patient selection criteria between the cohorts. In the KCH cohort, comprising unselected patients for constitutional BMF, frequencies of missense variants (CADD up to 15) were similar to those for ExAC.
Pathogenic/likely pathogenic RTEL1 variants associate with AA, idiopathic cytopenias, and hypoMDS
In silico analysis, pedigree data, and functional assays were used to assess a variant’s pathogenicity. Overall, 7 different likely pathogenic variants were identified in 9 unrelated patients (Figure 2A). Seven patients were heterozygous, and 2 carried biallelic variants (Table 1; Figure 2B). These variants associated with phenotypes across a wide age range (median 31 years, ranging from 6 to 54), but most patients were adults (n = 8; Table 1).
Likely pathogenic RTEL1 variants were identified in 8 patients suspected of constitutional BMF on the basis of their clinical features and TL (Figure 2). Clinical presentation varied from AA (NIH-1, moderate [MAA], and NIH-2, severe [SAA]), hypoMDS (KCH-1 and KCH-3), and ICUS (KCH-8, KCH-10, and NIH-4) to isolated red cell macrocytosis without anemia (KCH-11; Table 1). Most patients had a personal or family history consistent with a telomere disease or other inherited BMF. KCH-3 had a strong family history of cancer, and KCH-8 had a high arched palate and a family history of a cardiac anomaly. Likely pathogenic RTEL1 variants also were identified in a patient with adult-onset AA (KCH-15) that were not associated with inherited diseases (Figure 2A; Table 1). Paroxysmal nocturnal hemoglobinuria (PNH) clones were detected in 5 patients (a median PNH granulocyte clone size, 1.6%; range, 0.1% to 11%), but only 2 of them carried likely pathogenic variants: KCH-3 (with the R974X variant and a PNH granulocyte clone size, 0.1%) and KCH-15 (with a c.301+1 G>A variant and a PNH granulocyte clone size, 36.2%), as is shown in supplemental Table 1. Treatment with androgens, hematopoietic growth factors, or immunosuppressive (IST) agents was effective in 4 of 6 patients who required treatment (1 was lost to follow-up), and all remained alive with a median follow-up of 101.7 months (range of 76.5-206.7 months; Table 1).
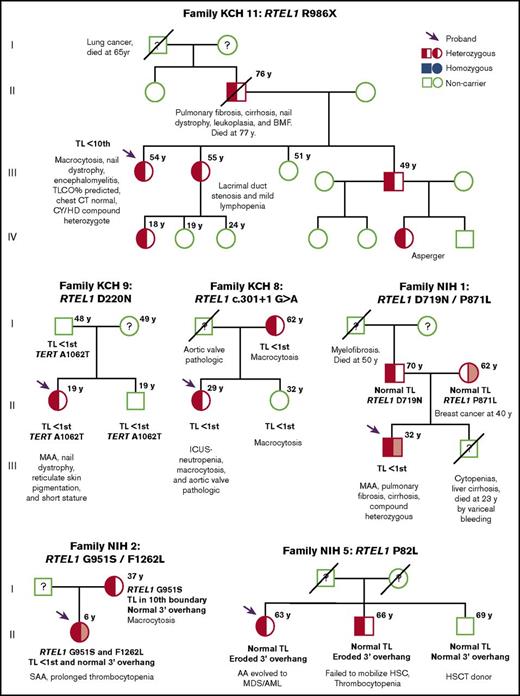
Family screening of 4 probands displayed variable penetrance of likely pathogenic RTEL1 variants (Figure 3). In the KCH-11 family, typical characteristics of telomere disease (pulmonary fibrosis and nail dystrophy) segregated with R986X in multiple generations. Two probands (NIH-1 and NIH-2) displayed inheritance of compound heterozygosity associated with AA; particularly severe disease was observed in the patient NIH-1 and his sibling, suggesting compound heterozygosity for the P871L and D719N variants to be particularly damaging. Proband’s parents were both asymptomatic, with normal TL and 3′ overhangs. Isolated red blood cell macrocytosis was a sole abnormality observed in affected family members heterozygous for c.301+1G>A (family KCH-8) and G951S (family NIH-2).
Pedigrees of 6 families identified with RTEL1 variants. Pedigrees from proband KCH-11, KCH-9, KCH-8, NIH-1, NIH-2, and NIH-5, respectively. The RTEL1 variant presented in the family is described above each pedigree. Arrows indicate the probands of each family. Open circles and squares show females and males who are noncarriers, respectively. Half-filled or solid circles and squares represent individuals who are heterozygous or homozygous for the RTEL1 variant, respectively. A line through the circle or square indicates individuals who are deceased. When tested, TL and clinical features are described in the figure under each individual. Relatives who lack clinical and mutational data are indicated by a question mark. For families NIH-1 and NIH-2, probands were identified as compound heterozygous (half-red, half–light red, square). In NIH-1, the proband’s father and mother carried the D719N (half-red square) and the P871L (half–light red circle) variants, respectively, both heterozygous. In NIH-2, the father was not screened, but the mother carried the G951S variant in heterozygosity. BMF, bone marrow failure; CY/HD, compound heterozygous for C282Y and H63D hereditary hemochromatosis genes; KCH, King’s College Hospital cohort; NIH, National Institutes of Health cohort; MAA, moderate aplastic anemia; TLCO, transfer factor of the lung for carbon monoxide.
Pedigrees of 6 families identified with RTEL1 variants. Pedigrees from proband KCH-11, KCH-9, KCH-8, NIH-1, NIH-2, and NIH-5, respectively. The RTEL1 variant presented in the family is described above each pedigree. Arrows indicate the probands of each family. Open circles and squares show females and males who are noncarriers, respectively. Half-filled or solid circles and squares represent individuals who are heterozygous or homozygous for the RTEL1 variant, respectively. A line through the circle or square indicates individuals who are deceased. When tested, TL and clinical features are described in the figure under each individual. Relatives who lack clinical and mutational data are indicated by a question mark. For families NIH-1 and NIH-2, probands were identified as compound heterozygous (half-red, half–light red, square). In NIH-1, the proband’s father and mother carried the D719N (half-red square) and the P871L (half–light red circle) variants, respectively, both heterozygous. In NIH-2, the father was not screened, but the mother carried the G951S variant in heterozygosity. BMF, bone marrow failure; CY/HD, compound heterozygous for C282Y and H63D hereditary hemochromatosis genes; KCH, King’s College Hospital cohort; NIH, National Institutes of Health cohort; MAA, moderate aplastic anemia; TLCO, transfer factor of the lung for carbon monoxide.
RTEL1 haploinsufficiency associates with telomere shortening and single-strand 3′ overhang erosion independent of length
We first assessed patients’ telomere integrity by measuring overall TL. Sixteen patients had very short telomeres (below first percentile of age-matched controls), and 5 had short telomeres (below 10th percentile). As was expected, all patients with likely pathogenic RTEL1 variants that led to protein LoF (R974X, R986X, and c.301+1G>A) or that were biallelic presented with very short telomeres (Table 1). However, 6 patients had normal TL for their age (supplemental Figure 2). We next assessed the single-strand 3′overhangs integrity in the NIH cohort and observed that 4 of 8 patients (NIH-3, -4, -5, and -6) had eroded 3′ overhangs, all of them independent of TL (Figure 4A). Despite their normal 3′ overhangs, the NIH-2, NIH-7, and NIH-8 had a high focal 3′ overhang signal in nondenaturing blotting and no signal in the lower part of the gel; thus, their range of 3′ overhang was not as broad as it was for healthy controls. Independent of length, overhang erosion associated with 1 likely pathogenic variant (M652T) in NIH-4 and 2 variants of uncertain significance and very low CADD phred score in NIH-3 and NIH-5 (P884L and P82L, respectively) (Table 1). Both NIH-3 and NIH-5 clonally evolved to monosomy 7 and MDS/AML, respectively, and demonstrated upregulation of genes related to DNA repair and DNA damage response. In contrast, the transcriptional profile of NIH-2 showed upregulation of genes related to homologous recombination, apoptosis, and senescence triggered by very short telomeres (supplemental Figure 7). The P82L variant segregated with disease in NIH-5 Family (Figure 3) but lacked adequate in silico prediction and genetic conservation (supplemental Table 2; supplemental Figure 6). NIH-5-affected brother failed stem-cell mobilization during attempted donation for transplant and had short 3′ overhangs, whereas the noncarrier brother was healthy and donated BM for transplantation (Figures 3 and 4A). Overhang erosion associated with a likely benign RTEL1 variant for NIH-6. In this case, RTEL1 may play a secondary role in the patient’s disease, because the R537C TERT variant may play a major role in patient’s telomere dysfunction.
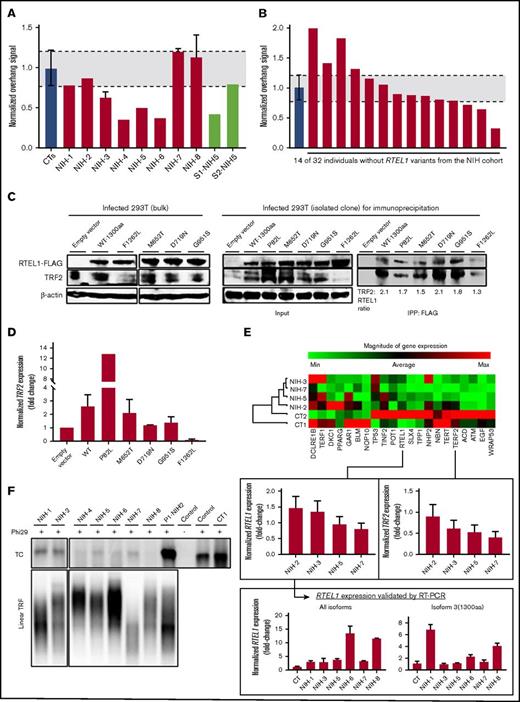
Impact of RTEL1 variants on telomere maintenance. (A) Normalized single-strand 3′ overhang measurement by nondenaturing Southern blot of patients from the NIH cohort and 2 siblings of NIH-5 (S1- and S2-NIH5). Relative 3′ overhang signals were determined by normalizing the sum of chemiluminescent signal (ΣODi) from each column in the nondenaturing membrane (overhang signals) by the telomeric signal in the denatured membrane (representing total genomic DNA). The background signal (sample treated with Exo1) was subtracted from total ΣODi for all the samples. Error bars represent the standard deviations of independent experiments. Patients’ 3′ overhang signals were then normalized by an average of 3′ overhang signals from healthy individuals used as a control in every experiment and plotted. The 3′ overhang signals of patients below a 95% confidence interval of 3′ overhang measurements from controls (gray interval in the graphic) were considered eroded. (B) Normalized single-strand 3′ overhang measurement of 14 patients from the NIH cohort without RTEL1 variants in which DNA was available. Three patients had telomeric overhangs below a 95% confidence interval of healthy controls that were considered short. (C) Western blot analysis. (Left) Whole extracts of bulk 293T cells stably expressing recombinant RTEL1-FLAG wild-type or one of the following RTEL1 variants: M652T, D719N, G951S, and F1262L. Control used was 293T infected with an empty vector. (Right) Whole extracts (input) of isolated RTEL1-FLAG WT or 293T clones with the P82L, M652T, D719N, G951S, and F1262L variants were immunoprecipitated with anti-FLAG to evaluate RTEL1 and TRF2 interaction. Protein expression was analyzed with antibodies as indicated. (D) TRF2 expression in both bulk-infected 293T cells and isolated clones normalized by the control (empty vector). Error bars represent the standard deviations between TRF2 expression in bulk 293T and isolated clones. (E) Clustergram of the telomere biology gene expression levels in 4 patients with RTEL1 variants from the NIH cohort and 2 controls using the RT2 Profiler PCR array system (Qiagen). Control samples were used as a reference for normalization. For comparison, the RTEL1 and TRF2 fold-change expression in relation to controls were plotted and shown in the graphic below the heatmap. The RTEL1 gene expression was validated by real-time PCR using a Taqman probe that detected the boundary of exon 7-8 in all isoforms and another probe that detected the RING domain in isoform 3. (F) Phi29-dependent t-circle amplification assay in patients’ peripheral blood. The t-circles were detected in a lower amount in patients with heterozygous variants than in biallelic patients. The asymptomatic mother of NIH-2 (P1-NIH2) and a healthy control (CT1) presented the highest number of t-circles. DNA extracted from VA13 cells was used as a control of the assay. IPP, immunoprecipitation; RT-PCR, reverse transcription polymerase chain reaction.
Impact of RTEL1 variants on telomere maintenance. (A) Normalized single-strand 3′ overhang measurement by nondenaturing Southern blot of patients from the NIH cohort and 2 siblings of NIH-5 (S1- and S2-NIH5). Relative 3′ overhang signals were determined by normalizing the sum of chemiluminescent signal (ΣODi) from each column in the nondenaturing membrane (overhang signals) by the telomeric signal in the denatured membrane (representing total genomic DNA). The background signal (sample treated with Exo1) was subtracted from total ΣODi for all the samples. Error bars represent the standard deviations of independent experiments. Patients’ 3′ overhang signals were then normalized by an average of 3′ overhang signals from healthy individuals used as a control in every experiment and plotted. The 3′ overhang signals of patients below a 95% confidence interval of 3′ overhang measurements from controls (gray interval in the graphic) were considered eroded. (B) Normalized single-strand 3′ overhang measurement of 14 patients from the NIH cohort without RTEL1 variants in which DNA was available. Three patients had telomeric overhangs below a 95% confidence interval of healthy controls that were considered short. (C) Western blot analysis. (Left) Whole extracts of bulk 293T cells stably expressing recombinant RTEL1-FLAG wild-type or one of the following RTEL1 variants: M652T, D719N, G951S, and F1262L. Control used was 293T infected with an empty vector. (Right) Whole extracts (input) of isolated RTEL1-FLAG WT or 293T clones with the P82L, M652T, D719N, G951S, and F1262L variants were immunoprecipitated with anti-FLAG to evaluate RTEL1 and TRF2 interaction. Protein expression was analyzed with antibodies as indicated. (D) TRF2 expression in both bulk-infected 293T cells and isolated clones normalized by the control (empty vector). Error bars represent the standard deviations between TRF2 expression in bulk 293T and isolated clones. (E) Clustergram of the telomere biology gene expression levels in 4 patients with RTEL1 variants from the NIH cohort and 2 controls using the RT2 Profiler PCR array system (Qiagen). Control samples were used as a reference for normalization. For comparison, the RTEL1 and TRF2 fold-change expression in relation to controls were plotted and shown in the graphic below the heatmap. The RTEL1 gene expression was validated by real-time PCR using a Taqman probe that detected the boundary of exon 7-8 in all isoforms and another probe that detected the RING domain in isoform 3. (F) Phi29-dependent t-circle amplification assay in patients’ peripheral blood. The t-circles were detected in a lower amount in patients with heterozygous variants than in biallelic patients. The asymptomatic mother of NIH-2 (P1-NIH2) and a healthy control (CT1) presented the highest number of t-circles. DNA extracted from VA13 cells was used as a control of the assay. IPP, immunoprecipitation; RT-PCR, reverse transcription polymerase chain reaction.
To assess whether 3′ overhang erosion was related to RTEL1 dysfunction, we assessed the overhangs from 14 individuals from the NIH cohort who did not carry a RTEL1 variant and in whom DNA was available. The frequency of patients with eroded 3′ overhang and RTEL1 variants (50%) was higher than was the frequency observed in patients without RTEL1 variants (21%), indicating that 3′ overhang shortening correlated with RTEL1 dysfunction specifically, rather than to BMF in general (Figure 4B).
RTEL1 modulates TRF2 expression in 293T cells
To assess the impact of RTEL1 variants in the helicase interaction with TRF2, we generated 293T cells stably expressing the RTEL1-FLAG WT, and with the F1262L, P82L, M652T, D719N, and G951S variants.
In bulk 293T-RTEL1-FLAG cell lines, we first evaluated recombinant RTEL1-FLAG and TRF2 expression (Figure 4C). Surprisingly, TRF2 expression was increased in the WT and the M652T-293T cell lines when compared with the 293T-empty vector, but it was significantly downregulated in 293T-F1262L cells (Figure 4C, left panel). To validate our results, we examined both FLAG and TRF2 expression in a single-cell isolated clone of 293T-RTEL1-FLAG and immunoprecipitated RTEL1-FLAG to check its binding with TRF2.
TRF2 expression was higher in cells stably expressing exogenous WT RTEL1 isoform 3 and the variants M652T, D719N, and G951S. The 293T-P82L cells had a 4-fold increase in TRF2 expression in comparison with WT (Figures 4D and 4C, middle panel). As is seen with bulk cells, the TRF2 expression in the single-cell isolated clone of 293T-F1262L cells was significantly downregulated in comparison with WT or the control (Figure 4C, middle panel). In our patients, NIH-2 did not display TRF2 downregulation, nor did NIH-5 display TRF2 upregulation, in PB cells assessed by PCR array (Figure 4E). The RTEL1 and TRF2 interaction was preserved in WT and 293T cells carrying RTEL1 variants (Figure 4C, right panel). A ratio of TRF2 immunoprecipitated with FLAG was similar between WT and mutants, but lowest in 293T-F1262L cells. In summary, the F1262L RING domain variant did not completely disrupt the RTEL1 and TRF2 interaction, but TRF2 downregulation perturbed the RTEL1 interaction with this shelterin.
RTEL1 patients display lower t-circle amount than controls
RTEL1 deficiency is linked to an aberrant accumulation of t-circles in cells.5 We checked t-circle formation in the PB samples from NIH patients by the TC assay. We did not observe any t-circle accumulation in patients’ cells; instead, all of them presented lower t-circle amounts when compared with the asymptomatic mother of NIH-2 (P1-NIH-2) and a healthy control (CT1). Among patients, biallelic patients were found with more t-circles than were heterozygous RTEL1 patients (Figure 4F).
Other germline variants
Six patients also carried variants in other telomere-associated genes. Co-occurrence in TERT was most frequent, observed in 4 patients, followed by TERC and SLX4 variants (Table 1). A novel TERT R756C variant identified in KCH-1 was classified as pathogenic and impaired telomerase activity assessed by TRAP assay (12% of wild-type activity) (supplemental Figure 4).
The polymorphisms TERT A279T and A1062T were found in KCH-6 and KCH-9, respectively, as well as in KCH-9 father and brother (Figure 3); both relatives lacked the RTEL1 D244N variant and had normal blood counts. A novel TERT R537C variant was identified in NIH-6 concomitant with the RTEL1 P867L. The TERC r.287 C>A identified in NIH-7, who had moderate AA, early graying of hair, and frequent miscarriages, was previously reported and predicted to be pathogenic.25 NIH-7 had very short telomeres but not 3′ overhang erosion. Additionally, we identified a variant in the SLX4 gene in NIH-4 that encodes an endonuclease that participates in t-loop excision and telomere shortening in RTEL1-deficient cells.4 SLX4 loss of function rescues the telomere loss phenotype in vitro,4 and its haploinsufficiency can account for normal TL even when RTEL1 is impaired.
Discussion
In our study, heterozygous RTEL1 variants classified as likely pathogenic are associated with AA, unexplained cytopenias, and hypoMDS, seen both at an early age and in adulthood. We classified variants using a combination of evidence that stringently assessed the pathogenicity of RTEL1 variants. We identified 7 likely pathogenic RTEL1 variants (of which 5 were novel) in 9 unrelated patients screened in 2 independent cohorts. Likely pathogenic RTEL1 variants were enriched in patients who were selected as possibly having inherited BMF (8 of 91), but it was also identified in 1 patient with adult-onset AA without typical features of the constitutional disease. In contrast to biallelic variants, RTEL1 haploinsufficiency was associated with clinical manifestations that did not meet criteria for any specific constitutional BMF syndromes, such as DC and HH syndromes. Constitutional BMF, especially in adults, is likely underreported, because of incomplete penetrance, reduced expressivity, and disease anticipation, as well as a lack of awareness by clinicians that it may present without mucocutaneous features and instead with pulmonary fibrosis or cirrhosis.26 The importance of the diagnosis of an inherited BMF disorder is highlighted in our study. First, there are implications for donor selection and choice of transplant-conditioning regimen in hematopoietic stem cell transplantation (HSCT). Family screening, including testing of siblings, is necessary to avoid using affected “silent” siblings as donors. Second, the risk of later cancers is higher in constitutional marrow failure than in acquired AA.16
As is seen with other telomere biology components, heterozygous RTEL1 variants had a variable penetrance, possibly acting as disease modulators in a complex genomic architecture. Penetrance of TERT or TERC mutations is very heterogeneous,27 and other genetic or epigenetic factors have been hypothesized to modulate disease manifestations. “Epigenetic-like” inheritance of short TL can result in human disease in the absence of a telomerase mutation.28 Biallelic TERT variants were shown to aggravate patient phenotype,29 and triallelic inheritance of homozygous TERT and heterozygous TERC variants was seen in a family member with severe DC phenotype.26 RTEL1 haploinsufficiency was not affected by downregulation of the helicase expression in patients’ cells, because RTEL1 expression seen in patients carrying heterozygous variants was higher or similar to that in controls (Figure 4E). Then it is unlikely that an epigenetic silencing, aberrant splicing, or deletion of the normal RTEL1 allele occurs, affecting disease phenotype.
We identified the co-occurrence of RTEL1 and TERT/TERC/SLX4 variants in 6 patients. However, RTEL1 coexisting variants were not associated with a more aggressive hematologic disease. Seven patients with RTEL1 variant had a coexisting PNH clone. Four of these patients carried RTEL1 variants classified as uncertain significance or likely benign and presented hypoMDS and AA. The presence of a PNH clone in these patients is in opposition to the pathogenic role of their RTEL1 variant. However, patient KCH-15 had a large PNH clone (36%; supplemental Table 1) and the c.301+1 G>A variant classified as pathogenic. This variant was identified in 2 unrelated patients and associated with a nonsevere phenotype in both cases. We previously reported that BMF patients with mutations in telomere biology genes might present a PNH clone, suggesting a coexisting immune component contributing to marrow failure.30 Indeed, some patients with telomere gene mutations and AA/BMF (including a patient in this study; Table 1) may respond to immunosuppressive therapy.
We also found that RTEL1 variants were associated with eroded 3′ overhang, independent of TL. Eroded 3′ overhangs are markers of cell senescence and telomere dysfunction31 and have been described in AA patients with TERT/TERC mutations.32 RTEL1 haploinsufficiency was associated with overhang shortening in only 1 previous paper: 2 siblings with HH syndrome and compound heterozygous with R974X and M492I variants presented with 3′ overhang erosion; but, unlike our study, they had very short telomeres. Their heterozygous parents, although asymptomatic, also presented with short telomeres and 3′ overhang erosion.9 In primary fibroblasts of other HH family, excessive 3′ overhang erosion appeared to impair cell proliferation and to promote extensive DNA damage response activation independent of TL.33 The only component found to be mutated in a patient with TL-independent telomere dysfunction was the Apollo enzyme.34 Similar to RTEL1, Apollo/SNM1B nuclease is required for telomere end-processing and replication. Apollo regulates 3′ overhang length and directly binds to TRF2 to maintain telomere homeostasis during cell replication in the S phase.35 A disturbance in the telomere replication and protection pathways, of which Apollo and RTEL1 are a part, may lead to telomere dysfunction independent of telomere shortening. Two patients with eroded 3′ overhangs and normal TL evolved to AML/MDS after IST treatment. Inability to suppress DNA-damage response combined with normal TL may predispose cells to genomic instability and leukemogenesis, rather than to drive pathways of senescence or apoptosis, as occurs in cells with critically short telomeres (Figure 5). Indeed, expression levels of DNA damage–related genes were different between patients with eroded 3′ overhangs and short telomeres (supplemental Figure 7). Also, MDS and AML were the primary diagnosis of 2 patients identified with variants that had strong evidence for pathogenicity, despite being classified as of uncertain significance. However, we infer from our data that RTEL1 variants classified as likely pathogenic were not solely implicated in excessive 3′ overhang erosion and inappropriate telomere capping.
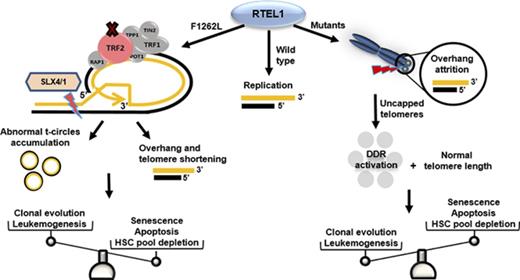
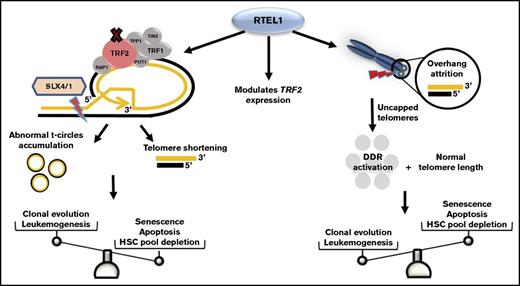
Schematic representation of RTEL1 roles in telomere dysfunction. In normal conditions, RTEL1 promotes G4 quadruplex and t-loop unwinding for DNA replication. The RING-domain mutations inhibit RTEL1-TRF2 interaction and then t-loop resolution.5 Thus, abnormal SXL4/1 activation persistently cuts t-loops, leading to telomere shortening as well as the accumulation of t-circles in cells. Short telomeres, commonly seen in patients with mutated RTEL1, trigger cell senescence and apoptosis via p53, leading to hematopoietic stem cell depletion in BM. However, some RTEL1 mutations were related to excessive 3′ overhang attrition in the absence of telomere shortening. In the context of cells maintaining their telomere lengths, the sustained DNA damage response activation caused by uncapped telomeres may promote leukemogenesis by molecular pathways distinct from typical accelerated telomere attrition associated with very short telomeres. Also, different RTEL1 variants modulated TRF2 expression in vitro, which may be an alternative mechanism related to RTEL1 dysfunction rather than impaired t-loop disassembly alone. DDR, DNA damage response; HSC, hematopoietic stem cell.
Schematic representation of RTEL1 roles in telomere dysfunction. In normal conditions, RTEL1 promotes G4 quadruplex and t-loop unwinding for DNA replication. The RING-domain mutations inhibit RTEL1-TRF2 interaction and then t-loop resolution.5 Thus, abnormal SXL4/1 activation persistently cuts t-loops, leading to telomere shortening as well as the accumulation of t-circles in cells. Short telomeres, commonly seen in patients with mutated RTEL1, trigger cell senescence and apoptosis via p53, leading to hematopoietic stem cell depletion in BM. However, some RTEL1 mutations were related to excessive 3′ overhang attrition in the absence of telomere shortening. In the context of cells maintaining their telomere lengths, the sustained DNA damage response activation caused by uncapped telomeres may promote leukemogenesis by molecular pathways distinct from typical accelerated telomere attrition associated with very short telomeres. Also, different RTEL1 variants modulated TRF2 expression in vitro, which may be an alternative mechanism related to RTEL1 dysfunction rather than impaired t-loop disassembly alone. DDR, DNA damage response; HSC, hematopoietic stem cell.
Assessment of the impact of variants in gene function and the use of frequency controls are critical. Pathogenicity assessment for a heterozygous RTEL1 variant is challenging, because approximately 1 of 100 individuals in the general population carries a rare variant in this gene with CADD score over 15 (Figure 1I). Consequently, detailed extended family history and functional data were imperative to classify heterozygous RTEL1 variants as pathogenic. It is important to point out that there is no single assay to assess the functional meaning of variants located outside the RING domain. The t-circle assay is used as a marker to identify RTEL1 deficient cells; however, as is seen in our results, other studies did not identify any accumulation of t-circles in patients’ cells.9,10 The interpretation of functional assays is still limited to what is known about the role of RTEL1 dysfunction in cell biology. Here, we describe RTEL1 as a modulator of TRF2 expression in vitro. Thus, TRF2 expression regulation may be an alternative mechanism related to RTEL1 dysfunction rather than impaired T-loop disassembly alone.
In summary, RTEL1 haploinsufficiency correlated with both accelerated telomere shortening and 3′ overhang attrition independent of length, and TL assessment alone may not identify all primary telomere defects. The pathogenicity of heterozygous RTEL1 variants must be evaluated with caution, because many very rare variants are present in population databases. In 5 patients, the RTEL1 variant (M344T, P867L, and P996H) was not etiologic. The number of patients in which RTEL1 variants are not pathogenic may be higher, because 13 variants were of uncertain significance. The combination of different tools for in silico prediction, well-established criteria to classify variants, a variant allele frequency control from ExAC, familial investigation, and functional data were critical in increasing the confidence of our analysis and in providing evidence that RTEL1 variants that were classified as likely pathogenic were etiologic.
The full-text version of this article contains a data supplement.
Acknowledgments
This work was funded by Bloodwise UK (program grants 10024 and 14017); by the Intramural Research Program of the National Heart, Lung, and Blood Institute, National Institutes of Health; and by the São Paulo Research Foundation (FAPESP; grant 13/08135-2). F.G.-R. was a recipient of a FAPESP scholarship (grant 2015/19074-0).
Authorship
Contribution: J.C.W.M. and F.G.-R. jointly wrote the article, performed data collection, experimental assays, data analysis, and interpretation, and contributed to study design; J.C. contributed bioinformatics data analysis and study design; J.J., S.K., X.F., M.d.P.F.I., F.S.D., J.P.L.d.S., Z.L., S.D., M.I., A.E.S., N.L., S.B., and R.I. contributed to experimental assays and data analysis; S.G., and I.C. performed data collection, data analysis, and interpretation; A.G.K. performed data analysis and contributed to patients’ recruitment; J.C., F.S.D., D.P.M., and A.P. performed data interpretation; R.T.C., N.S.Y., D.M.T., and G.J.M. contributed to study design, data interpretation, and patient recruitment; J.C.W.M., F.G.-R., J.C., S.K., I.C., R.T.C., N.S.Y., D.M.T., and G.J.M. critically reviewed the manuscript for intellectual content; and G.J.M. and D.M.T. had full access to all the data in this study and had final responsibility for the decision to submit for publication.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ghulam J. Mufti, Department of Haematological Medicine, King’s College Hospital, Denmark Hill, London SE5 9RS, United Kingdom; e-mail: ghulam.mufti@kcl.ac.uk.
References
Author notes
J.C.W.M. and F.G.-R. are joint first authors.
D.M.T. and G.J.M. are joint last authors.