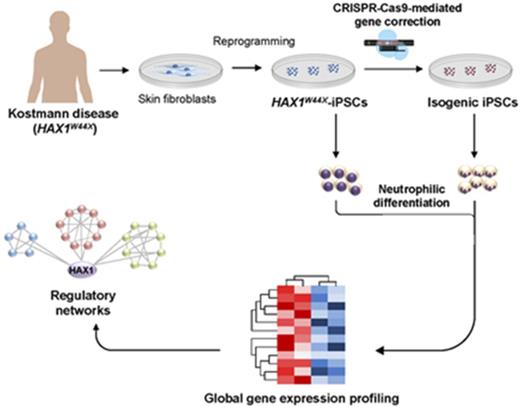
Key Points
HAX1W44X-iPSCs recapitulate Kostmann disease phenotype in vitro.
Genetic in situ correction of iPSCs reveals a dysregulated HAX1 and HCLS1-centered interaction network in Kostmann disease.
Abstract
Severe congenital neutropenia (SCN, Kostmann disease) is a heritable disorder characterized by a granulocytic maturation arrest. Biallelic mutations in HCLS1 associated protein X-1 (HAX1) are frequently detected in affected individuals, including those of the original pedigree described by Kostmann in 1956. To date, no faithful animal model has been established to study SCN mediated by HAX1 deficiency. Here we demonstrate defective neutrophilic differentiation and compensatory monocyte overproduction from patient-derived induced pluripotent stem cells (iPSCs) carrying the homozygous HAX1W44X nonsense mutation. Targeted correction of the HAX1 mutation using the CRISPR-Cas9 system and homologous recombination rescued neutrophil differentiation and reestablished an HAX1 and HCLS1-centered transcription network in immature myeloid progenitors, which is involved in the regulation of apoptosis, apoptotic mitochondrial changes, and myeloid differentiation. These findings made in isogenic iPSC-derived myeloid cells highlight the complex transcriptional changes underlying Kostmann disease. Thus, we show that patient-derived HAX1W44X-iPSCs recapitulate the Kostmann disease phenotype in vitro and confirm HAX1 mutations as the disease-causing monogenic lesion. Finally, our study paves the way for nonvirus-based gene therapy approaches in SCN.
Introduction
Severe congenital neutropenia (SCN) represents a rare hematologic disease entity that is clinically characterized by an absolute neutrophil count <0.5 × 109/L because of a granulocytic differentiation arrest at the promyelocyte stage.1 Patients may develop life-threatening bacterial infections if not treated with recombinant granulocyte colony-stimulating factor (G-CSF) to elevate neutrophil blood counts.1 Rolf Kostmann first characterized the disease in Swedish patients in 1956 as infantile genetic agranulocytosis.2 Multiple hereditary mutations have been reported in different forms of SCN (eg, in ELA2, G6PC3, GFI1, WASP SLC37A4).3 Autosomal recessive mutations in HCLS-associated protein X-1 (HAX1) were reported in 2007 as the genetic defect in the original Kostmann patients and many other families.4,5
Although the clinical phenotype of Kostmann disease is well-characterized, the pathogenesis is incompletely understood. HAX1 is a ubiquitously expressed protein.5 Its subcellular localization in the mitochondrial membrane, endoplasmic reticulum, and cytoplasm6-8 suggest a role in several cellular processes. HAX1 appears to be involved in antiapoptotic signaling through the activation of HTRA2 and the subsequent inhibition of mitochondrial outer membrane permeabilization by BAX accumulation.9 In addition, Skokowa et al suggested that HAX1, together with HCLS1, plays a role in Wnt/LEF1-mediated hematopoietic differentiation by transcriptional activation of CEBPα.10 However, Hax1 homozygous-null mice (Hax1−/−) have normal granulocyte counts in peripheral blood, but exhibit extensive apoptosis of neurons and lymphocytes and impaired B-cell development.11 Loss of motor neuron functions caused postnatal lethality in <14 weeks; thus, Hax1−/− mice do not model the phenotype of human Kostmann disease. As a result, preclinical research is limited to human in vitro disease models and formal proof for the HAX1 mutations because the causative and monogenic event in Kostmann disease is lacking.
Induced pluripotent stem cell (iPSC) technology provides an efficient and versatile platform to study rare hematologic disorders in vitro12-14 and allows fulfillment of Koch’s postulates in genetic diseases that cannot be recapitulated in animal models.15 Although a homozygous, 256C>T nonsense mutation in HAX1 (HAX1R86X) has been modeled previously using iPSCs,16 this variant has been described in only 2 patients and lies outside of exon 2a, the region where the majority of the reported mutations reside.4 Moreover, this variant has been linked to neurological deficits.4,17 In addition, viral-based overexpression of wild-type (WT) HAX1 complementary DNA (cDNA) was applied to revert the granulocytic differentiation arrest compared with iPSCs derived from a healthy individual.16
Here, we investigated iPSC-derived granulopoiesis from Kostmann patients (HAX1W44X-iPSCs) and gene-corrected isogenic lines generated by CRISPR-Cas9 gene editing. Using a new, robust neutrophilic differentiation assay, we have successfully modeled the Kostmann hematologic phenotype that is characterized by perturbed granulocytic differentiation and compensatory overproduction of monocytes.5 We demonstrate that targeted correction of the HAX1 mutation reestablished a HAX1- and HCLS1-centered transcription network in immature myeloid progenitors, which is involved in the regulation of apoptosis and also myeloid differentiation. Finally, our study paves the way for nonvirus-based gene therapy approaches in SCN.
Methods and materials
Cell culture
All experiments done with human embryonic stem cells (ESCs) and human iPSCs were approved by the German federal authorities (Robert Koch Institute: Aktenzeichen: 1710-79-1-4-41-E01). H9 ESCs were obtained from WiCell Research Institute (Madison, WI). CF1 mouse embryonic fibroblasts were kindly provided by the Max Planck Institute for Molecular Biomedicine, Münster, Germany. WT iPSCs were obtained and derived as described.18 The patient-derived skin fibroblasts were obtained and provided by the Severe Chronic Neutropenia International Registry. Informed consent was obtained from all patients and/or custodians, after institutional review board approval, according to national law and regulations and in accordance to the Declaration of Helsinki. For virus production, 293T cells were used (CRL-3216; ATCC, Manassas, VA).
Reprogramming
The RRL.PPT.SF.hOct34.hKlf4.hSox2.i2dTomato.pre lentiviral vector was used for cellular reprogramming.19 To enhance reprogramming efficiency, the microRNA 302/367 cluster was cloned into the backbone of the LeGO-G/BSD backbone (Addgene plasmid ID: 27354)20 for cotransduction with OCT4, KLF4, and SOX2.
Targeted gene correction
The CRISPR-Cas9–based gene correction was done as described by Ran et al21 using the pX330 plasmid (Addgene plasmid ID: 42230)22 and a 90-nt single-stranded oligodeoxynucleotide ([ssODN] produced by IDT, Coralville, IA; supplemental Table 1) for homology-directed repair. The ssODN was centered on the HAX1W44X mutation site. To test the cleavage efficiency of the targeting subgenomic RNA (sgRNA), a previously published reporter assay was used.23
Karyotype analysis
Classical chromosome banding analysis was carried out by the Department of Human Genetics of Hannover Medical School. Description of chromosomal aberrations and clone definition followed the recommendations of the International System for Cytogenetic Nomenclature, 2013.
Neutrophilic differentiation of iPSCs
Neutrophilic differentiation was performed as recently described.23 In short, iPSC colonies were detached as small fragments and shaken in suspension culture plates in basal embryonic stem cell medium at 100g for 5 days. Embryoid bodies were then collected and transferred to adherent culture plates in STEMdiff APEL medium (StemCell Technologies, Vancouver, BC, Canada) supplemented with 50 ng/mL G-CSF and 25 ng/mL interleukin-3 (Peprotech, Rocky Hill, NJ). Starting at day 15, suspension cells were collected and further cultured in RPMI 1640 supplemented with Glutamine (Merck Millipore), 10% fetal calf serum, and 50 ng/mL G-CSF for 7 to 14 days. Differentiated cells were then used for neutrophilic characterization.
Gene expression profiling
Isolated RNA samples of CD33+ myeloid progenitors were preamplified using Ovation RNA Amplification System V2 (Nugene, Irvine, CA). Expression profiles were generated on the Agilent SurePrint G3 platform in the Genome Analytics Department of Helmholtz Centre for Infection Research in Braunschweig, Germany. Expression data were analyzed with Genespring GX (Agilent, Santa Clara, CA), Gene Set Enrichment Analysis,24 and g:Profiler.25 Gene interaction networks were generated using GeneMANIA26 and Cytoscape 3.4.27
Results
Generation of Kostmann disease iPSCs
In 2007, Klein et al identified biallelic mutations in HAX1 in Kostmann patients.5 To obtain disease-specific iPSCs, we reprogrammed skin fibroblasts of 1 patient from the original pedigree5 using Cre-excisable lentiviral vectors (supplemental Figure 1A-C). The patient carried the homozygous 130_131insA nonsense mutation in HAX1 leading to a lack of WT HAX1 protein expression from a premature stop codon (W44X).5 Sequencing of HAX1 exon 2 confirmed that the 130_131insA mutation was retained on both alleles after reprogramming (Figure 1A). Morphology and alkaline phosphatase expression of HAX1W44X-iPSCs were comparable to the H9 ESC control (Figure 1B). Quantitative reverse transcriptase polymerase chain reaction (RT-PCR) and immunocytochemistry confirmed the expression of the pluripotency genes OCT4, SOX2, and NANOG at the messenger RNA and protein level (Figure 1C-D). Pluripotency surface marker staining of SSEA-4 showed a similar expression pattern of HAX1W44X-iPSCs and H9 ESCs (Figure 1E). No chromosomal aberrations were detected by karyotype analysis (Figure 1F). To functionally assess the quality of HAX1W44X-iPSCs, differentiation potential into ectoderm, mesoderm, and endoderm was tested using the scorecard assay.28 Following undirected differentiation, HAX1W44X-iPSCs showed upregulation of genes of all germ layers, whereas pluripotency genes were strongly downregulated (Figure 1G; supplemental Figure 2). Therefore, we successfully generated HAX1W44X-iPSCs from patient-derived skin fibroblasts.
Characterization of patient-derived HAX1W44X-iPSC. Generated HAX1W44X-iPSCs were compared with H9 ESCs and showed activation of their pluripotency network. (A) Sanger sequencing of HAX1W44X-iPSCs confirmed the c.130_131insA mutation leading to a stop codon in exon 2 of HAX1. (B) High expression pattern of alkaline phosphatase in ESCs was also detectable in generated iPSCs. Original magnification ×100; Naphthol/Fast Red Violet Solution. (C-D) Detection of pluripotency factors OCT4, SOX2, and NANOG by (C) immunohistochemistry (original magnification ×200) and (D) quantitative RT-PCR (n = 3, reference bar represents H9 ESC). (E) Flow cytometric quantification of ESC surface marker SSEA-4. (F) Karyotype analysis showed no chromosomal aberrations after reprogramming in HAX1W44X-iPSCs. (G) Upon undirected EB-based differentiation, scorecard assays demonstrated the capability of generated iPSCs to differentiate in all 3 germ layers. Pluripotency genes were strongly downregulated. EB, embryoid body.
Characterization of patient-derived HAX1W44X-iPSC. Generated HAX1W44X-iPSCs were compared with H9 ESCs and showed activation of their pluripotency network. (A) Sanger sequencing of HAX1W44X-iPSCs confirmed the c.130_131insA mutation leading to a stop codon in exon 2 of HAX1. (B) High expression pattern of alkaline phosphatase in ESCs was also detectable in generated iPSCs. Original magnification ×100; Naphthol/Fast Red Violet Solution. (C-D) Detection of pluripotency factors OCT4, SOX2, and NANOG by (C) immunohistochemistry (original magnification ×200) and (D) quantitative RT-PCR (n = 3, reference bar represents H9 ESC). (E) Flow cytometric quantification of ESC surface marker SSEA-4. (F) Karyotype analysis showed no chromosomal aberrations after reprogramming in HAX1W44X-iPSCs. (G) Upon undirected EB-based differentiation, scorecard assays demonstrated the capability of generated iPSCs to differentiate in all 3 germ layers. Pluripotency genes were strongly downregulated. EB, embryoid body.
Targeted correction of nonsense mutation in HAX1W44X-iPSCs using CRISPR-Cas9–induced homology directed repair
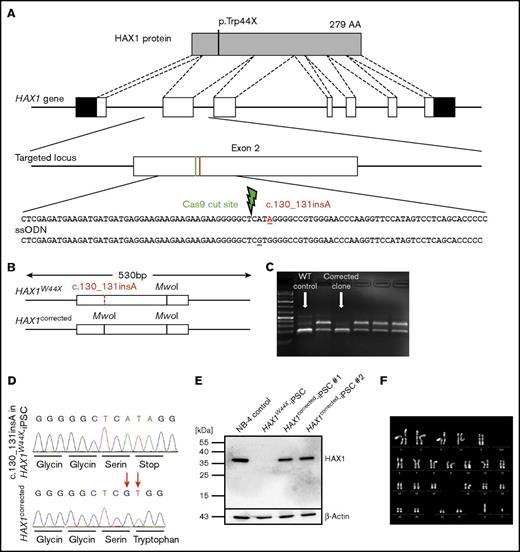
To provide a specific and efficient correction of the mutation in Kostmann disease iPSCs, we used the CRISPR-Cas9 technology (Figure 2A).21 For homology-directed repair, we designed a 90-nt ssODN as the corrected donor sequence. A guide was selected that cleaves the genomic DNA 3 base pairs upstream of the mutation site. To avoid ssODN cleavage by Cas9, we introduced a silent mutation in the serine codon upstream of the correction site (Figure 2A). Using a restriction-based screening approach followed by Sanger sequencing, we successfully identified 2 corrected clones of the 288 that were tested (Figure 2B-D). Our biallelic homology-directed repair efficiency of 0.7% is in the range of reported frequencies.29 Restoration of HAX1 protein expression after genetic correction was confirmed by western blotting (Figure 2E). The expression level was comparable to that in the promyelocytic cell line NB-4.
Targeted correction of c.130_131insA restored HAX1 protein expression. (A) Schematic overview of the HAX1 protein and the corresponding genomic locus including the location of c.130_131insA insertional mutation, which leads to a premature stop codon. The c.130_131insA mutation and the location of the double-strand break are indicated in red and green, respectively. The ssODN provided the corrected sequence and a silent mutation in the serine codon upstream of the mutation site. (B-C) Upon successful correction, a restriction site for MwoI was gained in exon 2 that was used to distinguish corrected clones from uncorrected after agarose gel electrophoresis (C). In corrected and WT clones, the additional MwoI site would cut the 530-bp amplicon into 2 fragments of ∼190 bp and 1 fragment of 144 bp. Digestion of uncorrected clone amplicons lead to bands of 336 and 194 bp. (D) The corrected sequence and the silent mutation in the serine codon were detected via Sanger sequencing. (E) The 2 healthy isogenic cell lines and the positive control NB-4 cells expressed HAX1 protein analyzed by western blotting. HAX1W44X-iPSCs showed no HAX1 expression. (F) Normal karyotype with no chromosomal aberrations could be detected in HAX1corrected-iPSCs.
Targeted correction of c.130_131insA restored HAX1 protein expression. (A) Schematic overview of the HAX1 protein and the corresponding genomic locus including the location of c.130_131insA insertional mutation, which leads to a premature stop codon. The c.130_131insA mutation and the location of the double-strand break are indicated in red and green, respectively. The ssODN provided the corrected sequence and a silent mutation in the serine codon upstream of the mutation site. (B-C) Upon successful correction, a restriction site for MwoI was gained in exon 2 that was used to distinguish corrected clones from uncorrected after agarose gel electrophoresis (C). In corrected and WT clones, the additional MwoI site would cut the 530-bp amplicon into 2 fragments of ∼190 bp and 1 fragment of 144 bp. Digestion of uncorrected clone amplicons lead to bands of 336 and 194 bp. (D) The corrected sequence and the silent mutation in the serine codon were detected via Sanger sequencing. (E) The 2 healthy isogenic cell lines and the positive control NB-4 cells expressed HAX1 protein analyzed by western blotting. HAX1W44X-iPSCs showed no HAX1 expression. (F) Normal karyotype with no chromosomal aberrations could be detected in HAX1corrected-iPSCs.
To exclude chromosomal instability caused by CRISPR-Cas9 editing, karyotype analysis was performed after genetic correction, and no chromosomal aberrations were detected in either corrected clone (Figure 2F). Furthermore, the sequences of the top 5 off-target sites30 that have 4 or more mismatches (TMEM255A, SHISA7, MARK4, PTH2, and DST; supplemental Table 2) showed no abnormalities when compared with its isogenic HAX1W44X-iPSC clone (supplemental Figure 3). Taken together, gene correction using CRISPR-Cas9 technology rescued HAX1 protein expression in HAX1W44X-iPSCs.
Correction of mutated HAX1 rescues Kostmann disease phenotype of patient-derived iPSCs
To assess the granulocytic differentiation potential of HAX1W44X iPSCs and isogenic gene-corrected iPSCs (HAX1corrected-iPSCs), a 2-step protocol was used (Figure 3A).23 iPSCs from a healthy donor served as the WT control (HAX1WT/WT-iPSC). The proliferation rates of HAX1W44X-iPSCs, gene-corrected iPSCs (HAX1corrected-iPSCs), and HAX1WT/WT-iPSCs did not significantly differ (supplemental Figure 4). In the first step, embryoid bodies (EBs) were generated from colony fragments. EBs were then seeded on attachment culture plates and incubated in hematopoietic specification medium containing interleukin-3 and G-CSF. In the second step, hematopoietic suspension cells were collected and further cultured in neutrophilic differentiation medium containing G-CSF. Using this protocol, granulocytes were derived from HAX1WT/WT-iPSCs with a purity of 91% to 99% as quantified by morphology (Figure 3B-C). In contrast, we observed that HAX1W44X-iPSCs predominantly differentiated into monocytes (Figure 3B-C), a finding consistent with the clinical phenotype.5 Few granulocytes (28%; range, 13% to 44%, P < .01) were present in HAX1W44X-iPSC–derived cells (Figure 3B-C). Similar findings were observed in 2 additional HAX1W44X-iPS clones (data not shown). After targeted gene correction, differentiation of iPSCs yielded a highly enriched granulocytic cell population almost at the level of HAX1WT/WT-iPSCs (91%; range, 83% to 99%; Figure 3B-C). By flow cytometry, CD11b+CD15+ neutrophilic cells reached a purity of 61% (range, 59% to 73%) in HAX1corrected and 75% (range, 61% to 83%) in HAX1WT/WT-iPSC clones, but only 28% (range, 21% to 35%) in the parental HAX1W44X clone (P < .01; Figure 3D-E). To further validate the function of the generated neutrophils, cells were stimulated with phorbol 12-myristate 13-acetate (PMA) to form neutrophil extracellular traps (NETs, Figure 3F). In primary neutrophilic granulocytes from the peripheral blood of a healthy donor, DNA staining showed NETs consisting of typical weblike structures. HAX1W44X-iPSC–derived cells did not form such NETs, whereas numerous NETs were observed in the corrected isogenic and the HAX1WT/WT clones (Figure 3F). An immortalized T-cell line (Jurkat) was used as a negative control and showed no activation following PMA treatment.
Neutrophilic in vitro differentiation showed recapitulation of Kostmann phenotype and rescue upon genetic correction. (A) Schematic overview of the hematopoietic in vitro differentiation protocol. Spin-EBs were generated, selected, and transferred into plates for adhesion. Attached EBs formed myeloid cell–forming complexes that released progenitors as suspension cells into the medium. After hematopoietic specification, step suspension cells were collected and applied to neutrophilic differentiation. IL-3, interleukin-3. (B-C) After neutrophilic differentiation, morphology was assessed by (B) May-Grünwald Giemsa staining of cytospins. Original magnification ×200 (insets, ×400). (C) Percentages of neutrophilic granulocytes (Gran), promyelocytes (Prom) and monocytes (Mono) are presented as mean ± standard deviation (SD) of n = 10 independent experiments. (D) Percentage (mean ± SD) of CD15+ (neutrophilic marker) HAX1WT/WT, HAX1W44X, and HAX1corrected cells after neutrophilic differentiation as assessed by flow cytometry (n = 4). (E) Representative fluorescence-activated cell sorted plots and gating strategy are shown. The values indicate mean ± SD of n = 4 experiments. (F) DNA staining after PMA stimulation showed NET formation in peripheral blood granulocytes, HAX1WT/WT and HAX1corrected-iPSC–derived cells. Original magnification ×100 (insets, ×400); SYTOX Green stain. (C-D) **P < .01 when compared with HAX1W44X-iPSCs.
Neutrophilic in vitro differentiation showed recapitulation of Kostmann phenotype and rescue upon genetic correction. (A) Schematic overview of the hematopoietic in vitro differentiation protocol. Spin-EBs were generated, selected, and transferred into plates for adhesion. Attached EBs formed myeloid cell–forming complexes that released progenitors as suspension cells into the medium. After hematopoietic specification, step suspension cells were collected and applied to neutrophilic differentiation. IL-3, interleukin-3. (B-C) After neutrophilic differentiation, morphology was assessed by (B) May-Grünwald Giemsa staining of cytospins. Original magnification ×200 (insets, ×400). (C) Percentages of neutrophilic granulocytes (Gran), promyelocytes (Prom) and monocytes (Mono) are presented as mean ± standard deviation (SD) of n = 10 independent experiments. (D) Percentage (mean ± SD) of CD15+ (neutrophilic marker) HAX1WT/WT, HAX1W44X, and HAX1corrected cells after neutrophilic differentiation as assessed by flow cytometry (n = 4). (E) Representative fluorescence-activated cell sorted plots and gating strategy are shown. The values indicate mean ± SD of n = 4 experiments. (F) DNA staining after PMA stimulation showed NET formation in peripheral blood granulocytes, HAX1WT/WT and HAX1corrected-iPSC–derived cells. Original magnification ×100 (insets, ×400); SYTOX Green stain. (C-D) **P < .01 when compared with HAX1W44X-iPSCs.
Taken together, this set of data supports recapitulation of the Kostmann phenotype using in vitro differentiation of patient-derived iPSCs, which was reversed by targeted gene correction. HAX1corrected-iPSCs yielded highly enriched neutrophilic populations that were able to form NETs upon PMA stimulation.
Genetic correction of HAX1W44X reestablishes HAX1/HCLS1-centered gene expression network
To investigate the transcriptional changes consequent to mutated HAX1, we performed global gene expression profiling of SSClow/CD33+ myeloid progenitors that were fluorescence-activated cell sorted after hematopoietic specification of the iPSC in vitro differentiation protocol (Figure 3A; supplemental Figure 5). We decided to use CD33+ myeloid progenitors for this analysis because the granulocytic differentiation arrest seen in Kostmann disease only becomes apparent after the promyelocyte stage. Comparison of HAX1W44X with HAX1WT/WT isogenic myeloid progenitors demonstrated the downregulation of a granulocyte expression program. Granulocyte-specific31 and myeloid-specific32 gene sets were negatively enriched (Figure 4A; supplemental Figure 6A, supplemental Table 3). Among the downregulated genes were well-characterized regulators of granulocyte differentiation, such as the lineage-determining transcription factor PU.1.33 These findings suggest perturbation of the granulocytic differentiation program already at the progenitor cell stage. Correction of mutated HAX1 in isogenic HAX1corrected-iPSCs led to a complete reversal of the perturbed expression (Figure 4B-C; supplemental Figure 6B-C; supplemental Table 4), providing molecular proof for the rescue of granulocytic differentiation.
Expression profiling of iPSC-derived CD33+myeloid progenitors identified a downregulated genetic interaction network in Kostmann disease. The granulocyte-specific gene set is positively enriched in (A) HAX1WT/WT and (B) HAX1corrected myeloid progenitors compared with HAX1W44X cells. (C) When comparing HAX1WT/WT and HAX1corrected cells, no significant enrichment could be observed, suggesting that those groups show similar granulocytic expression patterns. ES, enrichment score, FDR, false discovery rate; FWER, family-wise error rate; NES, normalized enrichment score. (D) Heatmap (left) of the 533 myeloid-specific genes (leading edge subsets) that were downregulated in HAX1W44X-iPSC–derived myeloid progenitors as compared with HAX1WT/WT- and HAX1corrected-iPSC–derived cells. Those were used to form a gene interaction network (supplemental Figure 9B). The downregulated gene interaction partners of HAX1 and HCLS1 in HAX1W44X-iPSC–derived CD33+ granulocytic progenitors are shown (right). (E-F) 3,3′-diethyloxacarbocyanine iodide (DiOC2(3)) mean fluorescence intensity (MFI) of (E) TRA-1-60+ iPSCs and (F) CD33+ cells during induced myeloid differentiation with or without addition of mitochondrial membrane potential disruptor carbonyl cyanide 3-chlorophenylhydrazone (CCCP). Mean ± SD of n = 4-5 experiments are shown. NS, not significant. *P < .05 when compared with HAX1wt/wt.
Expression profiling of iPSC-derived CD33+myeloid progenitors identified a downregulated genetic interaction network in Kostmann disease. The granulocyte-specific gene set is positively enriched in (A) HAX1WT/WT and (B) HAX1corrected myeloid progenitors compared with HAX1W44X cells. (C) When comparing HAX1WT/WT and HAX1corrected cells, no significant enrichment could be observed, suggesting that those groups show similar granulocytic expression patterns. ES, enrichment score, FDR, false discovery rate; FWER, family-wise error rate; NES, normalized enrichment score. (D) Heatmap (left) of the 533 myeloid-specific genes (leading edge subsets) that were downregulated in HAX1W44X-iPSC–derived myeloid progenitors as compared with HAX1WT/WT- and HAX1corrected-iPSC–derived cells. Those were used to form a gene interaction network (supplemental Figure 9B). The downregulated gene interaction partners of HAX1 and HCLS1 in HAX1W44X-iPSC–derived CD33+ granulocytic progenitors are shown (right). (E-F) 3,3′-diethyloxacarbocyanine iodide (DiOC2(3)) mean fluorescence intensity (MFI) of (E) TRA-1-60+ iPSCs and (F) CD33+ cells during induced myeloid differentiation with or without addition of mitochondrial membrane potential disruptor carbonyl cyanide 3-chlorophenylhydrazone (CCCP). Mean ± SD of n = 4-5 experiments are shown. NS, not significant. *P < .05 when compared with HAX1wt/wt.
To understand the cellular processes that are affected by the HAX1 mutation during granulocytic differentiation, we performed network analysis of the genes specifically downregulated in the HAX1W44X samples (supplemental Figures 7 and 8; for quantitative RT-PCR validation of selected genes, see supplemental Figure 9A). We observed that the downregulated genes are strongly interconnected by various known and predicted direct protein-protein interactions (Figure 4D; supplemental Figure 9B). The network analysis revealed nodes or clusters of interactions that could be attributed to gene ontology (GO) categories related to mitochondrial homeostasis (apoptotic mitochondrial changes, regulation of mitochondrial membrane potential) and the activation of apoptotic signaling pathways (regulation of apoptotic signaling pathway, regulation of leukocytic apoptosis, apoptotic mitochondrial changes). Accordingly, the mitochondrial membrane potential of HAX1W44X CD33+ myeloid progenitors but not of HAX1W44X-iPSCs is lower compared with the respective isogenic HAX1corrected or HAX1WT/WT samples (Figure 4E-F). This translates into a higher rate of apoptosis in HAX1W44X myeloid progenitors (supplemental Figure 10). However, the deregulation of clusters of genes involved in transcriptional control, myeloid cell behavior and reactive oxygen metabolism (GO terms: transcription factor binding, granulocytic migration, myeloid cell homeostasis, and reactive oxygen species [ROS] metabolic process) indicates functions of HAX1 beyond mitochondrial apoptosis. Inversely, the respective GO sets (containing all genes of the respective GO category) were negatively enriched in HAX1W44X samples confirming their regulation by HAX1. Most important, the interaction network of downregulated genes is centered around HAX1 and HCLS1 and includes several direct interaction partners of either protein (Figure 4D; supplemental Figure 9A-B). For example, the interaction of GSK3A and HCLS1 has been experimentally validated.34 GSK3A is a crucial player in Wnt signaling and interacts with CTNNB1/LEF1 as well as JUN.35 CTNNB1 is also among the downregulated genes in HAX1W44X myeloid progenitors. TMBIM6 is an inhibitor of BAX1-induced apoptosis and is directly related to the function of HAX1 in preventing mitochondrial outer membrane permeabilization by BAX accumulation.36 Central for this function of HAX1 is PARL,9 which is 1 of the downregulated genes in the HAX1W44X-iPSC–derived granulocytic progenitors.
Thus, mutation of HAX1 leads to global changes in transcription networks involved in the regulation of apoptosis and myeloid differentiation. Correction of mutated HAX1 reverses those changes, reestablishing normal myeloid cell homeostasis and differentiation.
Discussion
Here we demonstrate the successful establishment of an iPSC model for Kostmann disease that recapitulates the granulocytic differentiation arrest and compensatory monocyte overproduction seen in patients. We showed that targeted genetic correction of the HAX1 mutation reestablished a HAX1- and HCLS1-centered transcription network in immature myeloid progenitors, which is not only involved in the regulation of apoptosis, but also myeloid differentiation. Our study demonstrates that correction of the underlying genetic event in Kostmann patients using the CRISPR-Cas9 system might be used as a therapeutic intervention to cure the disease in the affected patients without causing genotoxicity introduced by virus-based strategies.37,38
Virus-based gene transfer has been successfully applied in current gene therapy trials39,40 ; however, viral gene therapy harbors the risk of inducing hematologic malignancies or, on the other hand, treatment failure resulting from silencing of the transgene.37,41-43 Transgene silencing is a particular problem upon use of lentivirally transduced iPSCs to derive hematologic cells.44-46 During the differentiation process, the open chromatin of iPSCs undergoes complex epigenomic changes,47 which can hamper transgene expression.48 In our hands, a rescue approach for the hematopoietic differentiation arrest of HAX1W44X-iPSCs by lentiviral overexpression of WT codon-optimized HAX1 cDNA was unsuccessful (supplemental Figure 11A-D). Even for disease-modeling approaches, the use of lentiviral-based cDNA overexpression for the correction of the respective disease phenotype is imprecise because of potentially occurring insertional mutagenesis and unknown effects of constitutively high transgene expression using viral promoters in cells, which normally do not express the protein.49 This can lead to a defective activation of cellular processes, which has also been observed in clinical trials for X-linked chronic granulomatous disease. Here, the overexpression of CYBB (gp90phox), a subunit of the reduced NAD phosphate oxidase enzyme complex, impaired long-term engraftment of HSCs after transplantation50 resulting from improper ROS production.51
The use of gene-editing techniques circumvents several of these issues. By precise genetic correction of the “disease-causing” mutation in patient-derived iPSCs, isogenic cell lines were derived that differ from the parental iPSC line only by the targeted sequence. This provides optimal experimental conditions for iPSC disease modeling as patient-specific or clonal variability between different iPSC lines can be largely excluded.52,53 Furthermore, this strategy allows for confidence in genetic causality without the necessity of animal models.54-56 Our CRISPR-Cas9– and homology-directed repair-based 1-step cell transfection and screening protocol allowed the generation and identification of corrected clones in less than 3 weeks with no detectable chromosomal aberrations or mutations at the sgRNA’s top predicted 5 off-target sites. Although whole genome sequencing is required to fully exclude off-target cleavage of the Cas9 endonuclease,57-59 careful selection of the sgRNAs and the subsequent data indicate that our system provides genetically defined conditions to investigate the net effect of the HAX1W44X mutation, as it was previously shown for ELANE mutations in SCN and X-linked chronic granulomatous disease.60,61 These isogenic iPSCs can facilitate drug or toxicity screenings to find the most advantageous therapies in this rare disease; furthermore, they create an ideal platform for the development of improved therapy in SCN.
HAX1 is primarily located in the mitochondrial membrane, endoplasmic reticulum, and cytoplasm.6-8 Since the identification of HAX1 mutations in Kostmann disease, it remained unclear why patients show selectively impaired neutrophilic differentiation. By genomewide expression profiling of iPSC-derived isogenic myeloid progenitor cells, we demonstrate that the HAX1W44X mutation not only affects the expression of genes involved in apoptosis or apoptotic mitochondrial changes, but also impaired expression of central transcriptional regulators of myeloid differentiation. Among the differentially repressed genes in the HAX1W44X-iPSC–derived myeloid progenitors was HCLS1, which is the direct interaction partner of HAX1.6 Skokowa et al highlighted the essential role of G-CSF–mediated HCLS1 activation and nuclear translocation via association of the Lyn and Syk kinases during granulopoiesis.10 In agreement with this observation, our findings demonstrate that this important axis for granulocytic differentiation was downregulated in HAX1W44X-iPSC–derived cells (Figure 4D). Similarly, the expression of key members of the Wnt-signaling cascade (GSK3A and CTNNB1) that is a crucial player in the pathogenesis of SCN,10 was affected by the HAX1 mutation. Interestingly, these observations also support previous findings that the SCN phenotype can be partially rescued in vitro by addition of Wnt3a.62 Noteworthy is that, by using purified myeloid progenitor cells, these defects already affect the specification process: an observation only feasible because of the generation of a fully isogenic-iPSC amendable to interrogate all stages of Kostmann disease. Furthermore, HAX1 appears to carry out antiapoptotic functions through the inhibition of MOMP by PARL-mediated activation of HTRA2.9 Recently, Abdelwahid et al demonstrated HAX1-mediated cell survival by protection from multiple apoptotic signals in a cardiomyocyte cell line.63 We also observed deregulation of genes involved in a multitude of apoptotic processes, such as PARL, FAIM2, and TMBIM6, suggesting that the role of HAX1 and HCLS1 in apoptosis of myeloid cells may be another key function in the Kostmann disease genetic network. In fact, we validated decreased mitochondrial membrane potential and increased apoptosis in those cells. Moreover, enhanced ROS production and subsequent DNA damage have been suggested to play a major role in the acquisition of neutropenia phenotype of Barth syndrome.64 We also identified downregulated genes in ROS metabolism, including CYBB, NCF1, and NCF2, which are mutated in chronic granulomatous disease.65-67 Together with other important neutrophilic genes such as MPO, NOX4, and SOD2, the ROS metabolism seems to be 1 of the central processes impaired in the myeloid cell differentiation of HAX1W44X-iPSCs.
Here we have demonstrated that targeted genetic correction of the HAX1 gene is applicable to repair patient-derived iPSCs. Moreover, we showed that the neutropenia phenotype observed upon in vitro differentiation could be reversed in corrected iPSCs, and we were able to demonstrate that monocytosis observed in patients5 is also recapitulated in culture. Taken together, we show that the iPSC differentiation system is suitable for study of the clinical phenotype of Kostmann SCN and to develop new therapeutic options. In addition, we suggest a Kostmann disease–specific gene signature in which HAX1 and HCLS1 act as the central players of a large dysregulated genetic network and that suggest further implications of HAX1 to other important cellular signaling pathways.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank M. Ballmaier for cell sorting, and K. Weber and B. Fehse for providing LeGO plasmids.
This work was supported by grants to J.-H.K. and T.C. from the José Carreras Leukemia Foundation. J.-H.K. is a fellow of the Emmy Noether-Programme of the German Research Foundation (DFG) (KL-2374/2-1). N.L. was supported by the DFG (LA 3680/2-1), the Else Kröner-Fresenius-Stiftung (2015_A92), and Hannover Medical School (young academy program). E.P. was supported by the Hannover Biomedical Research School. D.H. was supported by the Max-Eder program from the German Cancer Aid (no. 111743). A.S., G.G., N.L., E.P., and T.C. were supported by the DFG (Cluster of Excellence REBIRTH EXC 62/3 and/or SFB738). G.M. and S.H.O. were supported in part by National Institutes of Health, National Heart, Lung, and Blood Institute U01 HL100001 (Human Pluripotent Stem Cell and Progenitor Models of Cardiac and Blood Diseases, G. Q. Daley, principal investigator).
Authorship
Contribution: E.P. conducted all experiments, analyzed the data, and wrote the manuscript; J.-H.K. designed and supervised the project, planned the experiments, analyzed the data, and wrote the manuscript; T.C. designed and supervised the project and revised the manuscript; A.S. provided OCT4, KLF4, and SOX2 and codon-optimized HAX1 complementary DNA vectors, and cosupervised the project; D.H. designed experiments and revised the manuscript; S.E., M.A., and N.L. provided technical assistance and analyzed the data; G.M. and S.H.O. designed experiments, interpreted data, and revised the manuscript; G.G. and B.S. performed the cytogenetic analysis and interpreted the data; and K.W., codirector of the Severe Chronic Neutropenia International Registries, provided the human Kostmann disease sample and revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jan-Henning Klusmann, Department of Pediatric Hematology and Oncology, Hannover Medical School, Carl-Neuberg-Str 1, D-30625 Hannover, Germany; e-mail: klusmann.jan-henning@mh-hannover.de.
References
Author notes
T.C. and J.-H.K. contributed equally to this study.
The data reported in this article have been deposited in the Gene Expression Omnibus (accession number GSE97414).