Key Points
ST2, REG3α, and/or AREG at the time of acute GVHD diagnosis are excellent predictors of risk for 12-month NRM.
The best biomarker algorithm and threshold for risk stratification may depend on the target population.
Visual Abstract
Graft-versus-host disease (GVHD) is a major cause of nonrelapse mortality (NRM) after allogeneic hematopoietic cell transplantation. Algorithms containing either the gastrointestinal (GI) GVHD biomarker amphiregulin (AREG) or a combination of 2 GI GVHD biomarkers (suppressor of tumorigenicity-2 [ST2] + regenerating family member 3 alpha [REG3α]) when measured at GVHD diagnosis are validated predictors of NRM risk but have never been assessed in the same patients using identical statistical methods. We measured the serum concentrations of ST2, REG3α, and AREG by enzyme-linked immunosorbent assay at the time of GVHD diagnosis in 715 patients divided by the date of transplantation into training (2004-2015) and validation (2015-2017) cohorts. The training cohort (n = 341) was used to develop algorithms for predicting the probability of 12-month NRM that contained all possible combinations of 1 to 3 biomarkers and a threshold corresponding to the concordance probability was used to stratify patients for the risk of NRM. Algorithms were compared with each other based on several metrics, including the area under the receiver operating characteristics curve, proportion of patients correctly classified, sensitivity, and specificity using only the validation cohort (n = 374). All algorithms were strong discriminators of 12-month NRM, whether or not patients were systemically treated (n = 321). An algorithm containing only ST2 + REG3α had the highest area under the receiver operating characteristics curve (0.757), correctly classified the most patients (75%), and more accurately risk-stratified those who developed Minnesota standard-risk GVHD and for patients who received posttransplant cyclophosphamide-based prophylaxis. An algorithm containing only AREG more accurately risk-stratified patients with Minnesota high-risk GVHD. Combining ST2, REG3α, and AREG into a single algorithm did not improve performance.
Introduction
Acute graft-versus-host disease (GVHD) remains a major cause of morbidity and mortality after allogeneic hematopoietic stem cell transplantation (HCT), despite modern prophylaxis regimens such as posttransplant cyclophosphamide (PT-CY) that have reduced the maximum severity of GVHD but the overall incidence and the need for systemic treatment remain high.1-5 The maximum severity of acute GVHD is mainly driven by gastrointestinal (GI) tract damage and correlates well with nonrelapse mortality (NRM) and survival, but can only be determined in retrospect after treatment.6 The overall symptom severity at diagnosis, when the full extent of GI damage is not yet known, correlates modestly with response to treatment and long-term outcomes7,8 but the overall grade is used nonetheless to determine eligibility for treatment trials tailored for different risks, such as less toxic therapy for low-risk9-11 and more intensive therapy for high-risk GVHD12 (also NCT05263999, NCT04167514). Furthermore, clinical practice is heterogeneous, with some clinicians choosing topical therapy, whereas others prescribe systemic corticosteroids for patients with similar mild acute GVHD presentations. Laboratory measures of GVHD severity at the time of diagnosis that more accurately predict treatment outcomes than clinical symptoms, such as GI GVHD biomarkers, are needed to design more efficient clinical trials and may help guide treatment selection, including the decision to use topical rather than systemic treatment.
Three serum biomarkers, regenerating family member 3 alpha (REG3α), suppressor of tumorigenicity-2 (ST2), and amphiregulin (AREG), quantify GI damage in the context of acute GVHD.13-18 REG3α is an antimicrobial peptide secreted by Paneth cells that provides a key survival signal for intestinal stem cells necessary for the regeneration of GI crypts.16,17 ST2 is a ligand for interleukin-33 (IL-33), a protein secreted by damaged epithelial cells.19 The interaction between IL33 and ST2 is thought to be anti-inflammatory under normal conditions but it can potentiate GI tissue damage during the inflammation of GVHD.20 AREG, a weak epidermal growth factor ligand, promotes GI epithelial barrier repair and is secreted by effector cells, such as innate lymphoid cell-2s, gut-associated lymphoid tissue, and alloreactive T cells when stimulated by IL-33.21-25 ST2 and AREG are both strongly associated with the repair and inflammatory cascade known as the ST2-IL-33 axis, although the exact role of these biomarkers in GVHD pathogenesis is still an area of active study.26,27
Several groups have validated acute GI GVHD biomarker-based algorithms to predict short- and long-term outcomes at the time of diagnosis. The Mount Sinai Acute GVHD International Consortium (MAGIC) algorithm probability uses the concentrations of 2 biomarkers, ST2 and REG3α, to predict the response to systemic therapy, risk of NRM, and survival.28-30 This algorithm was recently validated as a prognostic tool that is superior to clinical prediction models, such as the Minnesota GVHD risk system.6,8,31 A group at the University of Minnesota developed AREG as a prognostic biomarker that predicts both the risk of NRM and OS and is superior to the Minnesota risk system.32,33 Both algorithms have been used to select patients with high- or low-risk GVHD for clinical trials testing primary treatment34-36 (also clinical trials NCT05123040, NCT02525029, NCT04291261, and NCT05090384).
A recent publication showed that the combination of ST2 and REG3α was the most accurate of a panel of 5 biomarkers in predicting GVHD outcomes but AREG was not included in that analysis.28 In this study, we expanded upon our prior work using the same large cohort of patients and identical statistical techniques to evaluate the combination of ST2, REG3α, and AREG that best stratifies patients with GVHD according to risk for 12-month NRM.
Methods
Study design and oversight
The MAGIC database and biorepository uses a PRoBE (prospective-specimen collection, retrospective-blinded-evaluation) design in which serum samples and clinical data are prospectively collected before clinical outcomes are known, biomarker concentrations are determined without knowledge of the patient’s clinical status or outcome, and unbiased methods (eg, random assignment) are used to include subjects in analyses.37 In this study, we included 715 patients from the MAGIC database and biorepository diagnosed with acute GVHD as defined by the MAGIC criteria,38 with sufficient remaining serum from a prior 730 patient study that compared GI and systemic biomarkers for predicting GVHD outcomes (supplemental Table 1).28 All the patients received topical and/or systemic therapy for acute GVHD upon diagnosis. Patients were divided into a training cohort (n = 341), who underwent allogeneic HCT between May 2004 and October 2015, and a validation cohort (n = 374), who underwent allogeneic HCT between November 2015 and April 2017, as previously reported (supplemental Table 2).28 PT-CY-based GVHD prophylaxis has become increasingly prevalent; thus, we supplemented the PT-CY subset with an additional 77 patients from the MAGIC database and biorepository who underwent allogeneic HCT between 2020 and 2022 and developed GVHD (supplemental Table 3). To avoid selection bias, we included patients who sequentially underwent transplantation in the reverse order from the most recent patient with 12 months follow-up. The size of the PT-CY subset (n = 133) relative to the total validation cohort (n = 451) approximates the proportion of patients (29%) in the MAGIC database and biorepository who received PT-CY prophylaxis between 2020 and 2022. All patients, parents, and legal guardians provided informed consent on an institutional review board-approved protocol.
Biomarker determination and algorithm development
We measured ST2 and REG3α concentrations at the Icahn School of Medicine at Mount Sinai28 and measured AREG concentrations at the University of Minnesota using enzyme-linked immunosorbent assay according to published protocols.32 ST2 and AREG were expressed in picograms per milliliter and REG3α was expressed in nanograms per milliliter. All biomarker values were log-10 transformed for use in algorithms. Competing risk regression that considered relapse and second transplant as competing risks was used in the training cohort to create biomarker algorithms for all 7 possible combinations of 1, 2, or 3 biomarkers to predict the probability of 12-month NRM from the time of diagnosis of GVHD. Each algorithm calculated the predicted probability of 12-month NRM as a value from 0.001 to 0.999 using the complementary log-log link for each individual patient in the training cohort and then identified the threshold to separate low and high risk according to the concordance probability (the value that maximizes sensitivity and specificity).39 Threshold performance was assessed using several metrics, including sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), balanced accuracy, and the proportion of patients correctly classified as high or low risk. Balanced accuracy was defined as the average sensitivity and specificity.40 Patients were deemed correctly classified as high-risk if they died from NRM within 12 months from the diagnosis of GVHD and as low-risk if they did not experience NRM. All assessments of performance and comparisons among algorithms used data from only the validation cohort.
NRM was defined as death within 12 months of GVHD onset from any cause other than relapse. Treatment response on day 28 of systemic therapy was defined as follows: a complete response required resolution of all GVHD symptoms; a partial response required improvement of at least 1 stage in at least 1 organ without worsening in any other organ; initiation of second-line systemic therapy, death before day 28, and all other responses were categorized as nonresponses.
Statistical methods
Patient characteristics between the training and validation cohorts were compared using the χ2 or Wilcoxon 2-sample tests as appropriate. Correlations between individual biomarker algorithms were evaluated using the Pearson correlation coefficient. Areas under the receiver operating characteristic curves were compared using DeLong test,41 and P value were adjusted for the false discovery rate.42 The cumulative incidences of NRM and relapse were calculated using Fine and Gray method.43 Differences in cumulative incidences were compared using Gray test44 except when the cumulative incidence curves crossed, in which case the cumulative incidence rates at 12 months were compared using χ2 tests.45 Overall survival (OS) was estimated using the Kaplan-Meier method, and the differences between groups were compared using the log-rank test. P-values were corrected for multiple comparisons using the Benjamini-Hochberg method.46 All tests were 2-sided and statistical significance was considered at P < .05. All analyses were performed using SAS version 9.4 and R statistical package version 4.0.3 (R Core Team 2020).
All patients, parents, and legal guardians provided informed consent on an institutional review board-approved protocol.
Results
Patient characteristics
Patient characteristics are shown in supplemental Table 2 (training and validation cohorts) and supplemental Table 3 (expanded PT-CY subset). The significant differences in age distribution, indication for transplantation, conditioning intensity, donor type, and GVHD prophylaxis between cohorts reflect changes in transplant practices between the earlier training (2004-2015) and later validation (2015-2017) cohorts. Despite these changes, there were no significant differences in GVHD characteristics, such as target organ involvement, severity at diagnosis, maximum severity, systemic treatment, or 12-month NRM. There were more patients with late acute GVHD in the validation cohort, whose median day of GVHD onset was 2 days later. When we applied the algorithms and thresholds for ST2 + REG3α and AREG that are currently in use in clinical trials29,32,34,36 (also NCT05123040 and NCT02525029) to the full cohort (n = 715), both algorithms performed similarly well (supplemental Figure 1).
Algorithm creation and validation
We used the training cohort to create algorithms that predicted the risk of 12-month NRM for all 7 possible combinations of ST2, REG3α, and AREG (supplemental Table 4). Each individual biomarker was an independent discriminator of the 12-month NRM, either alone or in a pairwise combination. When all 3 biomarkers were combined, REG3α and AREG remained significant predictors of 12-month NRM risk but ST2 was no longer significant, a finding that likely reflects that the correlation among biomarkers was the highest for AREG and ST2 (supplemental Figure 2). We used the validation cohort to calculate the area under the curve (AUC) of the receiver operating characteristic curves for each algorithm (Table 1). All algorithms were strong discriminators of the 12-month NRM but algorithms that combined biomarkers had greater AUCs than the single biomarker algorithms; the largest AUC belonged to the combination of ST2 + REG3α. There were no statistically significant differences among the algorithms (supplemental Table 5) and all algorithms produced 2 groups with significantly different risk of NRM. Similar findings were observed when the analyses were limited to the subset of validation cohort patients who underwent systemic treatment for acute GVHD (n = 321) (Table 1; supplemental Table 6), in which the ST2 + REG3α algorithm again had the largest AUC (0.739). Therefore, we focused our comparisons on the algorithms AREG and ST2 + REG3α, given their use in clinical trials, and the combination of ST2 + REG3α + AREG.
Risk stratification
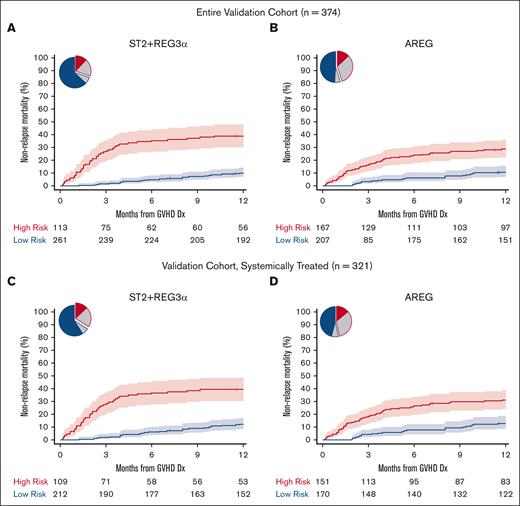
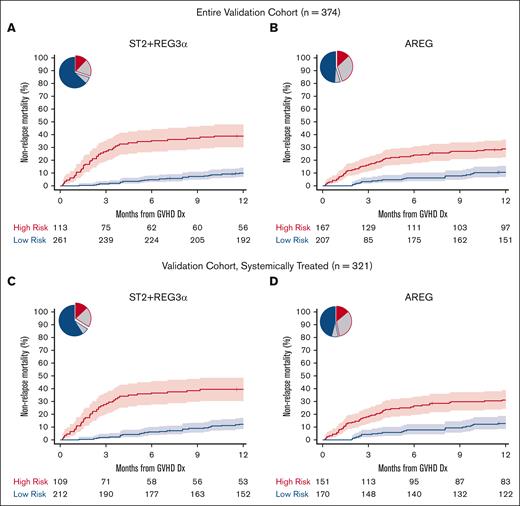
We identified thresholds for each algorithm in the training cohort corresponding to the concordance probability that maximized sensitivity and specificity and used those thresholds to risk stratify patients in the validation cohort (Table 2; supplemental Table 7). ST2 had the highest specificity, REG3α had the highest sensitivity, and the combination of the ST2 + REG3α algorithm had the highest balanced accuracy; these findings were unchanged when only patients who were systemically treated were analyzed (supplemental Table 7). All algorithms stratified patients into low- and high-risk groups with large and highly statistically significant differences in the 6-month and 12-month NRM (supplemental Table 8). The differences in 12-month NRM created by the ST2 + REG3α algorithm (29%) were considerably larger than that created by the AREG algorithm (18%) (Figure 1A-B) because of the higher specificity of the ST2 + REG3α algorithm, which correctly classified more patients as low risk than the AREG algorithm (70% vs 55%). Similar differences in 12-month NRM were observed when the algorithms were applied only to patients who were systemically treated (Figure 1C-D; supplemental Table 8). The inclusion of all 3 biomarkers in an algorithm resulted in risk groups that were close in size and PPV to the algorithm of AREG alone (Table 2). As expected from prior studies, the cumulative incidence of relapse was not significantly different with any algorithm; thus, all differences in the 12-month NRM translated into statistically and clinically significant differences in the 12-month OS (supplemental Table 8). As expected, patients at high risk for NRM were less likely to respond to systemic corticosteroid treatment than patients at low risk in the subset of the validation cohort patients who received systemic treatment (321/374, 86%), with the largest difference between groups again observed using the ST2 + REG3α algorithm (supplemental Table 9). When we chose a second threshold of 80% specificity as determined in the training cohort and applied to it the validation cohort, we found similar results in which the ST2 + REG3α algorithm correctly classified the greatest number of patients and produced the largest differences in NRM between groups for all patients as well as the subset of patients who were systemically treated (supplemental Tables 10 and 11).
Twelve-month NRM by risk classification for AREG and ST2 + REG3α biomarker algorithms (validation cohort). Pie charts show the proportion of patients classified as high-risk (HR, red border) and low-risk (LR, blue border). The proportions correctly classified as HR or LR are shaded in red and blue, respectively. The proportion incorrectly classified is shaded in gray. The cumulative incidence curves show the 12-month NRM, with shaded regions representing 95% confidence intervals. (A and B): All validation cohort patients (C and D): systemically treated subset. (A) ST2 + REG3α: NRM 39% vs 10%, P < .001; (B) AREG: NRM 29% vs 11%, P < .001; (C) ST2 + REG3α: NRM 39% vs 12%, P < .001; (D) AREG: NRM 31% vs 13%, P < .001.
Twelve-month NRM by risk classification for AREG and ST2 + REG3α biomarker algorithms (validation cohort). Pie charts show the proportion of patients classified as high-risk (HR, red border) and low-risk (LR, blue border). The proportions correctly classified as HR or LR are shaded in red and blue, respectively. The proportion incorrectly classified is shaded in gray. The cumulative incidence curves show the 12-month NRM, with shaded regions representing 95% confidence intervals. (A and B): All validation cohort patients (C and D): systemically treated subset. (A) ST2 + REG3α: NRM 39% vs 10%, P < .001; (B) AREG: NRM 29% vs 11%, P < .001; (C) ST2 + REG3α: NRM 39% vs 12%, P < .001; (D) AREG: NRM 31% vs 13%, P < .001.
Analyses of key subsets helped explain these modest differences in performance between the ST2 + REG3α and AREG algorithms, although it is important to note that there were no statistically significant differences among the AUCs (Table 3). Both algorithms successfully stratified patients who received systemic treatment for GVHD (n = 321; AREG 12% vs 30%, P < .001; ST2 + REG3α: 12% vs 39%, P < .001), and both algorithms stratified patients with lower GI GVHD at diagnosis effectively (n = 109, AREG: 8% vs 45%, P < .001; ST2 + REG3α: 12% vs 48%, P < .001). Minnesota risk classification stratifies patients for risk of NRM. AREG further risk stratified the Minnesota high-risk subset (Figure 2A-C; supplemental Table 12) and ST2 + REG3α more accurately classified the standard-risk subset (Figure 2D-F; supplemental Table 12). AREG correctly classified 7% more patients with Minnesota high-risk GVHD than ST2 + REG3α, but this group composed a small proportion of patients with GVHD (53/374, 14%); thus, the overall net effect was a correct classification of 1% more of the total population. In contrast, in patients with Minnesota standard-risk GVHD, which comprised most patients, ST2 + REG3α correctly classified 17% more patients than AREG (79% vs 62%); therefore, the overall net effect was a correct classification of 15% more patients in the total population. Further subset analysis showed that in patients with only skin rash at diagnosis (n = 199), ST2 + REG3α created distinct risk strata (8% vs 22%, P < .001) but AREG did not (10% vs 13%, P = .218). In addition, patients who received PT-CY-based GVHD prophylaxis (n = 133) were successfully stratified for risk of NRM using the ST2 + REG3α algorithm (12% vs 35%, P < .001) but not by the AREG algorithm (14% vs 21%, P = .22). These findings were consistent when the analyses were limited to patients who received systemic treatment for GVHD (Table 3). Given the increased use of PT-CY as GVHD prophylaxis and the large number of patients who presented only with rashes at the time of GVHD diagnosis, the ST2 + REG3α algorithm appears preferable for these important subgroups.
Twelve-month NRM by Minnesota risk and further stratification using ST2 + REG3α and AREG algorithms in the validation cohort. The cumulative incidence curves show 12-month NRM with shaded regions representing the 95% confidence intervals. (A) Minnesota high-risk acute GVHD: NRM, 42%. (B) Minnesota high-risk stratified by ST2 + REG3α: NRM 45% vs 24%, P = .083; (C) Minnesota high-risk stratified by AREG: 50% vs 8%, P = .013. (D) Minnesota standard-risk acute GVHD: NRM, 15%. (E) Minnesota standard risk stratified ST2 + REG3α: NRM 34% vs 9%, P < .001; (F) Minnesota standard risk-stratified by AREG: NRM 21% vs 11%, P = .003.
Twelve-month NRM by Minnesota risk and further stratification using ST2 + REG3α and AREG algorithms in the validation cohort. The cumulative incidence curves show 12-month NRM with shaded regions representing the 95% confidence intervals. (A) Minnesota high-risk acute GVHD: NRM, 42%. (B) Minnesota high-risk stratified by ST2 + REG3α: NRM 45% vs 24%, P = .083; (C) Minnesota high-risk stratified by AREG: 50% vs 8%, P = .013. (D) Minnesota standard-risk acute GVHD: NRM, 15%. (E) Minnesota standard risk stratified ST2 + REG3α: NRM 34% vs 9%, P < .001; (F) Minnesota standard risk-stratified by AREG: NRM 21% vs 11%, P = .003.
Discussion
Recent advances in GVHD prophylaxis have decreased the overall incidence of clinically severe (grade III/IV) GVHD but not the overall incidence of GVHD that requires treatment.5,47,48 High-performance laboratory tests that predict GVHD outcomes are needed to tailor therapy based on the risk. The 2 validated GVHD risk stratification algorithms used in clinical trials used different biomarkers and both predicted GVHD outcomes well. In this study, we used a large international multicenter cohort to directly compare these 2 algorithms and evaluate whether novel combinations of biomarkers would improve their performance. Each of the 2 algorithms was an excellent discriminator of the 6-month and 12-month NRM in these patients when assessed using identical statistical methods. Although other combinations of these GI GVHD biomarkers performed well, adding AREG to ST2 + REG3α did not improve upon AREG or ST2 + REG3α, perhaps because AREG is a downstream component of the IL-33/ST2 axis.
The ST2 + REG3α algorithm had several modest advantages over the other algorithms: it more accurately classified patients, identified greater differences in 12-month NRM and OS between the high- and low-risk groups and performed well in most patients (Minnesota standard risk GVHD), in which it was best at identifying patients who were at high risk of NRM despite the absence of high-risk clinical symptoms. This last finding may be because ST2 + REG3α identifies more patients as having a high risk for NRM before lower GI GVHD symptoms have manifested. Notably, the AREG algorithm was slightly better than ST2 + REG3α for patients with Minnesota high-risk GVHD. The ability to identify patients at low risk for NRM, despite the presence of high-risk clinical symptoms, is clinically important, as it may help avoid over-treatment. The addition of a third GI biomarker, AREG, did not improve the performance of ST2 + REG3α, which is likely due to the strong correlation between ST2 and AREG. The IL-33/ST2 axis plays a key role in the pathogenesis of GVHD, whereas IL-33 is both the ST2 ligand and an inducer of AREG secretion by type 2 innate lymphoid cells; however, the interactions surrounding these proteins in GVHD biology remain poorly described and an area of active investigation.19,23,27,49
Our study has several limitations. First, the training and validation cohorts were primarily obtained from a prior study.28 The training cohort differed from the validation cohort, reflecting the evolution of transplant practices: patients were younger, had different indications for transplant, had a different donor mix, received different GVHD prophylaxis, and were less likely to experience late-onset GVHD. The fact that the algorithms performed well in both cohorts is reassuring, but patients who underwent transplantation after 2017 were not included with the exception of the PT-CY subset; thus, these analyses do not fully reflect the most current transplant practices. It is noteworthy that the ST2 + REG3α algorithm successfully stratified patients who received PT-CY prophylaxis for risk of 12-month NRM, whereas the AREG algorithm did not. However, some other clinically relevant subsets were too small for analysis, such as patients whose GVHD treatment subsequently required treatment with ruxolitinib (n = 14). Second, previous ST2 + REG3α and AREG biomarker algorithms were developed from different data sets using different statistical methods and end points. In this study, we created new algorithms, including novel combinations, from the training set using identical statistical methods and 12-month NRM as the primary end point to compare algorithm performance with minimal bias. Thus, the new algorithms differ from the versions of the algorithms used in past clinical trials, although this new ST2 + REG3α algorithm yields the same results as the previously published algorithm (supplemental Figure 1). Furthermore, although categorical risk scores, such as high/low risk, are useful for separating patients into groups, the field would benefit from the development of calibrated risk scores that can be applied to individual patients. For example, it might be useful if a continuous value, such as the MAGIC Algorithm Probability or AREG concentration, accurately estimates an individual patient’s risk for NRM. Third, although there were clinically meaningful differences in performance among the algorithms in specific subgroups, no algorithm was statistically superior to any other. Fourth, the small number of patients with specific characteristics, such as Minnesota high-risk GVHD, raises the possibility that some subset analyses were underpowered and further study of these groups is needed. Fifth, our data set was not optimized to evaluate certain late complications such as chronic GVHD. Future studies are needed to prospectively collect late clinical events to evaluate the ability of different algorithms to predict long-term outcomes other than NRM and survival. Finally, a larger data set than the one used here would be needed to detect significant differences between algorithms and identify scenarios in which 1 algorithm might be preferred over another.
In conclusion, biomarkers enhance clinical risk stratification strategies by identifying patients at increased risk of NRM within the Minnesota standard-risk population and those at decreased risk of NRM within the Minnesota high-risk population. Thus, biomarkers will play an increasingly important role in GVHD clinical trial design and ultimately in clinical practice. Because no single biomarker is universally ideal, the choice of algorithm and threshold should be guided by the research or clinical aim. If the goal is to deescalate GVHD treatment, high NPV and sensitivity facilitate the identification of patients most likely to respond to standard treatment and survive long-term. For example, a recent study showed that patients with Minnesota standard-risk GVHD and low-risk biomarker scores by ST2 + REG3α could be successfully treated with inhibition of JAK1 by itacitinib monotherapy, thereby avoiding exposure to the toxicity of systemic corticosteroids.34 Conversely, high PPV and specificity are preferred for identifying a population at high risk of poor outcomes or for studying a potentially toxic intervention. For example, patients with Minnesota standard-risk GVHD who are at high risk for biomarkers may be appropriate for inclusion in clinical trials that intensify treatment given the risk of failure with standard treatment, even at the risk of more treatment-related toxicity. Similarly, patients with Minnesota high-risk GVHD who are at low risk for NRM by biomarkers might be excluded from clinical trials that intensify GVHD treatment. One could also consider adjusting thresholds to maximize NPV or PPV for different clinical scenarios and treatment goals. In summary, algorithms based on ST2 + REG3α and AREG, including previously published versions with their accompanying risk stratification thresholds, are suitable for identifying patients across a wide range of clinical presentations and are appropriate for use across a spectrum of clinical trial designs. Periodic re-examination of algorithms will be necessary as clinical practice evolves.
Acknowledgments
The authors thank the patients, their families, and the research staff for their participation. The authors also thank Gilbert Eng for the computer programming and database support.
This work was supported by the National Institutes of Health, National Cancer Institute grant PO1CA03942), University of Minnesota Foundation (GVHD Research Fund), Pediatric Cancer Foundation, and German Jose Carreras Leukemia Foundation grants DJCLS 01 GVHD 2016 and DJCLS 01 GVHD 2020.
Authorship
Contribution: A.E., D.K., J.L.M.F., J.E.L., and S.H. conceived and designed the study; A.E., P.A-H., F.A., J.B., C.C., Y-B.C., H.C., Z.D., S.G., E. Hexner, W.J.H., E. Holler, C.L.K., S.K., M.A.M., A.P., F.Q., M.Q., R.R., T.S., I.V., D.W., M.W., R.Y., R.N., and J.E.L. collected and reviewed the clinical data; S.G., G.M., S.K., and S.H. performed the laboratory analysis; A.E., N.K., N.S., and D.K. performed the statistical analysis; A.E., N.K., J.L.M.F., and J.E.L. wrote the report; and all authors reviewed and edited the manuscript.
Conflict-of-interest disclosure: B.C.B. is a coinventor of a CD83 CAR T cell licensed to CRISPR Therapeutics; received consulting fees from CTI BioPharma and Incyte; received research funding from Vitrac Therapeutics and CTI BioPharma; and is the current Director of Laboratory Science for American Society of Transplantation and Cellular Therapy. Y-B.C. received consulting fees from Incyte, Takeda, Vor Biopharma, Celularity, Equilium, and Pharmacosmos. H.C. received consulting fees from Incyte, Sanofi, Actinium, and REGiMMUNE, and research funding from Opna. C.L.K. received consulting fees from Horizon Therapeutics. M.A.M. received consulting fees from NexImmune, TScan, Hansa Biopharma, Stemline Therapeutics, CarDx, and Incyte; participated in a speakers’ bureau for Sanofi; and received research funding from NexImmune and Gilead. M.Q. received honoraria from Novartis and Vertex. R.R. received consulting fees from Atara Biotherapeutics, Allogene, Gilead Sciences, Takeda, Incyte, Instil Bio, TScan, Synthekine, Orca, Quell Therapeutics, Capstan, and Jasper; served in an expert witness role with Bayer; and received research funding from Atara Biotherapeutics, Incyte, Sanofi, Immatics, AbbVie, TCR2, Takeda, Gilead Sciences, CareDx, TScan, Synthekine, Bristol Myers Squibb, Johnson & Johnson, Genentech, and Precision BioSciences. T.S. received consulting fees from Moderna. J.L.M.F. and J.E.L. are coinventors of a GVHD biomarker patent and receive royalties from its licensure. J.E.L. received consulting fees from bluebird bio, Editas, Equillium, Incyte, Inhibrx, Kamada, Mesoblast, Sanofi, and X4 Pharmaceuticals, and research support from Genentech, Incyte, and Mesoblast. S.H. received consulting fees from Ossium Health; fees for clinical trial adjudication from CSL Behring; and research funding from Vitrac Therapeutics and Incyte. The remaining authors declare no competing financial interests.
Correspondence: John Levine, The Tisch Cancer Institute, Icahn School of Medicine at Mount Sinai, 1 Gustave L. Levy Pl, Box 1410, New York, NY 10029; email: john.levine@mssm.edu.
References
Author notes
J.L.M.F., J.E.L., and S.H. contributed equally to this study.
Data are available on request from the corresponding author, John Levine (john.levine@mssm.edu).
The full-text version of this article contains a data supplement.