Key Points
PARP1 is required for the maintenance of MLL-AF9 leukemias.
PARP1 inhibitors enhance the therapeutic effect of cytotoxic drugs against MLL-AF9 leukemias.
Introduction
Mixed-lineage leukemia (MLL)–rearranged hematological malignancies are found in >70% of infant acute lymphoblastic leukemias, ∼10% of acute myeloid leukemias (AMLs), and >30% of therapy-related leukemias.1 AML patients bearing MLL translocations have intermediate-to-poor prognoses (5-year disease-free survival of 30%-60%), and their leukemia cells are often resistant to conventional chemotherapies. MLL-rearranged leukemia is caused by chromosomal translocations that link the MLL gene (also known as histone-lysine N-methyltransferase 2A [KMT2A]) with >60 different partner genes to generate leukemia-inducing fusion proteins; the most common being MLL-AF4 (KMT2A-AFF1), MLL-AF9 (KMT2A-MLLT3), MLL-AF10 (KMT2A-MLLT10), MLL-AF6 (KMT2A-AFDN), and MLL-ENL (KMT2A-MLLT1).2 The expression of certain human MLL translocations (eg, MLL-AF9 or MLL-ENL) can transform murine hematopoietic progenitors and induce an aggressive leukemia that recapitulates aspects of human leukemia, including resistance to conventional chemotherapies, such as cytarabine and doxorubicin.3-7
MLL-AF9–positive leukemia cell proliferation and differentiation arrest appear to be highly dependent on the mechanism that prevents the accumulation of oxidative DNA damage, including DNA double-strand breaks (DSBs).8 We postulated that targeting DNA repair pathways would enhance the sensitivity of MLL-AF9–positive AML cells to the lethal effects of DSBs. In this article, we show that PARP1 inhibitors elevated the number of DSBs induced by cytarabine and doxorubicin in MLL-AF9 leukemia cells and exerted antileukemia effects in mice.
Methods
Primary cells
Peripheral blood samples from patients with newly diagnosed AML carrying t(9;11)(p22;q23), which encodes for MLL-AF9, were obtained from the Stem Cell and Leukemia Core Facility of the University of Pennsylvania (Philadelphia, PA). Samples of normal hematopoietic cells were purchased from Cambrex Bio Science (Walkersville, MD). Lin–CD34+ cells were obtained from mononuclear fractions by magnetic sorting by using the EasySep Negative Selection Human Progenitor Cell Enrichment Cocktail followed by Human CD34-Positive Selection Cocktail (StemCell Technologies). These studies were approved by the appropriate institutional review boards and met all requirements of the Declaration of Helsinki.
Transduction of Parp1−/− and Parp1+/+ cells
Parp1+/+ and Parp1−/− mice were provided by R. Caricchio (Temple University, Philadelphia, PA).9 Wild-type and knockout status were confirmed by polymerase chain reaction (PCR) of tail-snip DNA. Total murine bone marrow cells (mBMCs) recovered from healthy Parp1+/+ and Parp1−/− mice were subjected to either lineage depletion (MACS Miltenyi) to isolate Linlow mBMCs or to cell sorting to isolate Linlow, c-Kit+, Sca-1– cells. Cells were stimulated overnight in cytokine-enriched media (Iscove modified Dulbecco medium supplemented with 10% fetal bovine serum (FBS), 1% l-glutamine, 1% penicillin/streptomycin, 10 ng/mL murine stem cell factor (SCF), 6 ng/mL murine interleukin-3 [mIL-3] and 5 ng/mL mIL-6; cytokines were purchased from Peprotech). The following day, cells were transduced with the indicated retroviruses (MCSV-IRES-GFP [empty], MSCV-MLL-AF9-IRES-GFP, MSCV-HoxA9-IRES-Meis1, MSCV-AML1-ETO9a-IRES-GFP), as described previously.10,11 Following transduction, cells were plated in methylcellulose (M3434, Stem Cell Technologies) at 1000 cells/mL and grown for 5 to 7 days, as described previously.11 Following colony formation, colonies were enumerated and then homogenized and replated in methylcellulose at 10 000 cells/mL. Colonies were enumerated and replated a third time, however, the empty vector control did not form colonies after the third plating, and thus cells from the first replating were analyzed in experiments. For the generation of MLL-AF9+NRAS(G12D) and MLL-AF9+FLT3(ITD) cell lines, lineage-depleted cells were cotransduced with MSCV-MLL-AF9-Neor and MCSV-NRAS(G12D)-IRES-GFP or MSCV-FLT3(ITD)-IRES-GFP. Leukemia cell lines were generated by replating cells in methylcellulose 4 times, followed by long-term culture in cytokine-enriched media.
Real-time PCR
SuperScript II reverse transcriptase (Invitrogen) was used to generate randomly primed complementary DNA from 2 g of total RNA. A 50-ng sample of complementary DNA was analyzed by real-time PCR by using the threshold cycle method on an ABI StepOnePlus machine. Mouse Parp1 was detected by using the following primers: forward: 5′-aggccgcctactctatcctc-3′ and reverse: 5′-gattcactgctgccttgaga-3′. Mouse 18s RNA served as the internal control and was detected by using forward primer 5′-gcaattattccccatgaacg-3′ and reverse primer 5′-gggacttaatcaacgcaagc-3′.
Western blot
Nuclear protein lysates were obtained as described before12 and examined by western blot analysis by using antibodies against Parp1 (sc-7150) and anti-LaminB1 (sc-20682).
In vitro treatment
The cytotoxic drugs doxorubicin and cytarabine and the PARP1 inhibitors olaparib and BMN 673 (all from Selleckchem) were added to the indicated cells for 1 to 5 days. Murine hematopoietic/leukemia cells were maintained in RPMI 1640 supplemented with 10% FBS, 100 ng/mL SCF, and 20 ng/mL IL-3. Primary human hematopoietic cells were incubated in Iscove modified Dulbecco medium supplemented with 10% FBS and growth factors (100 ng/mL SCF, 20 ng/mL IL-3, 100 ng/mL Flt-3 ligand, 20 ng/mL granulocyte colony-stimulating factor, and 20 ng/mL IL-6), as previously described.13 Cells were plated in Methocult (Stemcell Technologies) and colonies were scored after 5 to 7 days. For quiescent/proliferating cells, Lin– cells were stained with 5(6)-carboxyfluorescein diacetate succinimidyl ester (CFSE), cell trace violet, or CellProliferation Dye eFluor670 (eBioscience) and incubated for 5 days in StemSpan serum-free expansion medium (Stem Cell Technologies, Vancouver, Canada) supplemented with the cocktail of growth factors (see above) and inhibitors when indicated. Quiescent (CFSEmax) and proliferating (CFSElow) leukemia and normal hematopoietic stem and progenitor cells were detected by flow cytometry using fluorochrome-conjugated anti-Lin, anti-CD34, and anti-CD38 antibodies (BD Pharmingen), as described previously.10,14 Cell viability was determined by Trypan blue exclusion. Cell death and γ-H2AX were measured by flow cytometry using Fixable Viability Dye eFluor 660 (eBioscience) and Alexa Fluor 488 Mouse anti-H2AX (phosphoserine 139) (BD Pharmingen) as described before.13
In vivo treatment
Syngeneic mice were injected IV with 1 × 105 GFP+MLL-AF9–transformed mBMCs (derived from a primary recipient mouse with frank leukemia). Seven days later, mice were treated with vehicle (control), BMN 673 (0.33 mg/kg per day by oral gavage for 7 days15 ), doxorubicin (1.5 mg/kg per day IV [days 1-3]) plus cytarabine (DA [50 mg/kg IV (days 1-5)3 ), and a combination of doxorubicin plus BMN 673. Leukemia burden (GFP+ cells) was analyzed by flow cytometry 7 days after the end of treatment. Median survival time was determined. Animal studies were approved by the Fox Chase Cancer Center Institutional Animal Care and Use Committee.
Statistical analyses
Data are expressed as means ± standard deviations (SDs) and were compared by using the unpaired Student t test; P < .05 was considered significant. Synergistic effect was evaluated by using the response additivity approach.16 Median survival time of the mice ± standard error was calculated by Kaplan-Meier log-rank survival analysis.
Results and discussion
Numerous reports indicate that AML cells accumulate high levels of spontaneous and genotoxic agent-induced DNA lesions, but are able to survive because of enhanced or altered DNA repair activities.17-21 Because DNA damage may constrain survival and proliferation of leukemia cells, transformed cells need to be protected from the detrimental effects of DNA damage, such as DSBs.8 Thus, leukemia cells may be highly dependent on specific DNA repair mechanisms, and targeting these pathways could sensitize them to DNA damage.22 This speculation is supported by reports, including our own, that AMLs carrying AML1-ETO (RUNX1-RUNXT1) and PML-RARα display a BRCA2 deficiency, which diminishes homologous recombination repair (HRR) activity and predisposes leukemia cells to synthetic lethality triggered by DNA repair inhibitors, such as PARP1 or RAD52 inhibitors.13,23
In this article, we show that Parp1−/− mBMCs expressing MLL-AF9 demonstrated reduced clonogenic activity in comparison with Parp1+/+ counterparts (Figure 1A). This effect was not caused by inherent defects of nontransformed Parp1−/− cells because their clonogenic activity was similar to that of their nontransformed Parp1+/+ counterparts. In addition, mBMCs expressing MLL-AF9, MLL-AF9+NRAS(G12D), and MLL-AF9+FLT3(ITD) were modestly sensitive to the PARP1 inhibitor olaparib when compared with cells transduced with an empty plasmid (Figure 1B). At the same time, mBMCs expressing AML1-ETO+FLT3(ITD) were exceptionally sensitive to olaparib treatment, whereas those expressing HOXA9+MEIS1 were resistant. These observations are in accordance with those of Esposito and colleagues,23 indicating that AML1-ETO–positive murine cells were highly sensitive to PARP1 inhibition, whereas those expressing MLL-AF9 display limited sensitivity.
PARP1 promotes the growth of MLL-AF9–positive leukemias. (A) Clonogenic activity of Parp1+/+ (+/+) and Parp1−/− (−/−) mBMCs expressing MLL-AF9 (MSCV-MLL-AF9-IRES-GFP, green bars) or empty plasmid (MCSV-IRES-GFP, purple bars) as described in the supplemental Materials and supplemental Results. Results represent mean number of colonies per 104 cells ± SD that formed after the second plating from 3 experiments. (B) Clonogenic activity of mBMCs expressing the indicated genes or empty plasmid (wild-type [wt]) and treated with increasing concentrations of olaparib for 72 hours following the plating in methylcellulose. Results represent mean percentage ± SD of surviving colonies compared with untreated counterparts from 3 experiments. (C) The effect of olaparib on Lin–CD34+, Lin–CD34+CD38–CFSEmax, and Lin–CD34+CD38–CFSElow cells obtained from AML primary cells expressing MLL-AF9 (3 patients, green symbols) and from healthy donors (3 donors, purple symbols). Results represent mean percentage ± SD of cells surviving after 5 days of treatment compared with untreated counterparts from 3 experiments per sample. *P = .03 when compared with the corresponding +/+ cells using Student t test.
PARP1 promotes the growth of MLL-AF9–positive leukemias. (A) Clonogenic activity of Parp1+/+ (+/+) and Parp1−/− (−/−) mBMCs expressing MLL-AF9 (MSCV-MLL-AF9-IRES-GFP, green bars) or empty plasmid (MCSV-IRES-GFP, purple bars) as described in the supplemental Materials and supplemental Results. Results represent mean number of colonies per 104 cells ± SD that formed after the second plating from 3 experiments. (B) Clonogenic activity of mBMCs expressing the indicated genes or empty plasmid (wild-type [wt]) and treated with increasing concentrations of olaparib for 72 hours following the plating in methylcellulose. Results represent mean percentage ± SD of surviving colonies compared with untreated counterparts from 3 experiments. (C) The effect of olaparib on Lin–CD34+, Lin–CD34+CD38–CFSEmax, and Lin–CD34+CD38–CFSElow cells obtained from AML primary cells expressing MLL-AF9 (3 patients, green symbols) and from healthy donors (3 donors, purple symbols). Results represent mean percentage ± SD of cells surviving after 5 days of treatment compared with untreated counterparts from 3 experiments per sample. *P = .03 when compared with the corresponding +/+ cells using Student t test.
Because rodent and human cells may display significant differences in DSB repair mechanisms,24 sensitivity to PARP1 inhibition of AML samples from patients carrying the MLL-AF9–encoding t(9;11)(p22;q23) was tested. Olaparib diminished the number of Lin–CD34+ immature primary leukemia cells, including leukemia stem cell–enriched Lin–CD34+CD38–CFSElow proliferating and Lin–CD34+CD38–CFSEmax quiescent cells by approximately twofold (Figure 1C). These results, together with other reports showing that olaparib reduced the number of primary AML cells bearing MLL fusions by ∼1.5 to 2.5-fold, suggest that PARP1 could be used as a therapeutic target in AMLs bearing MLL translocations.23
The sensitivity of MLL-AF9–positive cells to olaparib was not associated with a deficiency in HRR,23 and differences in cell cycle (supplemental Figure 1). We found that PARP1 messenger RNA levels were not altered, yet protein expression was elevated in untreated and doxorubicin and cytarabine–treated MLL-AF9–positive mBMCs compared with nontransformed wild-type mBMCs (Figure 2A), suggesting that PARP1 expression is subject to posttranscriptional regulation. We postulated that MLL-AF9 cells are reliant on PARP1 to cope with oxidative DNA damage (oxidized bases, single-strand breaks),8 which is supported by the reduced clonogenic activity of MLL-AF9–positive mBMCs in the absence of PARP1 (Figure 1A). In addition, MLL-AF9–mediated sensitivity to PARP1 inhibitors does not depend on its downstream effectors HOXA9 and MEIS1 because mBMCs overexpressing HOXA9+MEIS1 were resistant to olaparib and did not require PARP1 for proliferation.25
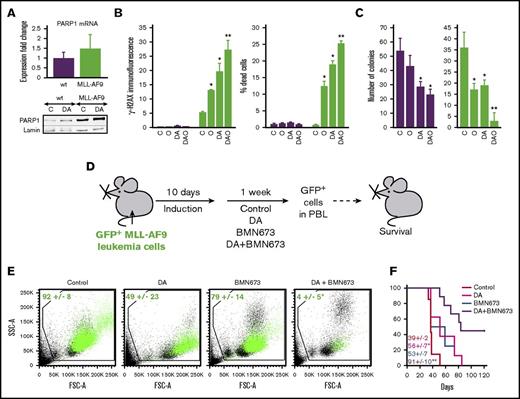
The effect of PARP1 inhibitor BMN 673 with or without cytotoxic drugs against MLL-AF9 leukemia in mice. (A-C) mBMCs expressing MLL-AF9 (MLL-AF9, green bars) or empty plasmid (wt, purple bars) were untreated (C) or treated with 10 nM doxorubicin plus 15 nM cytarabine (DA), 5 μM or 1 μM olaparib (O in B and C, respectively), and doxorubicin plus cytarabine plus olaparib (DAO) for 24 and 72 hours (A-B and C, respectively. (A) Real-time quantitative PCR analysis of PARP1 messenger RNA expression (top); western blot analysis of PARP1 and lamin (loading control) in nuclear cell lysates (bottom). (B) γ-H2AX fluorescence intensity (left) and percentage of dead cells (right). Results represent mean ± SD from 3 independent experiments. *P < .001 and **P ≤ .002 when compared with control and single treatment, respectively, using Student t test. (C) Mean number of colonies per 104 cells ± SD in methylcellulose (triplicate experiment). *P < .02 and ** P < .002 in comparison with control and individual treatment, respectively, using Student t test. (D) Experimental design: syngeneic mice were injected with 1 × 105 GFP+MLL-AF9 leukemia cells and were treated 10 days later with vehicle (Control), DA, BMN 673, or DA plus BMN 673. GFP+ leukemia cells in peripheral blood leukocytes and median survival time were scored. (E) GFP+MLL-AF9 leukemia cells (green dots) in representative plots (n = 3-4 per group). Results represent the mean percentage of GFP+MLL-AF9 leukemia cells ± SD. *P = .02 in comparison with other groups using the response additivity approach. (F) Survival curves and MST (7-9 mice/group). * P < .04 and ** P < .03 in comparison with Control and other groups, respectively, using Kaplan-Meier log-rank test.
The effect of PARP1 inhibitor BMN 673 with or without cytotoxic drugs against MLL-AF9 leukemia in mice. (A-C) mBMCs expressing MLL-AF9 (MLL-AF9, green bars) or empty plasmid (wt, purple bars) were untreated (C) or treated with 10 nM doxorubicin plus 15 nM cytarabine (DA), 5 μM or 1 μM olaparib (O in B and C, respectively), and doxorubicin plus cytarabine plus olaparib (DAO) for 24 and 72 hours (A-B and C, respectively. (A) Real-time quantitative PCR analysis of PARP1 messenger RNA expression (top); western blot analysis of PARP1 and lamin (loading control) in nuclear cell lysates (bottom). (B) γ-H2AX fluorescence intensity (left) and percentage of dead cells (right). Results represent mean ± SD from 3 independent experiments. *P < .001 and **P ≤ .002 when compared with control and single treatment, respectively, using Student t test. (C) Mean number of colonies per 104 cells ± SD in methylcellulose (triplicate experiment). *P < .02 and ** P < .002 in comparison with control and individual treatment, respectively, using Student t test. (D) Experimental design: syngeneic mice were injected with 1 × 105 GFP+MLL-AF9 leukemia cells and were treated 10 days later with vehicle (Control), DA, BMN 673, or DA plus BMN 673. GFP+ leukemia cells in peripheral blood leukocytes and median survival time were scored. (E) GFP+MLL-AF9 leukemia cells (green dots) in representative plots (n = 3-4 per group). Results represent the mean percentage of GFP+MLL-AF9 leukemia cells ± SD. *P = .02 in comparison with other groups using the response additivity approach. (F) Survival curves and MST (7-9 mice/group). * P < .04 and ** P < .03 in comparison with Control and other groups, respectively, using Kaplan-Meier log-rank test.
PARP1 inhibitors may cause massive entrapment of highly expressed PARP1 on oxidative DNA lesions, resulting in the accumulation of DSBs, which can induce apoptosis or cell cycle arrest in some cells, despite a fully active HRR. This speculation is supported by the observation that PARP1 inhibitor–treated MLL-AF9 cells accumulate potentially lethal DSBs (Figure 2B). In addition, a MTH1 inhibitor, which increases the pool of oxidized 2′-deoxynucleoside 5'-triphosphates, resulting in the accumulation of DNA damage,26,27 enhanced the effect of olaparib on MLL-AF9–positive AML cell lines and mBMCs (supplemental Figure 2A-B, respectively). In contrast, the antioxidant vitamin E, which reduces the number of reactive oxygen species–induced DSBs,28 diminished the inhibitory effect of olaparib on MLL-AF9–positive mBMCs (supplemental Figure 2C).
Next, we tested whether PARP1 inhibitors could enhance the effect of first-line cytotoxic drugs in MLL-AF9 cells. Comparing MLL-AF9–expressing mBMCs treated with olaparib or DA for 24 hours, cells treated with olaparib plus DA displayed significantly more DSBs, as detected by γ-H2AX (Figure 2B, left panel). Moreover, a significant increase of DSBs was not observed in similarly treated mBMCs transfected with empty plasmid. The accumulation of DSBs in MLL-AF9–positive mBMCs treated with olaparib + DA was associated with enhanced cell death compared with treatment with olaparib or cytotoxic drugs alone (Figure 2B, right panel). Olaparib or cytotoxic drug treatment reduced the clonogenic potential of MLL-AF9–positive mBMCs, whereas only cytotoxic drugs, but not olaparib alone, reduced the clonogenicity of mBMCs transfected with empty plasmid (Figure 2C). The combination of olaparib plus DA selectively enhanced the accumulation of DSBs and dead cells in MLL-AF9–positive mBMCs (Figure 2B) and resulted in a more than fourfold reduction of clonogenic activity when compared with olaparib or DA alone (Figure 2C).
To assess the therapeutic potential of targeting PARP1 in vivo, we assessed how the PARP1 inhibitor BMN 673, which displays superior pharmacokinetic parameters in mice than olaparib,15 either alone or in combination with DA, effects leukemia in a bone marrow transplant model of AML.11 Syngeneic mice transplanted with murine frank leukemia cells coexpressing GFP (GFP+) and MLL-AF9 were either left untreated or were treated with DA, BMN 673, or DA plus BMN 673, followed by examination of GFP+ cells in the peripheral blood and determination of median survival time (MST) (Figure 2D). Whereas treatment with cytotoxic drugs or BMN 673 alone did not significantly reduce GFP+ MLL-AF9 leukemia cells, BMN 673 synergistically enhanced the effect of the cytotoxic drugs and eliminated leukemia cells by >12-fold (Figure 2E). BMN 673 combined with DA also prolonged MST and 4 of 9 mice were alive 120 days posttransplant (Figure 2F).
In conclusion, olaparib dramatically enhances the in vitro toxic effect of doxorubicin and cytarabine on MLL-AF9–positive cells, which is associated with an increased accumulation of DSBs. Moreover, the combination of PARP1 inhibitors and cytotoxic drugs exerts strong and additive anti–MLL-AF9 leukemia effects in mice. Based on these results, we postulate that PARP1 inhibitors combined with standard cytotoxic drugs could be an effective therapeutic strategy against MLL-AF9–positive leukemias.
The full-text version of this article contains a data supplement.
Acknowledgments
This work was supported by National Institutes of Health, National Cancer Institute grants R01 CA134458 and CA186238 and the Leukemia and Lymphoma Society New Idea Award NIA-8999-14 (T.S.), and by National Institutes of Health, National Cancer Institute grant R00 CA158461, the W.W. Smith Foundation, and the American Society of Hematology Junior Scholar Award (S.M.S). B.V.L. was supported by the European Union's Horizon 2020 research and innovation program under Marie Sklodowska-Curie grant agreement no. 665735 and by funding from Polish Ministry of Science and Higher Education for the implementation of international projects, 2016-2020.
Authorship
Contribution: S.M., E.M., M.N.-S., M.H., B.V.L., and D.D.M. performed the experiments; S.M.S. designed the experiments and supervised E.M. and D.D.M.; I.T. designed the experiments and supervised M.H.; K.P. supervised B.V.L.; H.Z. performed statistical analyses; and T.S. designed the studies, supervised the experiments, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tomasz Skorski, Department of Microbiology and Immunology and Fels Institute for Cancer Research & Molecular Biology, Temple University School of Medicine, 3400 N. Broad St, MRB 548, Philadelphia, PA 19140; e-mail: tskorski@temple.edu.

![Figure 1. PARP1 promotes the growth of MLL-AF9–positive leukemias. (A) Clonogenic activity of Parp1+/+ (+/+) and Parp1−/− (−/−) mBMCs expressing MLL-AF9 (MSCV-MLL-AF9-IRES-GFP, green bars) or empty plasmid (MCSV-IRES-GFP, purple bars) as described in the supplemental Materials and supplemental Results. Results represent mean number of colonies per 104 cells ± SD that formed after the second plating from 3 experiments. (B) Clonogenic activity of mBMCs expressing the indicated genes or empty plasmid (wild-type [wt]) and treated with increasing concentrations of olaparib for 72 hours following the plating in methylcellulose. Results represent mean percentage ± SD of surviving colonies compared with untreated counterparts from 3 experiments. (C) The effect of olaparib on Lin–CD34+, Lin–CD34+CD38–CFSEmax, and Lin–CD34+CD38–CFSElow cells obtained from AML primary cells expressing MLL-AF9 (3 patients, green symbols) and from healthy donors (3 donors, purple symbols). Results represent mean percentage ± SD of cells surviving after 5 days of treatment compared with untreated counterparts from 3 experiments per sample. *P = .03 when compared with the corresponding +/+ cells using Student t test.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/1/19/10.1182_bloodadvances.2017006247/3/m_advances006247f1.jpeg?Expires=1767985086&Signature=qEuKIVfZ4H4~bZtZUmZkBhi7S275rXaYhuPzPpayIQ9cNkPadB4~OcC7UxEl8vdAemDhhC~07SYtxCNVeRp3CqYLYXmiiIVB58Uw1EesXCdMwRHuVKSgqc-P1K-CIga3eqCFK7o7LnjL7fBqRKI4gG6bAaqmrZwPfHXzpSw-2NxTPDcCsbjUK1DzPKubUw30Zr-fYk9EOTUuZA~03nXtLEWGWtAIMj8Nd~Kk7QQZr75WLOvwUjtr-fdbEJhmiImgL4xT1x6AnFRgzC3lXoTs9tUlj3969G7qHFxWIs-aAXFJ5uIOr1BZr5TE~dvoYTkISGCQk8XZ70aagRc4dLNR9Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)