Abstract
TAL-1 is a basic helix-loop-helix oncoprotein that is expressed in up to 30% of T-cell acute lymphoblastic leukemias but not in the T lineage. We have cloned a complementary DNA, called Human Immune Associated Nucleotide 1 (hIAN1), whose messenger RNA (mRNA) level expression is inversely correlated to the TAL-1 mRNA level in human leukemic T-cell lines. The hIAN1 encodes a 38-kd protein that belongs to a novel family of proteins conserved from plants to humans and characterized by motifs related to, but highly divergent from, the consensus motifs found in guanosine triphosphate (GTP)–binding proteins. Despite these divergent amino acids at positions involved in GTP/guanosine diphosphate (GDP) binding and guanosine triphosphatase (GTPase) activities, we found that hIAN1 specifically binds GDP (Kd = 0.47 μM) and GTP (Kd = 6 μM) and exhibits intrinsic GTPase activity. Among mature hematopoietic cells, hIAN1 is specifically expressed in resting T and B lymphocytes, and its expression level tremendously decreased at the protein but not the mRNA level during B- or T-lymphocyte activation, suggesting a specific role for this new type of GTPase during the immune response.
Introduction
The most common genetic alteration found in T-cell acute lymphoblastic leukemia (T-ALL) involves thetal-1/SCL/tcl-5 (hereafter referred as tal-1) gene, which codes for a basic helix-loop-helix transcription factor, TAL-1 (see Begley and Green1 for a review). In adults, TAL-1 expression is restricted to hematopoietic precursors; erythrocytic, megakaryocytic, and mastocytic cells; as well as some endothelial cells2; yet TAL-1 is never expressed in the T lineage. Gene-targeting experiments have established that TAL-1 is necessary for primitive and definitive hematopoiesis in mice and have also demonstrated a TAL-1 function in blood vessel formation.3-5
The role of TAL-1 in T-cell leukemogenesis is still poorly understood. In humans, activation of the tal-1 gene occurs by chromosomal translocations (5% of T-ALLs) or by an interstitial 90-kilobase (kb) deletion (25% of T-ALLs) (see Begley and Green1 for a review). In both cases, the TAL-1 coding sequence is not affected by these genetic rearrangements, which result in forced TAL-1 expression in the T-cell lineage. Indeed, unscheduled TAL-1 expression in the T-cell lineage of transgenic mice results in aggressive T-cell malignancies that appear relatively late in life and exhibit incomplete penetrance.6 However, concomitant expression of TAL-1 and LMO1 or LMO2, 2 proteins that interact with TAL-1 and are encoded by genes translocated in some T-ALLs, leads to leukemia early in life with a high degree of penetrance, suggesting a collaboration between these oncoproteins in the establishment of T-cell malignancies.7 8
The Jurkat T-cell line is derived from a human T-ALL and expresses high levels of the TAL-1 oncoprotein. We have previously derived a clonal subline of the Jurkat T-cell line, Jurkat-ΔCOOH, which produces only a mutant TAL-1 protein that exhibits a dramatic decrease of protein-binding activity to the TAL-1 DNA consensus sequence (E box).9 Growth curves indicate that the mutant subline exhibits a premature apoptosis upon medium depletion, and this phenomenon can be partially reverted by the expression of the wild-type TAL-1 protein in Jurkat-ΔCOOH.9 In this study, we used the Jurkat/Jurkat-ΔCOOH cell lines to isolate genes that might be regulated by TAL-1 during T-cell leukemogenesis. Using complementary DNA (cDNA) subtraction by representational difference analysis (RDA), we characterized a messenger RNA (mRNA) that is expressed only in Jurkat-ΔCOOH. This mRNA encodes a protein, Human Immune Associated Nucleotide 1 (hIAN1), which belongs to a novel family of proteins characterized by an unconventional guanosine diphosphate/guanosine triphosphate (GDP/GTP)–binding domain. Here we describe the biochemical properties and the expression pattern of hIAN1 in hematopoietic cells.
Materials and methods
Cell lines and primary human hematopoietic cells
Jurkat, Jurkat-ΔCOOH, Molt-4, RPMI 8402, CEM, and DU528 T-cell lines were grown in RPMI 1640 medium supplemented with 10% (20% for DU528) heat-inactivated fetal calf serum (FCS),l-glutamine, penicillin, and streptomycin at 37°C under 5% CO2. BaF3 cells were cultured in RPMI 1640 medium supplemented with 10% FCS, l-glutamine, penicillin, and streptomycin and 5% (vol/wt) WEHI-3B as a source of interleukin (IL)–3.
T-cell purification from human peripheral blood and activation were performed according to Costello et al.10 Primary T cells were maintained in RPMI 10% FCS. Stimulations were performed with anti-CD28 248 (mouse immunoglobulin (Ig)–M), and anti-CD3 289 (mouse IgG2a) obtained from Dr A. Moretta (Cancer Institute, Genoa, Italy) and used as ascites fluid (1/400 dilution) or as purified monoclonal antibody (mAb) (10 μg/mL), respectively. CD3 mAb was coated onto Petri tissue-culture dishes (CD3c). T-cell activation was controlled by proliferation assays and CD25/IL-2Rα expression.
Highly purified B cells were obtained from human peripheral blood monocyte cells (PBMCs) by positive selection by means of anti-CD19 magnetic microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany). B cells were activated for 4 days in flasks with irradiated (75 Gy) murine L cells transfected with human CD40 ligand and 10 mg/mL IL-4 (R&D Systems, Minneapolis, MN). Activated B cells were always more than 98% CD19+CD80+CD86+.
Jurkat-ΔCOOH was transfected by electroporation by a TAL-1–expressing vector previously described,9 and stable clones were established by limiting dilution. BaF3 cells were transfected by electroporation by an hIAN1-expressing vector allowing G418 resistance, and stable clones were also established by limiting dilution.
Cell cycle analysis
Cell cycle analysis was determined by fluorescence-activated cell sorting (FACS) following staining with propidium iodide. Cells were collected by centrifugation and washed with phosphate-buffered saline. The cells were permeabilized and incubated in a buffer containing 100 μg/mL propidium iodide, 50 μg/mL RNase, 0.1% Nonidet P-40, 5 mM NaCl, and 2 mM sodium citrate at 4°C for 2 hours prior to analysis with a Becton Dickinson FACSort analyzer (San Jose, CA). The cell cycle profile was analyzed by means of CellQuest software (Clearwater, FL).
Total RNA, polyA+ RNA, reverse transcriptase–polymerase chain reaction analysis and Northern blot
Total RNA was prepared by means of Trizol Reagent (GIBCO-BRL, Rockville, MD), and polyA+ RNA was obtained by means of the MPG RNA purification kit (Quantum, Vista, CA). First-strand DNA was reverse transcribed by means of 1 μg total RNA and 300 ng desoxyhexanucleotides, pd (N)6 (Pharmacia, Piscataway, NJ) and incubated for 1 hour at 37°C with Superscript II (GIBCO-BRL) in a buffer supplied by the manufacturer.
We used 1 μL reverse-transcriptase (RT) products for the S14 (ribosomal gene) polymerase chain reaction (PCR) (see below); the amount of cDNA used for each amplification was normalized by the amount of the S14 PCR product. The PCR reaction was performed in 50 μL containing 50 ng each oligonucleotide and 0.5 U ampli Taq Gold (Perkin Elmer, Branchburg, NJ). Reactions were performed in a 2400 Gene Amp PCR System (Perkin Elmer) under the following conditions: 10 minutes at 94°C; 29 cycles composed of 10 seconds at 94°C, 30 seconds at annealing temperature, and 30 seconds at 72°C; and finally 7 minutes at 72°C.
Primer sequences, annealing temperatures, and number of cycles were as follows: hIAN1 forward primer, 5′AGCCCAATACGGCAGTATGA3′; hIAN1 reverse primer, 5′AGTGTAACGGCCCAGTGGAA3′ (29 cycles, annealing at 54°C); S14 forward primer, 5′GGCAGACCGAGATGAATCCTCA3′; S14 reverse primer, 5′CAGGTCCAGGGGTCTTGGTCC3′ (29 cycles, annealing at 64°C); tal-1 forward primer, 5′TTGGGGAGCCGGATGCCTTC3′; and tal-1 reverse primer, 5′CTCCCGGCTGTTGGTGAA3′ (30 cycles, annealing at 58°C).
A Northern blot of several T-cell lines was performed with 15 μg total RNA and a Northern blot containing 2 μg polyA+ per lane. Selected RNA from each of several human tissues was purchased from Clontech Laboratories (Palo Alto, CA). Hybridizations were performed with the use of the Quick Express Hybrid solution (Clontech) and 32P-labeled probes corresponding to hIAN1, TAL-1, and glyceraldehyde phosphate dehydrogenase (GAPDH).
cDNA RDA, cloning, and sequencing of hIAN1 cDNA
RDA for cDNA was performed as described,11 starting from 2 μg polyA+ RNA from Jurkat and Jurkat-ΔCOOH. Only 2 successive subtractive hybridizations were performed, and the difference products (DP2s) were electroeluted, cloned in pBluescriptSK+ (pBSK+), and screened with a DP2-labeled probe. A 441–base pair (bp)DpnII-DpnII fragment that recognized the 2-kb mRNA present only in Jurkat-ΔCOOH was used as a probe to screen a Jurkat-ΔCOOH library previously constructed in pSPORT (GIBCO-BRL) in the laboratory.
The 3 positive clones were sequenced on both strands by means of the Applied Biosystems (Foster City, CA) PRISM ready reaction Dye-dideoxy terminator and Dye-Primer sequencing kits and samples were run on an ABI 373 A DNA sequencer (Applied Biosystems).
Production of recombinant glutathione-S-transferase–hIAN1 fusion protein
A cDNA containing the entire hIAN1 open reading frame (ORF) was obtained by PCR, with the use of primers containingEcoRI and XhoI sites, 5′ to the initiation methionine and 3′ from the stop codon, respectively.
The PCR product was purified, cloned in frame with glutathione-S-transferase (GST) in the pGEX4T3 vector, and sequenced. Expression of the GST-hIAN1 fusion protein was induced with 0.5 mM isopropyl β D-thiogalactoside for 3 hours at 37°C.
The fusion protein was affinity purified on glutathione beads in the presence of 5 mM MgCl2 and 0.1 mM GDP, dialyzed against 25 mM Tris (pH 7.5) and 1 mM dithiothreitol (DTT), quickly frozen in liquid nitrogen, and stored in multiple aliquots at −80°C. Independent preparations of GST-hIAN1 fusion protein ranged from 30% to 40% of active protein defined by their ability to bind GTP or GDP.
Polyclonal antibody production
Anti-hIAN1 antibody was raised against the full-length protein. Rabbits were inoculated with the purified GST-hIAN1 protein, after a preimmune serum sample had been taken. The animals were bled and reinoculated at the appropriate times. Serum was aliquoted and stored at −80°C.
In vitro transcription-translation and Western blot analysis
The cDNA fragments containing the hIAN1, hIAN2, and hIAN7 complete ORFs were subcloned into pBSK+ vector and sequenced. Then, 1 μg recombinant plasmids or 1 μg control plasmid encoding luciferase were transcribed and translated by means of the trinitrotoluene (T3/T7)–coupled reticulocyte lysate system (Promega, Madison, WI). Transcription/translation was performed for 90 minutes at 30°C.
Total protein extracts of Jurkat, Jurkat-ΔCOOH, BaF3, and B lymphocytes were obtained by means of a lysis buffer containing 50 mM Tris HCl (pH 8.0), 150 mM NaCl, 0.1% sodium dodecyl sulfate (SDS), and 1% NP40. Cytosolic and nuclear extracts of T lymphocytes were prepared according to Costello et al.10
Briefly, cells were harvested and washed in cold Tris-buffered saline, then resuspended in 0.4 mL (per 107 cells) of buffer A (10 mM Hepes [pH 7.8], 10 mM KCl, 2 mM MgCl2, 1 mM DTT, 0.1 mM EDTA, and 0.1 mM phenylmethylsulfonyl fluoride) supplemented with 10 μg/mL leupeptin (Sigma, St Louis, MO) and incubated on ice for 15 minutes. Next, 25 μL Nonidet P-40 solution (Sigma) was added, and cells were mixed vigorously for 15 seconds and then centrifuged (13 000 rpm, 15 seconds). Supernatants were used directly for immunoblotting. Protein concentration was measured with a commercial kit (Bio-Rad Laboratories, Hercules, CA) by the method of Bradford.
Translation products and total protein extracts were separated by electrophoresis in a 9% SDS–polyacrylamide gel electrophoresis (PAGE) according to Laemmli and transferred to Hybond-P polyvinylide difluoride membranes (Amersham, Buckinghamshire, United Kingdom). Immunoblot analysis was performed with the use of the anti-hIAN1 antiserum at 1/4000 dilution and revealed by the ECL detection system (Amersham).
Nucleotide-binding assays
Binding of GDP was assessed by incubating, at 25°C, 120 nM GST-hIAN1 fusion protein with 5 μM [3H]GDP (Amersham) (11.3 Ci/mmol [41.8 × 104 MBq/mmol]) in 50 mM Tris [pH 7.5], 1 mM DTT, 100 μg/mL bovine serum albumin (BSA), and various concentrations of MgCl2 (10 mM, 1 mM, or 1 μM) or 10 mM EDTA. At the indicated times, 50 μL was removed and immediately diluted in 2 mL washing buffer at 4°C (50 mM Tris [pH 7.5], 1 mM DTT, and 10 mM MgCl2), filtered through nitrocellulose filters (NC45; pore size, 0.45 mm) (Schleicher and Schuell, Dassel, Germany), and washed 3 times with 2 mL washing buffer. Radioactivity remaining on the filter corresponded to [3H]GDP bound to protein and was measured by liquid scintillation counting.
Specificity of nucleotide binding to hIAN1
The specificity of nucleotide binding to hIAN1 was assessed by a competition assay. First, 120 nM GST-hIAN1 was incubated at 25°C for 120 minutes with 5 μM [3H]GDP in binding buffer (50 mM Tris [pH 7.5], 1 mM DTT, 100 μg/mL BSA, and 10 mM MgCl2) and with various concentrations (0, 0.3, 3, 30, or 300 μM) of the following nucleotides: adenosine triphosphate (ATP), ribothymidine triphosphate (TTP), cytidine triphosphate (CTP), guanosine monophosphate (GMP), β γ imido guanosine triphosphate (GppNHp, a nonhydrolyzable analogue of GTP), and GDP. The residual amount of [3H]GDP bound was determined as above.
Determination of affinities for GDP and GppNHp
The purified GST-hIAN1 protein (120 nM) was incubated with increasing concentrations of [3H]GDP or [3H] GppNHp (25 Ci/mmol [92.5 × 104MBq/mmol]) (Amersham) in 50 μL binding buffer, at 25°C for 120 minutes. Bound [3H]GDP and [3H]GppNHp were measured as described above. The dissociation constants were calculated after performing a Scatchard analysis of the data.
Guanosine triphosphatase activity
Guanosine triphosphatase (GTPase) activity was assessed by thin-layer chromatography (TLC). First, 0, 5, or 10 μM GST-hIAN1 was incubated at 30°C with 50 μM [α32P]GTP (4000 cpm/pmol) in 50 mM Tris (pH 7.5), 1 mM DTT, 100 μg/mL BSA, and 10 mM MgCl2 in a total volume of 10 μL. At 0, 5, and 20 minutes, 2 μL reaction was removed and mixed with 2 μL solution containing 0.2% SDS, 5 mM EDTA, 50 mM GDP, and 50 mM GTP at 4°C. Samples were incubated at 70°C for 2 minutes to dissociate protein-bound nucleotides, and 1-μL aliquots were spotted onto polyethyleneimime-cellulose–covered TLC plates. They were developed in 0.6 M NaH2PO4 [pH 3.4], for 30 minutes, dried, and autoradiographed.
Results
Structural features of hIAN1, a protein translated from an mRNA selectively expressed in Jurkat-ΔCOOH
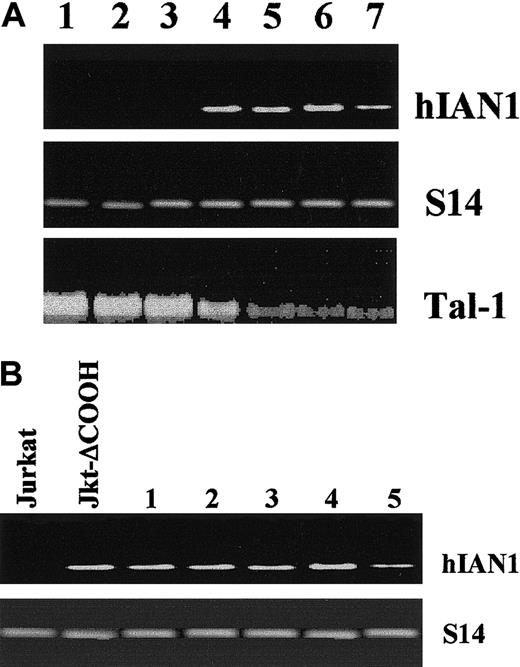
To identify genes potentially regulated by TAL-1 in leukemic cells, we performed a cDNA RDA between Jurkat and Jurkat-ΔCOOH at day 1 of culture, when the growth curves of the 2 cell lines start to differ (Leroy-Viard et al9 and Figure1A). RDA was performed with the Jurkat-ΔCOOH representations as “testers” and an excess amount of Jurkat representations as “drivers.” After 2 rounds of subtraction, the DP2 product showed discrete bands (data not shown). Random cloning of these differential products followed by screening with a labeled DP2 probe identified a 441-bpDpnII-DpnII DNA fragment that was used to hybridize a Northern blot containing total RNA isolated from Jurkat and Jurkat-ΔCOOH cells at different days of the growth curve. As shown in Figure 1B, the DpnII-DpnII fragment hybridized to a 2-kb RNA that was present only in Jurkat-ΔCOOH throughout the culture. This result clearly identified a Jurkat-ΔCOOH–specific mRNA and prompted us to clone the full-length cDNA.
hIAN1 mRNA expression in Jurkat and Jurkat-ΔCOOH.
(A) Growth analysis of Jurkat and Jurkat-ΔCOOH. The cells were cultured in RPMI 1640 containing 10% FCS and counted daily for 5 days. Viable cells represented trypan blue–negative cells that do not display any characteristic features of cells undergoing apoptosis. ● indicates Jurkat; ■, Jurkat-ΔCOOH. (B) Northern blot analysis of hIAN1 mRNA expression in Jurkat and Jurkat-ΔCOOH. The blot contained 5 μg total RNA isolated from Jurkat (odd lanes) and Jurkat-ΔCOOH (even lanes) at days 0 (lanes 1 and 2), 1 (lanes 3 and 4), and 3 (lanes 5 and 6) of culture and was hybridized with a radiolabeled 441-bpDpnII-DpnII fragment of the hIAN cDNA. The amount of RNA loaded in each lane was assessed by 18S and 28S staining.
hIAN1 mRNA expression in Jurkat and Jurkat-ΔCOOH.
(A) Growth analysis of Jurkat and Jurkat-ΔCOOH. The cells were cultured in RPMI 1640 containing 10% FCS and counted daily for 5 days. Viable cells represented trypan blue–negative cells that do not display any characteristic features of cells undergoing apoptosis. ● indicates Jurkat; ■, Jurkat-ΔCOOH. (B) Northern blot analysis of hIAN1 mRNA expression in Jurkat and Jurkat-ΔCOOH. The blot contained 5 μg total RNA isolated from Jurkat (odd lanes) and Jurkat-ΔCOOH (even lanes) at days 0 (lanes 1 and 2), 1 (lanes 3 and 4), and 3 (lanes 5 and 6) of culture and was hybridized with a radiolabeled 441-bpDpnII-DpnII fragment of the hIAN cDNA. The amount of RNA loaded in each lane was assessed by 18S and 28S staining.
Using this DpnII-DpnII DNA fragment as a probe, we screened a Jurkat-ΔCOOH cDNA library and isolated 3 positive clones that exhibited a similar restriction-enzyme pattern. Cloning and sequencing of the longest cDNA isolated revealed an ORF of 329 amino acids with the first ATG (nucleotide [nt] 77) lying in a favorable context for translation initiation (acaATGg)12 and preceded by an in-frame stop codon (nt 51) (Figure2).
Nucleotide and deduced amino acid sequence of hIAN1.
Sequences of different cDNAs were used to obtain this sequence. The first ATG (nt 77) is preceded by an in-frame stop codon (TGA, written in bold letters and underlined) at position 51. The amino acids potentially involved in GTP binding are in italic, bold, heavily underlined letters, and the predicted coiled-coil domain is doubly underlined. The polyadenylation site is in bold letters and underlined.
Nucleotide and deduced amino acid sequence of hIAN1.
Sequences of different cDNAs were used to obtain this sequence. The first ATG (nt 77) is preceded by an in-frame stop codon (TGA, written in bold letters and underlined) at position 51. The amino acids potentially involved in GTP binding are in italic, bold, heavily underlined letters, and the predicted coiled-coil domain is doubly underlined. The polyadenylation site is in bold letters and underlined.
The protein encoded by the ORF exhibited 3 motifs related to motifs found in GTP-binding proteins and a potential coiled-coil domain between amino acids 249 and 288 (Figure 2). Because the encoded protein shared significant homologies with murine Immune Associated Nucleotide 1 (IAN-1),13 it was called hIAN1.
Tal-1 and hIAN1 mRNAs are reciprocally expressed in human T-leukemic cells
To assess any relation between the expression of tal-1 and hIAN1 genes, we first analyzed the levels of hIAN1 mRNA in T-cell lines and in T leukemic blasts expressing different levels of TAL-1. The tal-1 gene expression in T-ALL results from 3 mechanisms: chromosomal translocations, interstitial deletion, or monoallelic activation without apparent gross abnormality of the tal-1 locus. Each mechanism leads to a different TAL-1 protein level. TAL-1 protein expression in T-ALLs harboring sil-tal interstitial deletions is often weaker than in T-ALLs harboring chromosomal translocations or monoallelic activation (our unpublished data, February, 2001). We measured the levels of hIAN1 mRNA in the following cell lines: 2 T-cell lines derived from T-ALL carrying a sil-tal interstitial deletion (CEM and RPMI 8402); 2 cell lines derived from monoallelic activation of the tal-1 locus without obvious abnormalities of the tal-1 gene (Jurkat and Molt-4); and 1 cell line (DU528) in which the tal-1 locus is activated by a chromosomal translocation.14-17 Northern blot analysis showed that the tal-1 mRNA level was higher in Jurkat, Molt-4, and DU528 than in CEM or RPMI 8402 (Figure 3A). Conversely, hIAN1 mRNA was present in CEM and RPMI 8402 and absent in Jurkat, Molt-4, and DU528 (Figure 3A).
hIAN1 and tal-1 mRNA expression in T-ALL.
(A) The hIAN1 and tal-1 mRNAs levels are reciprocally correlated. Northern blot analysis of hIAN1 and TAL-1 mRNA expression was performed on DU528 and Molt-4 (lanes 1 and 2), 2 T-ALL–derived cell lines that expressed TAL-1 after chromosomal rearrangement (DU528) or by a presently unknown mechanism (Molt-4); and on CEM and RPMI 8402 (lanes 5 and 6), and on 2 T-ALL–derived cell lines that harbor the sil-tal rearrangement. Jurkat and Jurkat-ΔCOOH cell lines (lanes 3 and 4) were used as controls. (B) Forced expression of TAL-1 in Jurkat-ΔCOOH does not modulate hIAN1 gene expression. Jurkat-ΔCOOH cells were transfected with a tal-1 expression vector, and 5 subclones (numbered 1 to 5) that expressed various levels of TAL-1 were selected. Total RNAs from Jurkat, Jurkat-ΔCOOH, and the 5 subclones were extracted, and hIAN1 mRNA level was studied by semiquantitative RT-PCR. The amount of cDNA used for PCR was normalized relative to the levels of the S14 gene product.
hIAN1 and tal-1 mRNA expression in T-ALL.
(A) The hIAN1 and tal-1 mRNAs levels are reciprocally correlated. Northern blot analysis of hIAN1 and TAL-1 mRNA expression was performed on DU528 and Molt-4 (lanes 1 and 2), 2 T-ALL–derived cell lines that expressed TAL-1 after chromosomal rearrangement (DU528) or by a presently unknown mechanism (Molt-4); and on CEM and RPMI 8402 (lanes 5 and 6), and on 2 T-ALL–derived cell lines that harbor the sil-tal rearrangement. Jurkat and Jurkat-ΔCOOH cell lines (lanes 3 and 4) were used as controls. (B) Forced expression of TAL-1 in Jurkat-ΔCOOH does not modulate hIAN1 gene expression. Jurkat-ΔCOOH cells were transfected with a tal-1 expression vector, and 5 subclones (numbered 1 to 5) that expressed various levels of TAL-1 were selected. Total RNAs from Jurkat, Jurkat-ΔCOOH, and the 5 subclones were extracted, and hIAN1 mRNA level was studied by semiquantitative RT-PCR. The amount of cDNA used for PCR was normalized relative to the levels of the S14 gene product.
Similar results were obtained with other T-cell lines and suggested a reciprocal expression of tal-1 and hIAN1 mRNAs in human T-cell lines expressing TAL-1. We then studied the expression of hIAN1 mRNA in blasts isolated from 4 human T-ALLs and showed that hIAN1 was highly expressed in T-ALL blasts that exhibit sil-tal interstitial deletions and weakly expressed in T-ALL blasts that exhibit chromosomal translocations involving the tal-1 gene (data not shown). These results are consistent with the reciprocal expression of tal-1 and hIAN1 mRNAs found in the leukemic T-cell lines.
To study whether TAL-1 directly regulates the hIAN1 gene, we studied hIAN1 mRNA level in 5 different clones of Jurkat-ΔCOOH stably transfected with a tal-1 expression vector and expressing various amounts of the wild-type TAL-1 protein (data not shown). Semiquantitative RT-PCR analysis on total RNA isolated from the 5 independent TAL-1–expressing clones was performed. Only one DNA fragment was obtained; its sequence showed that it corresponded to hIAN1 mRNA (data not shown). As shown in Figure 3B, the hIAN1 mRNA level was not affected by the expression of TAL-1 in Jurkat-ΔCOOH transfectants, suggesting that the hIAN1 gene is not a direct target of TAL-1.
hIAN1 belongs to a family of proteins encoded by genes clustered on human chromosome 7 band q36
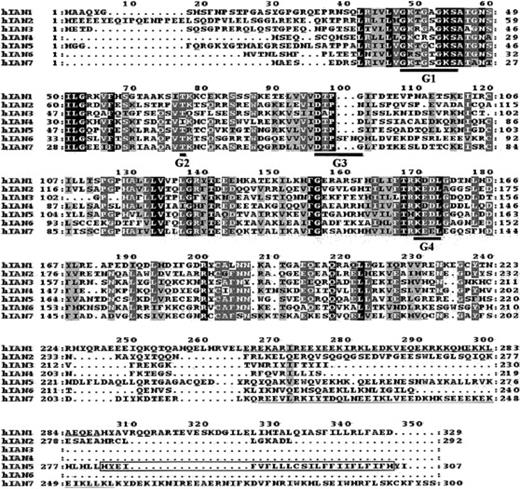
When we screened the Genbank database with the hIAN1 GTP/GDP–binding domain, we identified partial sequences of 5 new human coding sequences and 1 previously identified coding sequence, hIAN5.18 All the encoded proteins (named hIAN1 to hIAN7) showed a high degree of homology in their putative GTP/GDP–binding domain (Figure 4), indicating that hIAN1 defines a novel protein family.
Alignment of the different hIAN proteins found by screening the Genbank database with the hIAN1 GTP/GDP–binding domain.
Protein-sequence alignment was performed by means of ClustalX and displayed with Mac Box Shade 2.15 programs. Conserved amino acids are shaded in black while similar amino acids are shaded in gray. G1, G2, G3, and G4 motifs of the different hIAN proteins are heavily underlined; the coiled-coil domains of hIAN1 and hIAN7 are underlined; and the hydrophobic motif of the hIAN5 protein is boxed.
Alignment of the different hIAN proteins found by screening the Genbank database with the hIAN1 GTP/GDP–binding domain.
Protein-sequence alignment was performed by means of ClustalX and displayed with Mac Box Shade 2.15 programs. Conserved amino acids are shaded in black while similar amino acids are shaded in gray. G1, G2, G3, and G4 motifs of the different hIAN proteins are heavily underlined; the coiled-coil domains of hIAN1 and hIAN7 are underlined; and the hydrophobic motif of the hIAN5 protein is boxed.
No significant homology was found elsewhere in these proteins; only 2 members (hIAN1 and hIAN7) exhibited a coiled-coil motif, while hIAN5 has a hydrophobic domain at its COOH terminus.18
We then searched for the chromosomal localization for the hIAN genes and found, by means of the NCBI Map Viewer (National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD) that all of these genes were clustered on human chromosome 7 band q36. Careful analysis of the available sequences of this region revealed that the hIAN1, hIAN2, hIAN5, and hIAN7 genes were linked within 250 kb of genomic DNA and the hIAN3, hIAN4, hIAN6 genes were linked within 60 kb of genomic DNA (data not shown).
Biochemical characterization of hIAN1
As the hIAN members are characterized by a conserved unconventional GTP/GDP–binding motif, we studied the biochemical properties of hIAN1. GTP/GDP binding and GTPase activities usually require 5 motifs, G1, G2, G3, G4, and G5, that are highly conserved throughout the different families of GTP-binding proteins (see Valencia et al19 for a review). The G2 motif contains only a threonine residue involved in binding the Mg2+ ion complexed with GDP or GTP, whereas the other 4 domains share several amino acids. The G1, G3, and G4 motifs of hIAN1 share the consensus residues important for nucleotide binding with other known G proteins, but they contain, relative to Ras and Gα substitutions at 3 conserved positions, Gly12Ras→Thr39hIAN1, Gln61Ras→Ile89hIAN1, and Asn116Ras→Arg171hIAN1 (Figure5A). Finally, no sequence clearly corresponding to the G5 region can be found in the hIAN1; it should, however, be noted that this domain exhibits little strict conservation across G-protein families (Ras-related, Gα). Thus, hIAN1 protein, like the other members of the IAN protein family, contains notable substitutions of residues considered key in other GTPases.
hIAN1 is a GDP/GTP binding protein.
(A) Sequence of nucleotide-binding motifs G1, G3, and G4 from Ras, Gα, hIAN1, and mIAN-1. The amino acids written in bold letters have been shown to be conserved in all GTP-binding proteins; positions 12, 59, and 61 of Ras are the amino acids involved in GDP/GTP binding and GTPase activities. (B) Affinity purification of GST-hIAN1. After purification on glutathione-sepharose beads, material from a noninduced (lane 1) and induced culture of recombinant bacteria (lane 2) was loaded on a 10% SDS-PAGE, followed by staining with Coomassie blue. (C) Kinetics of GDP binding on hIAN1 protein in the presence of 10 mM MgCl2. First, 60 nM GST-hIAN1 was incubated with 5 μM [3H]GDP in 50 mM Tris (pH 7.5), 1 mM DTT, 100 μg/mL BSA, and 10 mM MgCl2 at 25°C. The binding of [3H]GDP was measured by filtration as described in “Materials and methods.” All measures were performed in triplicate. (D) Specificity of nucleotide binding to GST-hIAN1. First, 60 nM of GST-hIAN1 was incubated at 25°C for 120 minutes with 5 μM [3H]GDP in 50 mM Tris (pH 7.5), 1 mM DTT, 100 μg/mL BSA, and 10 mM MgCl2 in the presence of the indicated concentrations of various nucleotides. The amount of [3H]GDP-bound protein was measured as described in panel B. □, GppNHp; ●, GDP; ×, GMP; −, ATP; ⧫, TTP; ▵, CTP.
hIAN1 is a GDP/GTP binding protein.
(A) Sequence of nucleotide-binding motifs G1, G3, and G4 from Ras, Gα, hIAN1, and mIAN-1. The amino acids written in bold letters have been shown to be conserved in all GTP-binding proteins; positions 12, 59, and 61 of Ras are the amino acids involved in GDP/GTP binding and GTPase activities. (B) Affinity purification of GST-hIAN1. After purification on glutathione-sepharose beads, material from a noninduced (lane 1) and induced culture of recombinant bacteria (lane 2) was loaded on a 10% SDS-PAGE, followed by staining with Coomassie blue. (C) Kinetics of GDP binding on hIAN1 protein in the presence of 10 mM MgCl2. First, 60 nM GST-hIAN1 was incubated with 5 μM [3H]GDP in 50 mM Tris (pH 7.5), 1 mM DTT, 100 μg/mL BSA, and 10 mM MgCl2 at 25°C. The binding of [3H]GDP was measured by filtration as described in “Materials and methods.” All measures were performed in triplicate. (D) Specificity of nucleotide binding to GST-hIAN1. First, 60 nM of GST-hIAN1 was incubated at 25°C for 120 minutes with 5 μM [3H]GDP in 50 mM Tris (pH 7.5), 1 mM DTT, 100 μg/mL BSA, and 10 mM MgCl2 in the presence of the indicated concentrations of various nucleotides. The amount of [3H]GDP-bound protein was measured as described in panel B. □, GppNHp; ●, GDP; ×, GMP; −, ATP; ⧫, TTP; ▵, CTP.
Since the activity of GTP-binding proteins is regulated by their ability to bind GTP and/or GDP and to hydrolyze GTP, we first tested whether hIAN1 exhibited these properties. The hIAN1 was expressed as a GST fusion protein in Escherichia coli and purified on glutathione-sepharose beads to near homogeneity, as revealed by Coomassie blue staining after SDS-PAGE (Figure 5B). The ability of hIAN1 to bind guanine nucleotides was first studied by incubating the recombinant fusion protein with [3H]GDP and analyzing its association kinetics by means of a filter-binding assay. GST-hIAN1 was able to efficiently bind GDP (Figure 5C) with saturable kinetics; the proportion of active fusion protein calculated from such saturation curves was found to be from 30% to 38% depending on the protein preparation used in the experiments. The ability of the protein to bind GDP was highly dependent on the concentration of Mg2+ since it dramatically decreased at low (10 μM and 1 mM) free concentrations of MgCl2 or in the presence of 10 mM EDTA (data not shown).
The specificity of nucleotide binding to hIAN1 was examined in a competition–binding assay (Figure 5D). Both GDP and the nonhydrolyzable analogue of GTP, GppNHp, were efficient competitors of [3H]GDP binding to hIAN1, though with different potencies: approximately 12-fold more GppNHp than GDP was required to inhibit 50% binding of [3H]GDP, indicating a large difference in affinities for the diphosphate and triphosphate guanine nucleotides. In contrast, GMP was unable to compete with [3H]GDP; neither were the other nucleotide triphosphates, ATP, CTP, and TTP (Figure 5D).
After establishing that the binding of GDP and GppNHp at low concentrations (0.7 and 2 μM, respectively) reached a plateau at 90 minutes (data not shown), we further characterized the respective affinities of hIAN1 for GDP and GppNHp. Thus, the direct binding of [3H]GDP and [3H]GppNHp were measured after 120 minutes at various nucleotide concentrations (Figure6A,C).
Determination of the hIAN1 affinity for GDP and GppNHp.
First, 60 nM GST-hIAN1 protein was incubated for 120 minutes at 25°C with various concentrations of GDP (0.175, 0.3, 0.7, 1.5, 3, 5, and 15 μM) or GppNHp (2, 4, 8, 10, 16, 30, 60, 80, 100, and 120 μM); then, the amounts of bound nucleotide were determined by filter-binding assays. All measures were performed in triplicate. Panels A and C show the concentration-dependent binding of GDP (panel A) and GppNHp (panel C) to GST-hIAN1. Scatchard representations are shown in panel B (GDP) and panel D (GppNHp). r indicates bound nucleotide/total protein (pmoles); F is the concentration of free nucleotide (micromolar).
Determination of the hIAN1 affinity for GDP and GppNHp.
First, 60 nM GST-hIAN1 protein was incubated for 120 minutes at 25°C with various concentrations of GDP (0.175, 0.3, 0.7, 1.5, 3, 5, and 15 μM) or GppNHp (2, 4, 8, 10, 16, 30, 60, 80, 100, and 120 μM); then, the amounts of bound nucleotide were determined by filter-binding assays. All measures were performed in triplicate. Panels A and C show the concentration-dependent binding of GDP (panel A) and GppNHp (panel C) to GST-hIAN1. Scatchard representations are shown in panel B (GDP) and panel D (GppNHp). r indicates bound nucleotide/total protein (pmoles); F is the concentration of free nucleotide (micromolar).
Scatchard analysis of the data (Figure 6B,D) revealed that hIAN1 indeed bound both nucleotides to a similar extent (0.3 to 0.38 site per molecule protein, according to the preparation used) but with widely different affinities, since the dissociation constants calculated from these experiments were 0.47 μM for GDP and 6 μM for GppNHp, consistent with the competition experiment described above.
Since GTP hydrolysis plays an important role by regulating the activity of GTP-binding proteins, we investigated the intrinsic GTPase activity of hIAN1. Different amounts of GST-hIAN1 were incubated with [α32P]GTP, and GTP hydrolysis was analyzed by TLC. As shown in Figure 7, a radioactive spot corresponding to GDP appeared; its intensity increased with time and with the amount of GST-hIAN1 protein in the reaction. No spot corresponding to GMP was detected, showing that GDP formation was not due to a contaminating phosphatase activity in the reaction.
GTPase activity of GST-hIAN1.
Intrinsic GTPase activity was assessed by TLC analysis. Different amounts of GST-hIAN1 were incubated at 30°C with 50 μM [α32P]GTP in the presence of 10 mM MgCl2. Aliquots were removed from the reaction at the indicated times and analyzed by TLC as described in “Materials and methods.”
GTPase activity of GST-hIAN1.
Intrinsic GTPase activity was assessed by TLC analysis. Different amounts of GST-hIAN1 were incubated at 30°C with 50 μM [α32P]GTP in the presence of 10 mM MgCl2. Aliquots were removed from the reaction at the indicated times and analyzed by TLC as described in “Materials and methods.”
Thus, this experiment demonstrated that hIAN1 exhibits an intrinsic GTPase activity, despite the divergent amino acids thought to be involved in the mechanisms of the GTPase activity of Gα and Ras.
Expression of hIAN1 mRNA in normal tissues
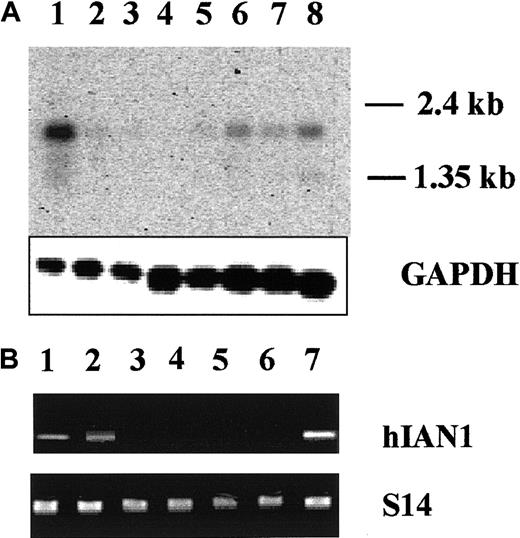
We studied hIAN1 mRNA expression in several normal tissues by Northern blot analysis (Figure 8A). The hIAN1 mRNA was highly expressed in spleen and peripheral blood leukocytes that contain mostly B and T lymphocytes. The hIAN1 mRNA was also detected, but at a lower level, in thymus, small intestine, colon, and ovary, but not detected in prostate or testis.
hIAN1 mRNA expression pattern.
(A) Expression of hIAN1 mRNA in various human tissues. Clontech poly A+ Multiple Tissues Blot was hybridized with an hIAN1 probe. Each lane contains 2 μg poly A+ RNA. Lane 1, spleen; lane 2, thymus; lane 3, prostate; lane 4, testis; lane 5, ovary; lane 6, small intestine; lane 7, colon; lane 8, peripheral blood lymphocytes. The amounts of RNA loaded and transferred to the membrane were assessed by a GAPDH hybridization. (B) Expression of hIAN1 mRNA in mature hematopoietic cells. RT-PCR analysis of hIAN1 mRNA expression was performed on B lymphocytes (lane 1), T lymphocytes (lane 2), mature erythroid cells (lane 3), mature myeloid cells (lane 4), and mature megakaryocytic (lane 5) cells. Jurkat and Jurkat-ΔCOOH cells (lanes 6 and 7) were used as controls. The amount of cDNA used for PCR was normalized relative to the levels of the S14 gene product.
hIAN1 mRNA expression pattern.
(A) Expression of hIAN1 mRNA in various human tissues. Clontech poly A+ Multiple Tissues Blot was hybridized with an hIAN1 probe. Each lane contains 2 μg poly A+ RNA. Lane 1, spleen; lane 2, thymus; lane 3, prostate; lane 4, testis; lane 5, ovary; lane 6, small intestine; lane 7, colon; lane 8, peripheral blood lymphocytes. The amounts of RNA loaded and transferred to the membrane were assessed by a GAPDH hybridization. (B) Expression of hIAN1 mRNA in mature hematopoietic cells. RT-PCR analysis of hIAN1 mRNA expression was performed on B lymphocytes (lane 1), T lymphocytes (lane 2), mature erythroid cells (lane 3), mature myeloid cells (lane 4), and mature megakaryocytic (lane 5) cells. Jurkat and Jurkat-ΔCOOH cells (lanes 6 and 7) were used as controls. The amount of cDNA used for PCR was normalized relative to the levels of the S14 gene product.
To precisely determine the hIAN1 mRNA expression in mature hematopoietic cells, we performed semiquantitative RT-PCR on total RNAs isolated from highly purified hematopoietic cell populations. As shown in Figure 8B, hIAN1 mRNA was not found in myeloid, erythroid, or megakaryocytic cells but was present in mature T and B cells. Interestingly, we also found that, at the mRNA level, all the known members of the hIAN family were expressed in mature T and B cells (data not shown).
During T- and B-lymphocyte activation, hIAN1 protein level is highly regulated at the posttranscriptional level
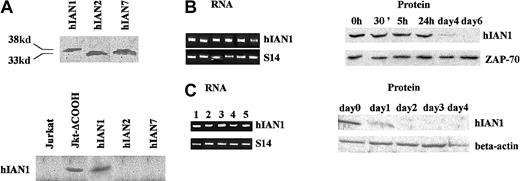
Among mature hematopoietic cells, hIAN1 mRNA was found only in B and T cells; therefore, we studied the hIAN1 mRNA and protein levels during T- and B-cell activation. First, we generated an anti-hIAN1 antiserum and studied its specificity, as hIAN members share a common GTP/GDP–binding domain. The cDNA fragments containing the entire hIAN1, hIAN2, and hIAN7 coding sequences were obtained from fresh human T lymphocytes, subcloned in the pBSK vector, and sequenced. In vitro transcription/translation of these plasmids was then performed in the presence of 35S methionine. As shown in Figure9A, hIAN1 ORF encoded only one protein product that migrates on SDS-PAGE as a 38-kd polypeptide while hIAN 2 and hIAN7 ORFs encoded a 33-kd and a 35-kd polypeptide, respectively. Using the anti-hIAN1 antiserum, we detected, by Western blotting, the hIAN1 product but not the hIAN2 or hIAN7 products (Figure 9A). Indeed, no product was detected when we used the preimmune serum (data not shown). Finally, the anti-hIAN1 antiserum did not detect any protein in Jurkat, while it detected only the 38-kd protein in Jurkat-ΔCOOH (Figure 9A). Taken together, these data demonstrate the specificity of hIAN1 antiserum.
Modulation of hIAN1 protein level during T- and B-lymphocyte activation.
(A) The hIAN1, hIAN2, and hIAN7 cDNAs were in vitro transcribed/translated as described in “Materials and methods,” and the 35S-labeled protein products were run on a 10% SDS-PAGE. The same in vitro–translated products together with total protein extracts of Jurkat and Jurkat-ΔCOOH were run on a 10% SDS-PAGE, blotted, and revealed by the anti-hIAN1 antiserum. (B) T lymphocytes were purified from human peripheral blood and activated with anti-CD28 and anti-CD3. Before activation (lane 1), and 30 minutes (lane 2), 5 hours (lane 3), 1 day (lane 4), 4 days (lane 5), and 6 days (lane 6) after activation, total RNA was extracted from half of the culture and protein extract was performed on the other half. Semiquantitative RT-PCR analysis was performed in the linear range of amplification, and Western blot analysis was performed by means of the anti-hIAN1 antiserum. The loading control for the Western blot was obtained with the use of an anti-ZAP70 antibody.25 (C) B lymphocytes were purified from human PBMCs and activated with 10 mg/mL IL-4 and irradiated (75 Gy) murine L cells that expressed the human CD40 ligand. RNAs and proteins were studied before activation (lane 1) and 1 day (lane 2), 2 days (lane 3), 3 days (lane 4), and 4 days (lane 5) after activation as described in panel B; the loading control for the Western blot was an anti–β-actin antibody.
Modulation of hIAN1 protein level during T- and B-lymphocyte activation.
(A) The hIAN1, hIAN2, and hIAN7 cDNAs were in vitro transcribed/translated as described in “Materials and methods,” and the 35S-labeled protein products were run on a 10% SDS-PAGE. The same in vitro–translated products together with total protein extracts of Jurkat and Jurkat-ΔCOOH were run on a 10% SDS-PAGE, blotted, and revealed by the anti-hIAN1 antiserum. (B) T lymphocytes were purified from human peripheral blood and activated with anti-CD28 and anti-CD3. Before activation (lane 1), and 30 minutes (lane 2), 5 hours (lane 3), 1 day (lane 4), 4 days (lane 5), and 6 days (lane 6) after activation, total RNA was extracted from half of the culture and protein extract was performed on the other half. Semiquantitative RT-PCR analysis was performed in the linear range of amplification, and Western blot analysis was performed by means of the anti-hIAN1 antiserum. The loading control for the Western blot was obtained with the use of an anti-ZAP70 antibody.25 (C) B lymphocytes were purified from human PBMCs and activated with 10 mg/mL IL-4 and irradiated (75 Gy) murine L cells that expressed the human CD40 ligand. RNAs and proteins were studied before activation (lane 1) and 1 day (lane 2), 2 days (lane 3), 3 days (lane 4), and 4 days (lane 5) after activation as described in panel B; the loading control for the Western blot was an anti–β-actin antibody.
We then studied the hIAN1 mRNA and protein levels during T- and B-cell activation. A CD3/CD28 T-lymphocyte activation was performed, and hIAN1 mRNA and protein were studied at 0 hours (lane 1), 30 minutes (lane 2), 5 hours (lane 3), 1 day (lane 4), 4 days (lane 5), and 6 days (lane 6). Using a semiquantitative PCR assay, we showed that the hIAN1 mRNA level remained constant during this CD3/CD28 T-lymphocyte activation while the hIAN1 protein level started to decrease at day 4 and was undetectable at day 6. A similar study was performed on a CD40-ligand/IL-4 B-lymphocyte activation and showed a similar discrepancy between hIAN1 mRNA and protein levels (Figure 9C), with the decrease of hIAN1 protein level starting earlier during this B-lymphocyte activation than in the CD3/CD28 T-lymphocyte activation. These results indicate that hIAN1 gene expression is highly regulated at the posttranscriptional level during B- and T-cell activation.
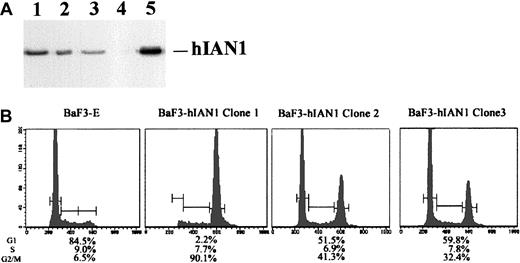
Following IL-3 withdrawal, hIAN1 expression in BaF3 cells led to a G2/M cell cycle arrest
The murine pre-B hematopoietic cell line BaF3 is completely dependent on IL-3 for cell proliferation and survival. These cells did not express mIAN1 mRNA (data not shown) and were used to study a possible role for hIAN1 in cell proliferation and/or survival. We established 3 stable BaF3 clones that expressed various levels of the hIAN1 protein (Figure 10A) and studied these cells in the presence or absence of IL-3. Both the parental BaF3 cells transfected with the empty vector (BaF3-E) and the 3 clones proliferated normally with no difference in cell cycle or apoptosis in the presence of IL-3 (data not shown). After 14 hours of IL-3 deprivation, the BaF3-E and the 3 clones displayed similar signs of apoptosis (30% to 40% of apoptotic cells), and longer deprivation led to cell death in both BaF3-E and BaF3-hIAN1 cells.
Cell cycle analysis of BaF3-hIAN1 subclones following IL-3 withdrawal.
(A) Western blot analysis of 3 BaF3 subclones that expressed different amounts of the hIAN1 protein. Western blot was performed as described in “Materials and methods.” Lanes 1, 2, and 3 show 3 independent BaF3 subclones that contained various amounts of the hIAN1 protein (BaF3-hIAN1 clone 1, BaF3-hIAN1 clone 2, and BaF3-hIAN1 clone 3). Lane 4 shows the BaF3 parental cell line transfected with an empty vector (BaF3-E), and lane 5 is the hIAN1 protein detected in the Jurkat-ΔCOOH cells. (B) Cell cycle analysis of BaF3-E and BaF3-hIAN1 cells in the absence of IL-3. BaF3-E and BaF3-hIAN1 cells were cultured 14 hours in the absence of IL-3. The cells were then collected, permeabilized, and stained with propidium iodide. The cell cycle profile is expressed as number of cells against DNA content and shows the percentage of cells in each phase (G1, S, G2/M) as indicated.
Cell cycle analysis of BaF3-hIAN1 subclones following IL-3 withdrawal.
(A) Western blot analysis of 3 BaF3 subclones that expressed different amounts of the hIAN1 protein. Western blot was performed as described in “Materials and methods.” Lanes 1, 2, and 3 show 3 independent BaF3 subclones that contained various amounts of the hIAN1 protein (BaF3-hIAN1 clone 1, BaF3-hIAN1 clone 2, and BaF3-hIAN1 clone 3). Lane 4 shows the BaF3 parental cell line transfected with an empty vector (BaF3-E), and lane 5 is the hIAN1 protein detected in the Jurkat-ΔCOOH cells. (B) Cell cycle analysis of BaF3-E and BaF3-hIAN1 cells in the absence of IL-3. BaF3-E and BaF3-hIAN1 cells were cultured 14 hours in the absence of IL-3. The cells were then collected, permeabilized, and stained with propidium iodide. The cell cycle profile is expressed as number of cells against DNA content and shows the percentage of cells in each phase (G1, S, G2/M) as indicated.
However, while the BaF3-E cells were arrested at the G1phase of the cell cycle, the 3 BaF3-hIAN1 clones were arrested at the G1 and the G2/M phases of the cell cycle. Interestingly, the ratio between the G2/M and the G1 phase arrest seemed to be correlated with the hIAN1 expression level, suggesting that hIAN1 might be involved in the G1-to-S transition in BaF3 cells deprived of IL-3.
Discussion
Although TAL-1 is expressed in more than 30% of human T-ALLs, most of the pathways regulated by this expression are currently not defined. To characterize genes potentially regulated by TAL-1 during T-lymphocyte leukemogenesis, we compared mRNAs expressed in Jurkat, a human T-cell line that expresses TAL-1, with those expressed in Jurkat-ΔCOOH, a Jurkat subclone that expresses a mutated TAL-1 protein that cannot bind DNA. Phenotypically, Jurkat and Jurkat-ΔCOOH growth curves started to diverge at day 1 of culture; thus, this time point was chosen for RDA analysis. Although we might have missed genes turned on or off by TAL-1 later in the culture, most of the mRNAs we have characterized showed the same expression pattern throughout the 5 days of culture (unpublished data, December, 1998). Using Jurkat-ΔCOOH representations as “testers” and Jurkat representations as “drivers,” we have characterized a human gene, hIAN1, that is specifically expressed in Jurkat-ΔCOOH and thus might be negatively regulated by TAL-1. We then showed a reciprocal expression of tal-1 and hIAN1 mRNAs in T-cell lines and in leukemic blasts from T-ALL patients that expressed different levels of TAL-1 protein, and these results extended our preliminary finding. However, the expression of the wild-type TAL-1 protein in Jurkat-ΔCOOH cells had no effect on hIAN1 gene expression, suggesting that hIAN1 is not a direct TAL-1 target gene.
The reciprocal expression of tal-1 and hIAN1 mRNAs in T-cell lines and T leukemic blasts that expressed different levels of TAL-1 protein can be linked to recent studies20,21 showing that the presence of oncogenic transcription factors in T-ALL induces a blockade at specific stages of thymocyte differentiation and suggests that the expression of one of these factors during T-lymphocyte leukemogenesis is correlated to a specific pattern of gene expression that reflects the stage in which the disruption of T-cell development has occurred. As the presence of TAL-1 in T-ALL leads to an arrest at the double-positive stage and as mIAN-1 started to be expressed at the double-positive to single-positive stage during T-cell differentiation,13 our data on hIAN1 and tal-1 mRNA expression in T-ALL suggest there might be a slight delay in the blockade of T-cell differentiation, depending on whether the tal-1 gene is activated by chromosomal translocations or by sil-tal rearrangements. The hIAN1 therefore appeared as a new marker of T-cell differentiation blockade in TAL-1–dependent T-ALL.
The study of the hIAN1 primary structure showed that it contains motifs related to those found in G proteins. However, these motifs displayed amino acid substitutions at 3 highly conserved positions (corresponding to the positions 12, 61, and 116 in the Ras protein) that are important for nucleotide binding as well as intrinsic and GTPase-activating protein–stimulated GTPase activities.22-24 Using this putative GTP/GDP–binding domain as a probe, we searched, in silico, for human proteins that contain a similar domain and found 6 other proteins encoded by genes clustered on human chromosome 7 band q36. Apart from this putative GTP/GDP–binding domain, these proteins do not display any significant homology, suggesting that this family of proteins is defined only by this domain. A recent study has shown that mIAN-4, a member of the mouse IAN family, is located in the central region of mouse chromosome 6, which is proximal to a region of synteny to human chromosome 7p15-14.18
However, as noticed by the authors, there is a break in synteny between human chromosome 7p15-14 and 7q34-35 in a region where the mIAN-4 gene lies. Here, we showed that the human IAN gene family is located on chromosome 7q36, and this clarifies the region of synteny between these 2 chromosomes.
Despite the amino acid substitutions present in the putative GTP/GDP domain of hIAN1, our biochemical study showed that hIAN1 is a true GTPase and thus defines a new family of G proteins. An interesting finding of the current study concerned the different binding affinities of hIAN1 for GDP and GppNHp (about 12-fold higher for GDP) and the absolute requirement of Mg2+ for GDP and GppNHp binding. These properties are not found for Ras-like proteins and might be related to the particular nucleotide-binding site of hIAN1. Despite these unusual characteristics, equilibrium-binding calculations show that, given the generally accepted cellular concentrations for GDP (100 μM) and GTP (1 mM), hIAN1 should be 50% bound to GTP in the absence of GTPase activity and auxiliary proteins. This prediction is supported by a recent study on mIAN-4 indicating that part of this protein is associated with GTP in vitro.18 This strongly suggested that despite its difference in affinity for GDP and GTP, hIAN1, like most other G proteins (eg, EF-Tu, Ras-related, G heterotrimeric) could act as a molecular switch cycling between biochemically distinct GDP- and GTP-bound forms. Thus, the IAN family seems to define a novel type of GTPase proteins, and the identification of proteins regulating their nucleotide-binding state and GTPase activity will shed light on the signal transduction pathways involving hIAN proteins.
As the hIAN1 or mIAN-1 expression pattern suggests a function of this protein in the immune system, we first studied the regulation of hIAN1 gene expression during B- and T-lymphocyte activation. We showed that the hIAN1 protein level decreased to an undectectable level during a CD40-ligand/IL-4 B-cell activation or a CD3/CD28 T-cell activation while the steady-state hIAN1 mRNA levels are largely unaffected during these activations. These results suggested a regulation of the hIAN1 mRNA translation and/or a specific degradation of the hIAN1 protein during B- or T-lymphocyte activation, and we are currently studying the posttranscriptional regulation of hIAN1 gene expression in mature B and T lymphocytes. Finally, we showed that, following IL-3 withdrawal, the expression of hIAN1 in BaF3 cells could lead to a G2/M arrest, while parental BaF3 cells were arrested at the G1phase. This result, together with the expression pattern of hIAN1 protein during the B- and T-cell activation, suggested that hIAN1 might be involved at some steps of the G1 checkpoint of the B- and T-lymphocyte cell cycle.
In conclusion, the highly regulated expression of the hIAN1 protein during B- and T-lymphocyte activation, together with the coordinated expression of all the genes coding for the hIAN members in resting T and B lymphocytes, suggested the presence of currently unknown coordinated functions of this protein family during the immune response. The biochemical characterization of hIAN1 shown in this study, together with previous work on the GTPase structure/function,22-24 will provide a molecular basis for designating gain or loss of function hIAN proteins, which represent powerful tools for studying the function of these proteins in the immune response.
We thank J. P. Farcet and J. Marquet (EA 2348, Hôpital Henri Mondor, Créteil, France) for providing us with B cells, and Nathalie de Preobrajensky and Sebastien L'Hoste for their help in the databank screening.
Supported by the Institut National de la Santé et de la Recherche Médicale and by the Ligue Nationale Contre le Cancer (Equipe labellisée); Sandra Aresta is a fellow of PraxisXXI (FCT, Ministério para a Ciência e Tecnologia, Portugal).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Paul-Henri Roméo, INSERM U474, Maternité Port-Royal, 75014 Paris, France; e-mail:romeo@cochin.inserm.fr.



![Fig. 5. hIAN1 is a GDP/GTP binding protein. / (A) Sequence of nucleotide-binding motifs G1, G3, and G4 from Ras, Gα, hIAN1, and mIAN-1. The amino acids written in bold letters have been shown to be conserved in all GTP-binding proteins; positions 12, 59, and 61 of Ras are the amino acids involved in GDP/GTP binding and GTPase activities. (B) Affinity purification of GST-hIAN1. After purification on glutathione-sepharose beads, material from a noninduced (lane 1) and induced culture of recombinant bacteria (lane 2) was loaded on a 10% SDS-PAGE, followed by staining with Coomassie blue. (C) Kinetics of GDP binding on hIAN1 protein in the presence of 10 mM MgCl2. First, 60 nM GST-hIAN1 was incubated with 5 μM [3H]GDP in 50 mM Tris (pH 7.5), 1 mM DTT, 100 μg/mL BSA, and 10 mM MgCl2 at 25°C. The binding of [3H]GDP was measured by filtration as described in “Materials and methods.” All measures were performed in triplicate. (D) Specificity of nucleotide binding to GST-hIAN1. First, 60 nM of GST-hIAN1 was incubated at 25°C for 120 minutes with 5 μM [3H]GDP in 50 mM Tris (pH 7.5), 1 mM DTT, 100 μg/mL BSA, and 10 mM MgCl2 in the presence of the indicated concentrations of various nucleotides. The amount of [3H]GDP-bound protein was measured as described in panel B. □, GppNHp; ●, GDP; ×, GMP; −, ATP; ⧫, TTP; ▵, CTP.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/99/9/10.1182_blood.v99.9.3293/6/m_h80922474005.jpeg?Expires=1767704455&Signature=Uv8KRjwIJ2bOVAc1ocoGCmkUbbnMC7-qhi~WGy0KwzcQqk1CWnD7M6tUDpFK-5bKCWSWoadd-jz59tQPcKdxh1Kpk3f0R-CiKSePHv-RWKdN9SXWgBxcifBIMlAfCQnxyxZtjCyFGXdqf8xc~ly7z48LAJQoKQybS-z4uXr1gdHQKO4hQk6KW9~i7C9Q-LH49Ha-LMWKEFQEExQggGAbCZhzuj~OVgit11eE6Cyo~7AE~1XEfDaA-TPBPsiwO9Is~JJ-ijWBkFfztgrCwpB5~f9qow~4AUmEY~5hh9L7WGsFubYLqtHcXLo~I0xvgETeLFDQrkieCciYK6kAFJs5~w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 7. GTPase activity of GST-hIAN1. / Intrinsic GTPase activity was assessed by TLC analysis. Different amounts of GST-hIAN1 were incubated at 30°C with 50 μM [α32P]GTP in the presence of 10 mM MgCl2. Aliquots were removed from the reaction at the indicated times and analyzed by TLC as described in “Materials and methods.”](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/99/9/10.1182_blood.v99.9.3293/6/m_h80922474007.jpeg?Expires=1767704455&Signature=HEhnu1o1XADLnf4fo4H9IIUP4KkiwR1ZzktiFsBigMqhPcqNMV3A4qx7VLlOl0HqArLer8kCMel0W6b2aub~K2le~BJXjKuGF1PKfqbgxzc8A4BWJiVgoz4Qe7x7461Zx0jPrZesMnYhgOaWtIwLgFUfP1aLMqfdcz8ZLPe~EumbEqRObbGpVoT3c0moFskEuyTplcKYzzkx79pKYH4gbsuKifHr3LtK53m0aSfVMGd1He3jAvpIR0I9MR9zzRQ8E0V3QBaXnCgSKQgu3J8HB7EuagWBdNuRFQakO7V9Jbwlxb0PNmtYVm5z3mGstOVu0HXvny6H~1gXEIwoeWjVNw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal