We investigated the prognostic value of p16INK4aimmunocytochemistry (ICC) analysis in 126 cases of newly diagnosed childhood acute lymphoblastic leukemia (ALL). The incidence of negative p16INK4a ICC was 38.1% and was more frequent in T-lineage ALL. Overall survival (OS) and event-free survival (EFS) were significantly higher in patients with positive p16INK4a ICC than in patients with negative ICC (6 years OS, 90% versus 63%,P = .0014; 6 years EFS, 77.8% versus 55%,P = .0033). The p16INK4a ICC remained a significant prognostic factor within the subgroup of B-precursor ALL. Multivariate analysis showed that negative p16INK4a ICC was an independent prognostic factor for OS (relative risk [RR], 3.38;P = .02) and EFS (RR, 2.49; P = .018). Sequential study showed that p16INK4a expression remained stable during first relapse in most patients. These findings indicate that p16INK4a ICC is an independent factor of outcome in childhood ALL.
Introduction
The p15INK4b and p16INK4aproteins are cell-cycle regulators involved in the inhibition of G1 phase progression. The p16INK4aand p15INK4b genes are homozygously deleted in many tumor types, including childhood acute lymphoblastic leukemia (ALL), but p16INK4a expression in leukemic cells may vary in the absence of gene deletion or point mutation.1-12 We recently reported the results of p16INK4a protein analysis by immunocytochemistry (ICC) in a limited cohort of adult ALL patients.13 The technique of p16INK4a ICC requires only bone marrow or blood smears; many samples can be processed in the same day, and this technique allows direct identification of leukemic cells, leading to an easier interpretation. We observed that negative p16INK4a ICC conferred an adverse outcome in adult ALL with standard-risk karyotype, but the small number of samples did not allow us to perform multivariate analysis. The low rate of high-risk karyotypic abnormalities in childhood ALL allowed us to expect a specific prognostic value of p16INK4a ICC in childhood ALL.
In a large, homogenously treated cohort of childhood ALL patients, we investigated the influence of negative p16INK4a ICC on overall survival (OS) and event-free survival (EFS) in univariate and multivariate analysis. Additionally, we sequentially investigated p16INK4a ICC in a cohort of those patients at relapse.
Study design
We analyzed p16INK4a gene expression by p16INK4a ICC in 126 childhood ALLs (89 B-precursor ALLs, 28 T-ALLs, and 9 acute undifferentiated leukemias). Patient bone marrow (n = 87) or blood (n = 39) smears were collected from May 1992 to December 2000. Median age was 4.9 years (range, 0.5-15). There were 73 males and 53 females. Of these patients, 80 were treated according to the European Organization for Research and Treatment of Cancer (EORTC) protocol 58881 between 1991 and December 1998, and 46 were treated by EORTC protocol 58951 between January 1998 and December 2000.14 The median observation time for all patients was 4.8 years (range, 1.4-9.2 years). There were 31 other patients studied by p16INK4a ICC in first relapse. Among them, 20 were also studied sequentially (14 at diagnosis and first relapse, 6 in first and second or third relapse).
Immunocytochemical detection of p16INK4a protein was performed with the immunoglobulin G1-κ mouse monoclonal antibody antihuman p16INK4a (clone DCS-50.1/H4) (Oncogene Research Products, Cambridge, United Kingdom), as previously described.13 The ICC reaction was performed with avidin-biotin-peroxidase technique by means of Vector reagents (Vector Laboratories, Burlingame, CA). Positive cells appeared with brownish granules. Samples were considered ICC-positive when more than 5% of cells showed p16INK4a protein, according to our previous study in adult ALL.13
The chi-square and Fisher exact test were used for comparison between initial parameters. OS and EFS were estimated according to the Kaplan-Meier method. Events were defined as induction death, relapse, and death in complete remission (CR). Multivariate analysis was based on the Cox proportional hazards regression model. Statistical analyses were performed on SPSS 9.1 analysis software (SPSS, Chicago, IL).
Results and discussion
As previously observed in adult ALL, a great variation in the percentage of p16INK4a ICC-positive cells was seen (median, 20%; range, 0-100).13 We found 51 samples (38.1%) to be p16INK4a ICC–negative. All patients achieved CR. No difference for sex, white blood cell count, presence of a bulky mass, central nervous system disease, hemoglobin level, chromosome 9p abnormalities, or karyotype could be observed between p16INK4a ICC–positive and p16INK4aICC–negative cases. However, positive p16INK4a ICC was found significantly more frequently in B-precursor ALL (70.7%) than in T-ALL (43%) (P = .006). Positive p16INK4a ICC ALL patients were also more likely to be older than age 9 years than were negative p16INK4a ICC ALL patients, but significance (P = .046) disappeared after stratification by phenotype (P = .205).
Increased sensitivity threshold for glucocorticoid-induced apoptosis induced by forced p16INKa expression in lymphoblastic leukemia cell line has been reported.15 However, we did not find significant association in our cohort of childhood ALL patients between p16INK4a ICC status and prednisone response at day 8 (P = .434). Hence, it remains unclear whether p16INK4a expression influences in vivo glucocorticoid-induced apoptosis of ALL cells.
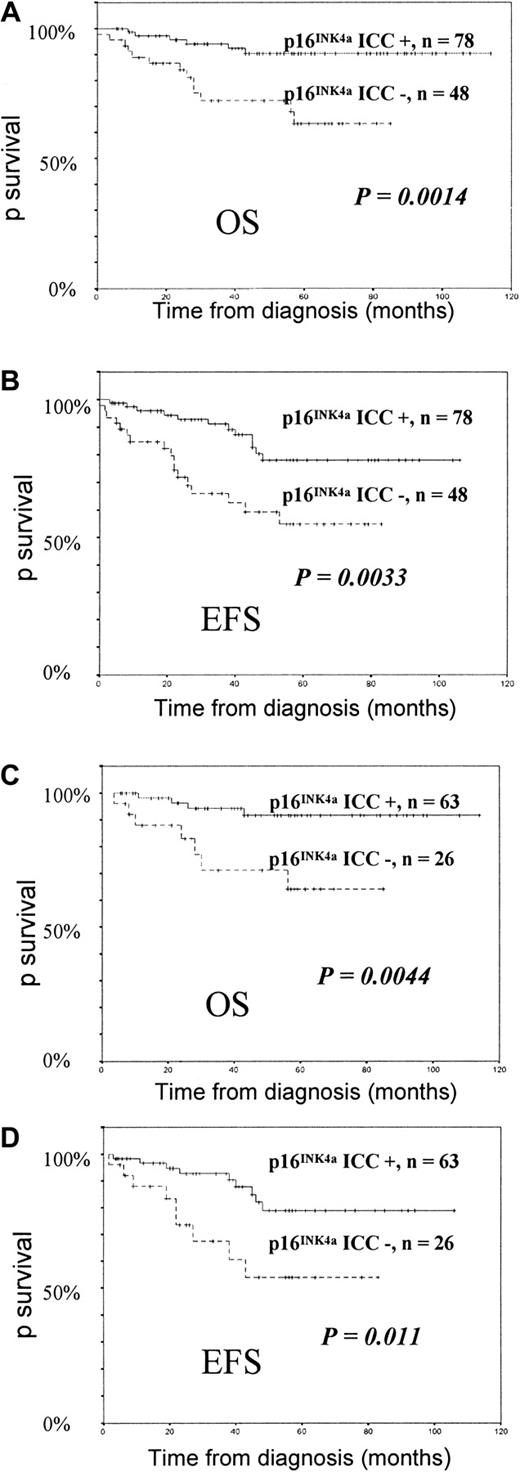
Univariate analysis showed significantly better OS and EFS in patients with positive p16INK4a ICC at diagnosis (Figure1). OS estimates at 6 years for patients with or without positive p16INK4a ICC at diagnosis were 90% (SE = 3.8%) and 63% (SE = 8.7%), respectively (P = .0014, log-rank test). EFS estimates at 6 years were 78% (SE = 5.8%) and 54% (SE = 8.4%) for p16INK4aICC–positive and p16INK4a ICC–negative subgroups, respectively (P = .0033, log-rank test). When cutoff values of 0% and 10%, rather than 5%, for negative versus positive p16INK4a ICC were used, results were less significant. These findings indicate that, as in our previous analysis in adult ALL, 5% is a valid cutoff value for prognostic studies in ALL.13 Despite the association between negative ICC and T phenotype at diagnosis, p16INK4a ICC remained a significant prognostic factor in the subgroup of B-precursor ALLs for both OS (P = .0044) and EFS (P = .011) (Figure 1). This was not the case for T-ALL (OS, P = .23; EFS,P = .19).
Survival of childhood ALL patients according to their p16INK4a ICC status.
OS (panel A) and EFS (panel B) of the 126 childhood ALL patients. OS (panel C) and EFS (panel D) of childhood B-precursor ALL patients according to their p16INK4a ICC status.
Survival of childhood ALL patients according to their p16INK4a ICC status.
OS (panel A) and EFS (panel B) of the 126 childhood ALL patients. OS (panel C) and EFS (panel D) of childhood B-precursor ALL patients according to their p16INK4a ICC status.
Final models of multivariate analysis showed that p16INK4a ICC remained an independent prognostic factor for both OS (P = .02) and EFS (P = .0188) in the whole cohort (Table1). However, karyotype remained the main prognostic factor for OS (relative risk = 3.92 versus 3.38). Incorporation of immunophenotype into the model did not modify the results. It has been suggested in previous studies that the significance of p16INK4a gene deletions would disappear within the subgroups of T-ALL and B-precursor ALL.16,17Our data show that T phenotype does not account for the poorer outcome of p16INK4a ICC–negative patients. However, the results of gene deletion studies and protein expression analyses by ICC may differ. Indeed, several studies have shown that p16INK4aprotein expression in leukemic cells is a complex phenomenon and can be altered not only by gene deletion but also by promoter methylation and other, unknown mechanisms.18-21 We also observed, both in adult ALL and in the present pediatric study, that a few samples of leukemic cells with no detectable p16INK4a protein at diagnosis showed p16INK4a expression at relapse (Table2, patient I120).13 These findings might explain why p16INK4a ICC provides prognostic information distinct from that derived throughp16INK4a gene deletion analysis. The lower proportion of high-risk karyotypes such as t(9;22)(q34;q11) in children may explain why p16INK4a ICC has a specific prognostic value in childhood ALL as compared with adult ALL.
Multivariate Cox model analysis for overall and event-free survival of the 126 childhood acute lymphoblastic leukemia patients
| Variable . | Univariate analysis . | Multivariate analysis . | ||
|---|---|---|---|---|
| P . | P . | RR . | 95% CI . | |
| OS | ||||
| p16INK4aICC* | .0014 | .0204 | 3.38 | 1.20 -9.48 |
| Karyotype† | .0032 | .0227 | 3.92 | 1.21 -12.72 |
| Age‡ | .0165 | .12 | 2.30 | 0.81 -6.61 |
| WBC1-153 | .0425 | .0719 | 2.55 | 0.92 -7.09 |
| Prednisone response day 81-155 | .069 | — | — | — |
| Hemoglobin1-154 | .964 | — | — | — |
| Bulky tumor mass | .27 | — | — | — |
| Immunophenotype1-159 | .25 | — | — | — |
| Sex | .8425 | — | — | — |
| EFS | ||||
| p16INK4aICC* | .0033 | .0188 | 2.49 | 1.16 -5.32 |
| Karyotype† | .0609 | — | — | — |
| Age‡ | .0374 | .0735 | 2.06 | 0.93 -4.55 |
| WBC1-153 | .297 | — | — | — |
| Prednisone response day 81-155 | .4 | — | — | — |
| Hemoglobin1-154 | .73 | — | — | — |
| Bulky tumor mass | .77 | — | — | — |
| Immunophenotype1-159 | .28 | — | — | — |
| Sex | .52 | — | — | — |
| Variable . | Univariate analysis . | Multivariate analysis . | ||
|---|---|---|---|---|
| P . | P . | RR . | 95% CI . | |
| OS | ||||
| p16INK4aICC* | .0014 | .0204 | 3.38 | 1.20 -9.48 |
| Karyotype† | .0032 | .0227 | 3.92 | 1.21 -12.72 |
| Age‡ | .0165 | .12 | 2.30 | 0.81 -6.61 |
| WBC1-153 | .0425 | .0719 | 2.55 | 0.92 -7.09 |
| Prednisone response day 81-155 | .069 | — | — | — |
| Hemoglobin1-154 | .964 | — | — | — |
| Bulky tumor mass | .27 | — | — | — |
| Immunophenotype1-159 | .25 | — | — | — |
| Sex | .8425 | — | — | — |
| EFS | ||||
| p16INK4aICC* | .0033 | .0188 | 2.49 | 1.16 -5.32 |
| Karyotype† | .0609 | — | — | — |
| Age‡ | .0374 | .0735 | 2.06 | 0.93 -4.55 |
| WBC1-153 | .297 | — | — | — |
| Prednisone response day 81-155 | .4 | — | — | — |
| Hemoglobin1-154 | .73 | — | — | — |
| Bulky tumor mass | .77 | — | — | — |
| Immunophenotype1-159 | .28 | — | — | — |
| Sex | .52 | — | — | — |
RR indicates relative risk; CI, confidence interval; OS, event-free survival; ICC, immunocytochemistry; WBC, white blood count; EFS, event-free survival.
Greater than 5% acute lymphoblastic leukemia cells; 5% or fewer acute lymphoblastic leukemia cells.
High risk, including t(9; 22), t(4; 11), t(1; 19) and hypodiploid; intermediate and low risk.
Younger than 1 and older than 9 years; 1 year or older and 9 years or younger.
Lower than 50 000/μL; 50 000/μL or higher.
Circulating blasts at 1000/μL or more; fewer than 1000/μL circulating blasts.
More than 11 g/dL; lower than 11 g/dL.
T-acute lymphoblastic leukemia; B-precursor acute lymphoblastic leukemia.
Sequential analysis of p16INK4aimmunocytochemistry in patients with acute lymphoblastic leukemia
| Patient no. . | Immunophenotype . | Positive leukemic cells by p16INK4a ICC, % . | |||
|---|---|---|---|---|---|
| Diagnosis . | First relapse . | Second relapse . | Third relapse . | ||
| I101 | B | ND | 80 | 95 | 100 |
| I102 | B | ND | 90 | 0 | — |
| I103 | B | ND | 100 | 0 | — |
| I104 | B | 70 | 100 | — | — |
| I105 | B | 100 | 25 | — | — |
| I106 | AUL | 3 | 2 | — | — |
| I107 | T | 0 | 1 | 0 | 0 |
| I108 | B | ND | 100 | ND | 100 |
| I109 | T | 75 | 40 | — | |
| I110 | B | 2 | 2 | — | — |
| I111 | B | 16 | 100 | — | — |
| I112 | B | 95 | 20 | — | — |
| I113 | AUL | 100 | 30 | — | — |
| I114 | T | ND | 2 | 0 | — |
| I115 | B | 1 | 1 | ND | — |
| I116 | B | ND | 1 | 0 | — |
| I117 | B | 100 | 70 | — | — |
| I118 | T | 0 | 1 | — | — |
| I119 | B | 80 | 1 | 0 | — |
| I120 | B | 2.5 | 16 | — | — |
| I121 | T | ND | 40 | — | — |
| I122 | B | ND | 35 | — | — |
| I123 | B | ND | 25 | — | — |
| I124 | B | ND | 50 | — | — |
| I125 | B | ND | 100 | — | — |
| I125 | AUL | ND | 100 | — | — |
| I127 | B | ND | 1 | — | — |
| I128 | B | ND | 2 | — | — |
| I129 | T | ND | 1 | — | — |
| I130 | B | ND | 35 | — | — |
| I131 | B | ND | 7 | — | — |
| Patient no. . | Immunophenotype . | Positive leukemic cells by p16INK4a ICC, % . | |||
|---|---|---|---|---|---|
| Diagnosis . | First relapse . | Second relapse . | Third relapse . | ||
| I101 | B | ND | 80 | 95 | 100 |
| I102 | B | ND | 90 | 0 | — |
| I103 | B | ND | 100 | 0 | — |
| I104 | B | 70 | 100 | — | — |
| I105 | B | 100 | 25 | — | — |
| I106 | AUL | 3 | 2 | — | — |
| I107 | T | 0 | 1 | 0 | 0 |
| I108 | B | ND | 100 | ND | 100 |
| I109 | T | 75 | 40 | — | |
| I110 | B | 2 | 2 | — | — |
| I111 | B | 16 | 100 | — | — |
| I112 | B | 95 | 20 | — | — |
| I113 | AUL | 100 | 30 | — | — |
| I114 | T | ND | 2 | 0 | — |
| I115 | B | 1 | 1 | ND | — |
| I116 | B | ND | 1 | 0 | — |
| I117 | B | 100 | 70 | — | — |
| I118 | T | 0 | 1 | — | — |
| I119 | B | 80 | 1 | 0 | — |
| I120 | B | 2.5 | 16 | — | — |
| I121 | T | ND | 40 | — | — |
| I122 | B | ND | 35 | — | — |
| I123 | B | ND | 25 | — | — |
| I124 | B | ND | 50 | — | — |
| I125 | B | ND | 100 | — | — |
| I125 | AUL | ND | 100 | — | — |
| I127 | B | ND | 1 | — | — |
| I128 | B | ND | 2 | — | — |
| I129 | T | ND | 1 | — | — |
| I130 | B | ND | 35 | — | — |
| I131 | B | ND | 7 | — | — |
ICC indicates immunocytochemistry; AUL, acute undifferentiated leukemia; ND, not done.
Of the children with ALL analyzed at first relapse, 11 (35.5%) showed negative p16INK4a ICC (Table 2). Previous studies had shown that p16INK4a gene deletion could be acquired during evolution of the disease, but these findings have not been observed by other groups.3,5,16,22-24 In our study, ICC status became negative between diagnosis and first relapse in one patient (I119), and between first and second relapse in two others (I102, I103). Substantial variations of the percentage of p16INK4a-positive cells were also observed in 5 patients, suggesting that p16INK4a expression varies widely among both patients and varieties of disease progression. Thus, p16INK4a ICC should be tested when drugs targeting p16INK4a protein are tested.20 These findings indicate that absence of p16INK4a expression is an early phenomenon in the evolution of ALL. Some patients may lose p16INK4a expression during further relapse, but the impact of p16INK4a inactivation in those leukemic cells, which probably have accumulated numerous other gene alterations, remains to be determined.
Thus, the simple and reproducible p16INK4a ICC method should provide important prognostic information in large-scale prospective therapeutic studies.
Supported by the Ligue Contre le Cancer (Comité du Nord and Comité du Pas de Calais).
J.H.D. and M.F. contributed equally to this work as first authors.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Bruno Quesnel, Service des Maladies du Sang, CHU Lille, 1 Place de Verdun, 59037, Lille, France; e-mail:bquesnel@nordnet.fr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal