Cyclic adenosine monophosphate response-element binding protein (CREB) is a nuclear protein that regulates expression of genes that control cell proliferation, differentiation, and survival. To analyze CREB expression in leukemia cells, we conducted Western blot analysis of bone marrow cells obtained from patients with acute lymphoblastic leukemia, patients with acute myeloid leukemia, and patients without active leukemia. CREB was expressed at a higher frequency in bone marrow cells from patients with acute lymphoid or myeloid leukemia than in patients with leukemia remission or without leukemia. Our results indicate that CREB expression could be a useful marker for leukemia in patients with acute disease and suggest a role for CREB in leukemogenesis.
Introduction
Cyclic adenosine monophosphate (cAMP) response-element binding protein (CREB) is a nuclear protein that regulates gene expression on activation of cAMP-dependent or cAMP-independent signal-transduction pathways in cells.1-3CREB binds the cAMP response element in the promoter regions of target genes that regulate cell proliferation and survival, such asbcl-2 and egr-1.4-6 Phosphorylation of CREB at serine 133 in response to growth factors modulates the function of CREB.1,7,8 Thus, CREB was proposed to be a critical regulator of growth factor–induced gene expression leading to cell proliferation, differentiation, and survival.1 7-10
In studying signaling pathways activated by the hematopoietic growth factor granulocyte-macrophage colony-stimulating factor (GM-CSF), we previously found that CREB was phosphorylated in response to GM-CSF stimulation of myeloid leukemia cells.6,11 12 In the current study, to determine whether CREB is overexpressed in leukemia cell lines and bone marrow specimens from patients with acute leukemia, we conducted Western blot analysis to detect the presence of CREB protein. We found that CREB was expressed in most leukemia cell lines and in bone marrow from patients with acute leukemia but that CREB levels were below the detection limit in normal bone marrow samples and in bone marrow samples from patients without active leukemia. Our results suggest that CREB could be a marker for leukemia.
Study design
Patient selection
We analyzed bone marrow samples from 26 patients with newly diagnosed acute myeloid leukemia (AML), 20 patients with acute lymphoblastic leukemia (ALL), and 25 patients without active leukemia. Included in the cohort of patients without active leukemia were 9 patients with ALL and 4 patients with AML in documented remission. The nonleukemia bone marrow samples were from 4 patients with neutropenia, 4 patients with idiopathic thrombocytopenic purpura (ITP), 2 healthy volunteers (donors of bone marrow for transplantation), and 2 patients with lymphoma who underwent bone marrow aspiration as part of an evaluation for metastasis. The patients' age ranged from 4 months to 80 years. The diagnoses of leukemia were made according to standard morphologic criteria on the basis of bone marrow aspirations and biopsies and immunohistochemical, immunophenotyping, and cytogenetic studies. Among the patients analyzed, one patient with ALL had t(12;2). Among the patients with AML, 5 had abnormal cytogenetic findings, including 5q−, inv(16), t(15;17), extra chromosome 11, and t(8;21). Informed consent to participation in the study was obtained from all patients before bone marrow samples were obtained, in compliance with the Helsinki protocol.
Western blot analysis
Mononuclear cells from bone marrow specimens were extracted by Ficoll-Hypaque density gradient separation. Cells were lysed and Western blotting was done with rabbit polyclonal anti-CREB antiserum (0.5 μg/mL; UBI, Lake Placid, NY) and antiactin antiserum (60 μg/mL; Santa Cruz Biotechnology, Santa Cruz, CA) as described previously.11
Immunohistochemical studies
Tissue cores were obtained from tumor areas of each donor block and transferred to the recipient paraffin block by using a custom-made precision instrument.13 Sections (5 μm) were cut with a microtome and placed on positively charged slides. The slides were baked, deparaffinized in xylene, and rehydrated through graded alcohols to water. Antigen retrieval was done by immersing the slides in target retrieval solution (DAKO, Carpinteria, CA). Slides were blocked by using protein blocking solution (DAKO). Washes were done in Tris-buffered saline and 0.05% Tween 20 (pH 7.4). Slides were incubated with 10 μg/mL rabbit polyclonal anti-CREB antibody (UBI). A biotinylated link antibody and a streptavidin-horseradish peroxidase kit (LSAB2; DAKO) were used, along with a diaminobenzidine chromogen and peroxide substrate, to detect the bound antibody complexes. Counterstaining was done with hematoxylin. Light microscopy was used to evaluate the intensity and localization of the staining.
Results and discussion
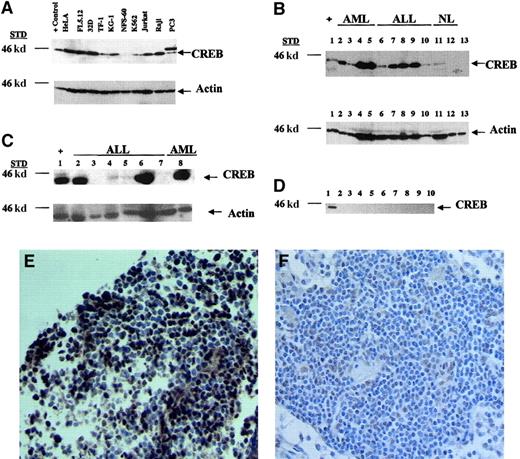
To determine whether CREB expression was increased in leukemia cell lines, we first examined CREB protein levels in the myeloid leukemia cell lines 32D, TF-1, KG-1, NFS-60, and K562 and the lymphoid leukemia cell lines Jurkat and Raji. Nonleukemia cell lines were also analyzed, including HeLa, PC3, and FL5.12, which are cervical cancer, prostate cancer, and myeloid cell lines, respectively. CREB was expressed in 6 of the 7 leukemia cell lines, with NFS-60 cells expressing decreased levels of CREB (Figure1A). PC3 cells expressed a higher-molecular-weight band of 48 kd, which is most likely a protein that cross-reacts with the CREB antibody. Nonhematopoietic cell lines, including HeLa, also had high levels of CREB expression. Thus, we found that most myeloid and lymphoid leukemia cell lines overexpress CREB.
Expression of CREB in leukemia cell lines and bone marrow samples from patients with acute leukemia.
Shown are results of Western blot analyses of cell lines and of bone marrow specimens from patients with and without leukemia. (A) Leukemia and nonleukemia cell lines were assessed for CREB expression by Western blot analysis. Cells were grown to a density of 1 × 106 cells/mL, and lysates were prepared from 1 × 106 cells. Twenty micrograms of protein from cell lysates was loaded on a 10% sodium dodecyl sulfate (SDS)–polyacrylamide gel. Blots were probed with anti-CREB antiserum (0.5 μg/mL), stripped, and reprobed with actin (60 μg/mL) as the loading control. The arrows represent CREB (43 kd) or the actin control (43 kd). (B) Mononuclear cells from bone marrow obtained from patients with leukemia or without active disease were processed with the Ficoll-Hypaque method. Lysates were prepared from 1 × 106 mononuclear cells. Twenty micrograms of protein was loaded on a 10% SDS-polyacrylamide gel. Western blot analysis was done with anti-CREB antiserum. Shown are results with positive controls (32Dcl3 cell lysates; lane 1), results with the AML (lanes 2-5) and ALL (lanes 6-7) samples, and results with samples from patients without active leukemia or with normal bone marrow (lanes 11-13). (C) Western blot analysis of lysates from bone marrow from patients with ALL and AML at diagnosis (lanes 2, 5, and 8), remission (lanes 3 and 7), and relapse (lane 4); lanes 2-4 are results from the same patient with ALL at diagnosis, remission, and relapse, respectively. Lanes 6 and 7 are results from the same patient at diagnosis and remission, respectively. (D) Bone marrow from 9 patients without leukemia was obtained and isolated as described above. Lane 1 shows results with the positive (plus sign) control (32Dcl3 cell lysates). All lanes had equal protein loading on Ponceau S staining (data not shown). Densitometry measurements showed that the CREB-to-actin ratio ranged from 0.1 to 3.0 and the fold difference between leukemia and nonleukemia samples ranged from 3.5 fold to 10 fold. (E) Immunohistochemical analysis with CREB antiserum (10 μg/mL) was done on bone marrow biopsy specimens from (E) a representative patient with ALL and (F) a normal lymph node (× 40 magnification). Nuclear staining of CREB was observed in leukemia blast cells but not in normal lymphocytes. Staining was not detected with rabbit IgG (data not shown).
Expression of CREB in leukemia cell lines and bone marrow samples from patients with acute leukemia.
Shown are results of Western blot analyses of cell lines and of bone marrow specimens from patients with and without leukemia. (A) Leukemia and nonleukemia cell lines were assessed for CREB expression by Western blot analysis. Cells were grown to a density of 1 × 106 cells/mL, and lysates were prepared from 1 × 106 cells. Twenty micrograms of protein from cell lysates was loaded on a 10% sodium dodecyl sulfate (SDS)–polyacrylamide gel. Blots were probed with anti-CREB antiserum (0.5 μg/mL), stripped, and reprobed with actin (60 μg/mL) as the loading control. The arrows represent CREB (43 kd) or the actin control (43 kd). (B) Mononuclear cells from bone marrow obtained from patients with leukemia or without active disease were processed with the Ficoll-Hypaque method. Lysates were prepared from 1 × 106 mononuclear cells. Twenty micrograms of protein was loaded on a 10% SDS-polyacrylamide gel. Western blot analysis was done with anti-CREB antiserum. Shown are results with positive controls (32Dcl3 cell lysates; lane 1), results with the AML (lanes 2-5) and ALL (lanes 6-7) samples, and results with samples from patients without active leukemia or with normal bone marrow (lanes 11-13). (C) Western blot analysis of lysates from bone marrow from patients with ALL and AML at diagnosis (lanes 2, 5, and 8), remission (lanes 3 and 7), and relapse (lane 4); lanes 2-4 are results from the same patient with ALL at diagnosis, remission, and relapse, respectively. Lanes 6 and 7 are results from the same patient at diagnosis and remission, respectively. (D) Bone marrow from 9 patients without leukemia was obtained and isolated as described above. Lane 1 shows results with the positive (plus sign) control (32Dcl3 cell lysates). All lanes had equal protein loading on Ponceau S staining (data not shown). Densitometry measurements showed that the CREB-to-actin ratio ranged from 0.1 to 3.0 and the fold difference between leukemia and nonleukemia samples ranged from 3.5 fold to 10 fold. (E) Immunohistochemical analysis with CREB antiserum (10 μg/mL) was done on bone marrow biopsy specimens from (E) a representative patient with ALL and (F) a normal lymph node (× 40 magnification). Nuclear staining of CREB was observed in leukemia blast cells but not in normal lymphocytes. Staining was not detected with rabbit IgG (data not shown).
To assess CREB protein levels in patients with acute leukemia, we conducted Western blot analysis with cell lysates prepared from bone marrow samples. We observed CREB expression in bone marrow samples from 16 of 20 patients with ALL (80%) and 17 of 26 with AML (65%) (Table1). Figure 1B shows a representative Western blot of lysates from ALL or AML patients with expression of CREB. In contrast, CREB expression was detected in only 1 of 25 patients without active leukemia (4%; Figure 1B and 1D). The increased frequency of CREB expression in leukemias compared to controls was statistically significant (P < .001 for both AML and ALL by Fisher exact test). Thus, CREB was expressed at detectable levels more frequently in bone marrow from patients with acute leukemia. The one patient in the nonleukemia group with CREB expression had ITP. We also immunoblotted membranes with activating transcription factor (ATF)–1 antibody. Both CREB and ATF-1 are members of the CREB-ATF family of transcription factors.1 ATF-1 was not expressed in bone marrow from patients with leukemia, suggesting that CREB expression was specific and not due to cross-reactivity with a related protein family member (data not shown).
CREB expression in bone marrow samples studied
| Diagnosis . | Positive for CREB (no. of patients) . | Negative for CREB (no. of patients) . |
|---|---|---|
| Leukemia | ||
| AML | 17 | 9 |
| ALL | 16 | 4 |
| No leukemia | ||
| AML remission* | 0 | 4 |
| ALL remission† | 0 | 9 |
| Neutropenia | 0 | 4 |
| ITP | 1 | 3 |
| Lymphoma | 0 | 2 |
| Healthy | 0 | 2 |
| Diagnosis . | Positive for CREB (no. of patients) . | Negative for CREB (no. of patients) . |
|---|---|---|
| Leukemia | ||
| AML | 17 | 9 |
| ALL | 16 | 4 |
| No leukemia | ||
| AML remission* | 0 | 4 |
| ALL remission† | 0 | 9 |
| Neutropenia | 0 | 4 |
| ITP | 1 | 3 |
| Lymphoma | 0 | 2 |
| Healthy | 0 | 2 |
CREB indicates cyclic adenosine monophosphate response-element binding protein; AML, acute myeloid leukemia; ALL, acute lymphoblastic leukemia; and ITP, idiopathic thrombocytopenic purpura.
Includes remission samples from 2 patients with AML who were positive for CREB at diagnosis.
Includes remission samples from 3 patients with ALL who were positive for CREB at diagnosis.
To assess CREB levels during therapy, we conducted Western blot analysis with lysates from the same patients at diagnosis, remission, and relapse. In 2 patients with ALL, CREB expression was elevated at diagnosis but below detectable levels at remission (Figure 1C, lanes 2-7). In one of these patients, CREB expression was observed again at relapse, although levels were lower than those at diagnosis (Figure 1C, lane 4). In one patient with AML, CREB expression was also increased at relapse but not during remission (data not shown).
To determine whether CREB was expressed specifically in the blast cells from patients with acute leukemia, bone marrow biopsy specimens were analyzed by immunohistochemistry. A representative biopsy sample (Figure 1E,F) showed nuclear staining with CREB antiserum in more than 90% of lymphoblasts in the bone marrow but less frequently in normal lymphocytes in the lymph node. Control antibody (rabbit IgG) did not show CREB expression (data not shown).
CREB has been reported to play a role in the proliferation, differentiation, and survival of cells. Antisense oligonucleotides to CREB were found to induce death of human leukemia cells and bone marrow cells from patients with AML and chronic myeloid leukemia.14 In the current study, we did not observe translocations involving the CREB locus at 2q32.3-q34, except in one ALL patient with t(12;2) who also had positive CREB results.15 This finding suggests that other mechanisms are involved in increased CREB expression in acute leukemia. Understanding the role of CREB in leukemogenesis will provide new insights into how transcription factors regulate cell proliferation during normal and neoplastic hematopoiesis.
We thank Richard Pan and Phuong Kim Vu for technical assistance and Jorge Vargas, Patricia Mora-Garcia, Deepa Shankar, and Mike Lin for critical reading of the manuscript.
Supported by National Institutes of Health (NIH), National Cancer Institute (NCI), grant CA68221; the Leukemia and Lymphoma Society of America; and American Cancer Society grant RPG-99-081-01-LBC (K.M.S.). H.N.C.-V. is supported by NIH Clinical and Fundamental Immunology Training Grant AI07126-25. E.M.L. is supported in part by NCI grant CA-16042.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Kathleen M. Sakamoto, Department of Pediatrics, Mattel Children's Hospital at UCLA, 10833 LeConte Ave, Los Angeles, CA 90095-1752; e-mail: kms@ucla.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal