Recently we have identified subgroups of de novo primary diffuse large B-cell lymphoma (DLBCL) based on complementary DNA microarray-generated gene expression profiles. To correlate the gene expression profiles with cytogenetic abnormalities in these DLBCLs, we examined the occurrence of the t(14;18)(q32;q21) in the 2 distinctive subgroups of DLBCL: one with the germinal center B-cell gene expression signature and the other with the activated B cell–like gene expression signature. The t(14;18) was detected in 7 of 35 cases (20%). All 7 t(14;18)-positive cases had a germinal center B-cell gene expression profile, representing 35% of the cases in this subgroup, and 6 of these 7 cases had very similar gene expression profiles. The expression of bcl-2 and bcl-6 proteins was not significantly different between the t(14;18)-positive and -negative cases, whereas CD10 was detected only in the group with the germinal center B-cell expression profile, and CD10 was most frequently expressed in the t(14;18)-positive cases. This study supports the validity of subdividing DLBCL into 2 major subgroups by gene expression profiling, with the t(14;18) being an important event in the pathogenesis of a subset of DLBCL arising from germinal center B cells. CD10 protein expression is useful in identifying cases of DLBCL with a germinal center B-cell gene expression profile and is often expressed in cases with the t(14;18).
Introduction
The t(14;18)(q32;q21) is believed to play a crucial role in the pathogenesis of follicular lymphoma because it deregulates the expression of the antiapoptotic geneBCL2 by bringing it into proximity of the immunoglobulin heavy chain gene enhancer.1-3 The t(14;18)(q32;q21), with its associated BCL2gene rearrangement, has also been detected in up to one third of cases of primary diffuse large B-cell lymphoma (DLBCL).4-8However, it is not known whether cases with the t(14;18) represent a unique subset of DLBCL or what role this translocation plays in their pathogenesis. We have recently shown that gene expression patterns can delineate 2 major subgroups of DLBCL.9 One subgroup has an expression profile resembling that of normal germinal center (GC) B cells, the GC B cell–like (GCBL) type. The other subgroup has a profile resembling that of peripheral blood B cells activated by mitogenic stimuli, the activated B cell–like (ABL) type. If the t(14;18) is the initiating pathogenic event in primary DLBCL, we hypothesized that these lymphomas will have a GC B-cell gene expression profile. Therefore, we examined our patients with DLBCL with gene expression data to determine the role of the t(14;18) in the pathobiology of DLBCL.
Patients, materials, and methods
Patient population, tumor samples, and gene expression profiling by complementary DNA microarray
We studied 35 of the 42 DLBCL tissue samples with gene expression profiles determined previously by complementary DNA (cDNA) microarray.9 Thirty-two cases were from the University of Nebraska Medical Center and 3 cases were from the Stanford University Medical Center. All cases have been reviewed by the pathologists to confirm the diagnosis and exclude the presence of a follicular lymphoma.
Detection of the t(14;18)(q32;q21) by fluorescence in situ hybridization
Of the original 42 cases of DLBCL previously studied by cDNA microarray, 35 cases were studied by fluorescence in situ hybridization (FISH). These included 32 cases from Nebraska and 3 cases (DLBCL 0037, 0040, and 0042) from Stanford. Formalin-fixed, paraffin-embedded tissue cores were used in 34 cases, whereas frozen sections were used in case DLBCL-0042. The stained sections of the 35 cases were examined to confirm the presence of DLBCL and to select areas for FISH analysis. For paraffin-embedded tissue, 3 or 4 tissue cores (0.6 mm in diameter) were taken directly from the marked areas using a tissue microarrayer (Beecher Instruments, Silver Spring, MD). The tissue cores were dewaxed in xylene followed by rehydration. They were then minced and digested at 37°C for 30 minutes using 100 μL of a 0.05% proteinase K solution in 0.05 M Tris-HCl, 0.01 M EDTA, and 0.01 M NaCl. The cell pellets were rinsed and resuspended in 50 to 100 μL phosphate-buffered saline (PBS). From each suspension, 3 to 5 μL was placed in a 12-mm round area on a hybridization slide and heated at 60°C in an oven for 10 to 30 minutes. Prior to hybridization, the slides were pretreated with a 0.05% pepsin solution at 37°C for 1 to 2 hours for paraffin tissue preparations, 30 minutes for frozen sections. They were postfixed in 0.95% formaldehyde/PBS solution with 0.45% MgCl2 for 5 minutes at room temperature and then rinsed in PBS and dehydrated in ethanol. The dual color probe, LSI IGH Spectrum Green/LSI BCL2 Spectrum Orange (Vysis, Downers Grove, IL) was used to detect the t(14;18)(q32;q21), and the CEP 18 Spectrum Aqua (Vysis) probe was used to confirm the chromosome 18 copy number. Hybridization was performed as recommended by the manufacturer using a HYBrite instrument (Vysis). The cells were counterstained using 0.01 mg/mL 4′,6-diamidino-2-phenylindole. Analysis was performed on an Olympus BX51 microscope equipped with appropriate filters, and the images were captured with CytoVision Image Capture software (Applied Imaging, Santa Clara, CA).
Detection of BCL2 gene rearrangement by polymerase chain reaction
Of 35 cased studied by FISH, 30 cases had high-molecular-weight DNA available for polymerase chain reaction (PCR) analysis. Amplification of the BCL2/JH translocation at the major breakpoint region (mbr) and the minor cluster region (mcr) was performed as described by Sharp et al10 using an OminiGene thermocycler (Hybaid, Middlesex, United Kingdom). Positive controls consisted of DNA extracted from the human B-cell lymphoma line RL-7 for the mbr of BCL2, and DHL-16 for the mcr of BCL2. Negative controls consisted of sterile water instead of DNA, and DNA from normal peripheral blood mononuclear cells obtained from healthy volunteer donors. Standard precautions were taken to guard against PCR contamination, and amplified materials were analyzed in a laboratory physically separated from the one where unamplified materials were handled.11
Immunohistochemistry for CD10, bcl-2, and bcl-6
Formalin-fixed, paraffin-embedded 5-μm sections were stained with the avidin-biotin-peroxidase complex (ABC) method.12 Antigen retrieval was performed by incubation for 30 to 60 minutes in 10 mM citrate buffer (pH 6.0) in a 95°C hot water bath followed by 20 minutes of cooling at room temperature (32 cases from Nebraska), or using EDTA buffer at pH 8.0 for CD10 and bcl-6 (3 cases from Stanford). Antibodies to the following antigens were used: CD10 (clone 56C6, Ventana, Tucson, AZ, for cases from Nebraska; or Novocastra, Newcastle upon Tyne, United Kingdom, for cases from Stanford), bcl-2 (clone 24; Dako, Carpinteria, CA), and bcl-6 (polyclonal, Santa Cruz Biotechnology, Santa Cruz, CA, for cases from Nebraska; or PG-B6p, Dako, for cases from Stanford). A tumor was considered to be positive for expression of a specific antigen if more than 10% of the tumor cells showed appropriate immunoreactivity.
Statistical analysis
The Fisher exact test was used to compare the frequencies of the t(14;18) in the GCBL and ABL subgroups of DLBCL. This test was also used to compare the frequencies of CD10, bcl-2, and bcl-6 protein expression in the t(14;18)-positive and -negative subgroups of DLBCL. The survival analysis was performed on cases with and without t(14;18) using log-rank test. The Student t test was used to compare the microarray measurements of gene expression levels between the t(14;18)-positive and -negative subgroups of DLBCL. Genes differentially expressed between the 2 groups were selected with a cutoff of P < .05. The ratio of specific gene expression in the t(14;18)-positive cases versus the t(14;18)-negative cases of the GCBL subgroup was calculated by dividing the mean fluorescence intensity of the t(14;18)-positive by the mean fluorescence intensity of the t(14;18)-negative GCBL cases. Agglomerative hierarchical clustering was performed using the CLUSTER program (M. Eisen:http://www.microarrays.org/software).13
Results
Occurrence of the t(14;18) in DLBCL
To define the subset of DLBCL with the t(14;18), we applied interphase FISH to nuclei isolated from tumor tissues (Figure1). Of the 35 cases studied, a t(14;18) was detected in 7 of 20 cases (35%) of GCBL-DLBCL but in none of 15 cases of ABL-DLBCL (P = .01, Fisher exact test; Figure2). Using PCR, we foundBCL2/JH gene rearrangements in 3 of 5 t(14;18)-positive cases by FISH, which is similar to our detection rate in follicular lymphoma. None of the t(14;18)-negative cases by FISH had a BCL2/JH rearrangement by PCR.
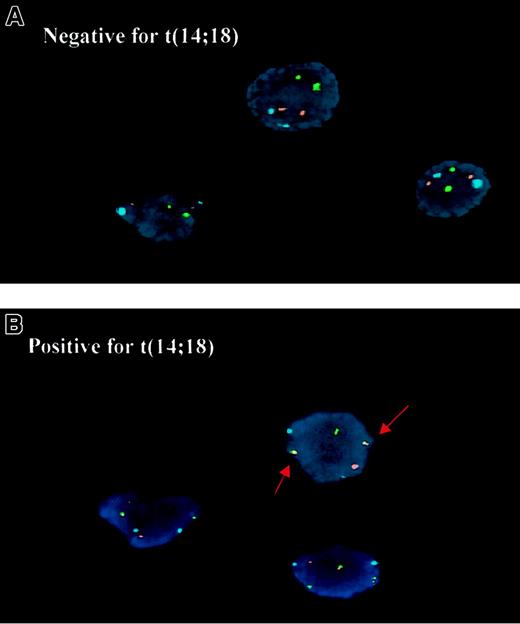
Detection of the t(14;18) by interphase FISH.
(A) The t(14;18)-negative cells show one hybridization signal in the IgH region on each chromosome 14 (green), each BCL2 region on chromosome 18 (red), and the centromere of chromosome 18 (aqua). (B) The t(14;18)-positive cells show 2 abnormal yellow fusion signals.
Detection of the t(14;18) by interphase FISH.
(A) The t(14;18)-negative cells show one hybridization signal in the IgH region on each chromosome 14 (green), each BCL2 region on chromosome 18 (red), and the centromere of chromosome 18 (aqua). (B) The t(14;18)-positive cells show 2 abnormal yellow fusion signals.
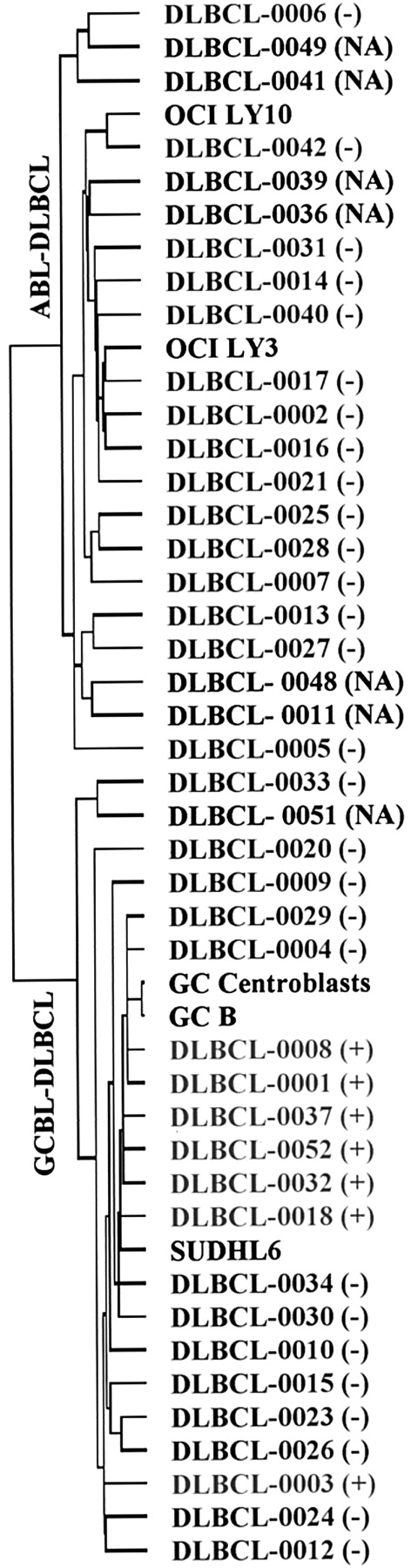
Dendrogram showing the distribution of t(14;18)-positive cases of DLBCL (red).
This hierarchical clustering dendrogram was generated based on the genes in the GC B-cell gene expression signature previously described by Alizadeh et al.9 The positive and negative results of FISH for the t(14;18) are indicated as (+) or (−), whereas (NA) indicates that no material was available for FISH study.
Dendrogram showing the distribution of t(14;18)-positive cases of DLBCL (red).
This hierarchical clustering dendrogram was generated based on the genes in the GC B-cell gene expression signature previously described by Alizadeh et al.9 The positive and negative results of FISH for the t(14;18) are indicated as (+) or (−), whereas (NA) indicates that no material was available for FISH study.
Patients with the t(14;18) included 6 women and 1 man ranging from 59 to 81 years of age at the time of diagnosis. There was no significant difference in overall survival when these 7 patients were compared with the t(14; 18)-negative patients in the entire group (P = .35) or in the GCBL group (P = .99), but the number of cases studied is small.
Bcl-2, bcl-6, and CD10 protein expression
To further characterize the cases, we performed immunohistochemistry to determine the expression of bcl-2, bcl-6, CD10, and CD20 proteins. These results are summarized in Table1. The frequencies of bcl-2 protein expression were not significantly different between the GCBL (15 of 20, 75%) and ABL (10 of 15, 67%) subgroups of DLBCL (P = .46). Four of 7 t(14;18)-positive cases (57%) and 21 of 28 t(14;18)-negative cases (75%) of DLBCL showed bcl-2 protein expression in the tumor cells. This finding indicates that the t(14;18) does not always lead to bcl-2 protein expression and that bcl-2 expression is often present in DLBCL in the absence of the t(14;18). Expression of bcl-6 protein was detected in 30 of 35 cases (86%) of DLBCL and showed no significant association with any subgroup. However, CD10 protein was detected in 11 of 20 cases (55%) in the GCBL subgroup, including 6 of 7 with the t(14;18) (86%), and none in the ABL subgroup (P < .001). Protein expression data for the 7 cases with the t(14;18) is shown in Table2. All 7 cases with the t(14;18) showed expression of bcl-6 protein, but not always detectable bcl-2 or CD10 protein by immunohistochemistry.
Expression of bcl-2, bcl-6, and CD10 proteins in subgroups of DLBCL
| Protein . | GCBL . | P* . | ABL . | P† . | ||
|---|---|---|---|---|---|---|
| t(14; 18)+ . | t(14; 18)− . | Subtotal . | ||||
| Bcl-2 | 4/7 | 11/13 | 15/20 (75%) | .29 | 10/15 (67%) | .46 |
| Bcl-6 | 7/7 | 12/13 | 19/20 (85%) | 1.00 | 11/15 (73%) | .14 |
| CD10 | 6/7 | 5/13 | 11/20 (55%) | .07 | 0/15 (0%) | < .001 |
| Protein . | GCBL . | P* . | ABL . | P† . | ||
|---|---|---|---|---|---|---|
| t(14; 18)+ . | t(14; 18)− . | Subtotal . | ||||
| Bcl-2 | 4/7 | 11/13 | 15/20 (75%) | .29 | 10/15 (67%) | .46 |
| Bcl-6 | 7/7 | 12/13 | 19/20 (85%) | 1.00 | 11/15 (73%) | .14 |
| CD10 | 6/7 | 5/13 | 11/20 (55%) | .07 | 0/15 (0%) | < .001 |
Probability based on Fisher exact test for comparisons between t(14; 18)-positive and t(14; 18)-negative cases within GCBL subgroup.
Probability based on Fisher exact test for comparisons between GCBL and ABL.
Expression of bcl-2, bcl-6, and CD10 proteins in the cases of DLBCL with the t(14; 18)
| Case . | bcl-2 . | bcl-6 . | CD10 . |
|---|---|---|---|
| 0001 | Pos | Pos | Pos |
| 0003 | Neg | Pos | Neg |
| 0008 | Pos | Pos | Pos |
| 0018 | Pos | Pos | Pos |
| 0032 | Neg | Pos | Pos |
| 0037 | Pos | Pos | Pos |
| 0052 | Neg | Pos | Pos |
| Case . | bcl-2 . | bcl-6 . | CD10 . |
|---|---|---|---|
| 0001 | Pos | Pos | Pos |
| 0003 | Neg | Pos | Neg |
| 0008 | Pos | Pos | Pos |
| 0018 | Pos | Pos | Pos |
| 0032 | Neg | Pos | Pos |
| 0037 | Pos | Pos | Pos |
| 0052 | Neg | Pos | Pos |
Pos indicates positive; neg, negative.
Gene expression profile associated with the t(14;18) in DLBCL
All 7 cases with the t(14;18) by FISH had the GC gene expression signature. Six of these cases, all positive for CD10 protein expression, had very similar gene expression patterns and formed a tight cluster together with normal GC B cells and the t(14;18)-positive cell line SUDHL-6 in the dendrogram (Figure 2). However, one t(14;18)-positive case (DLCL-0003), which was negative for CD10 and bcl-2 protein expression, was located outside of this tight cluster, indicating a substantial difference in its overall gene expression pattern. BCL2 gene expression was quite variable and generally low in GCBL-DLBCL as compared to the reference pool when hybridization to clones 1336385 and 342181 were examined (Figure3). When the sequences of the 3 different bcl-2 cDNA clones (232714, 342181, 1336385 ) used for microarray analysis were examined, only clone 232714 was able to consistently detect the bcl-2 transcripts in t(14;18)-positive lymphomas and the t(14;18) lymphoma cell line SUDHL-6. This clone contained a cDNA fragment from the middle portion of BCL2 gene and can hybridize to truncated transcripts from cases with bcl-2 translocation at the mbr. When we examined BCL2 gene expression as indicated by cDNA clone 232714, we found that 6 of 7 t(14;18)-positive DLBCL cases show overexpression (the exception being the outlying case DLCL-0003). The case (DLCL-0018) with translocation at the mcr showed concordant results with all the clones. A group of t(14;18)-negative cases in both the GCBL and ABL groups also had high levels of bcl-2 transcripts, which are detected by all 3 cDNA clones as expected.
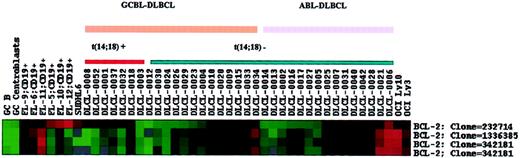
BCL2 gene expression in DLBCL.
Each horizontal row represents a separate cDNA clone on the microarray and each vertical column a separate case. The results are represented as the ratios of the hybridization signals on each spot on the microarray generated by the experimental mRNA samples versus the reference mRNA sample. These ratios are a measure of the relative gene expression of the experimental sample versus the reference standard and are depicted according to the color scale shown at the bottom, from a fluorescence ratio of 0.25 to 4 (−2 to +2 in log-base 2 units). GC B and GC centroblasts indicate GC B cells and GC centroblasts from tonsil; FL, follicular lymphoma. Note that normal GC cells have very low levels of bcl-2 expression. All follicular lymphoma cases, the t(14;18)-positive SUDHL-6 cell line, and 6 of the 7 DLBCL cases with the t(14;18) translocation have overexpression of bcl-2 when assessed with clone 232714, which detects bcl-2 mRNA truncated at the 3′ end due to breakage at the mbr. The case DLCL-0018 has a breakpoint at the mcr by PCR assay and the intact message is detectable with all cDNA clones. The t(14;18)-negative DLBCLs that overexpress bcl-2 have bcl-2 mRNA detectable by clones other than 232714, indicating that the message is not truncated.
BCL2 gene expression in DLBCL.
Each horizontal row represents a separate cDNA clone on the microarray and each vertical column a separate case. The results are represented as the ratios of the hybridization signals on each spot on the microarray generated by the experimental mRNA samples versus the reference mRNA sample. These ratios are a measure of the relative gene expression of the experimental sample versus the reference standard and are depicted according to the color scale shown at the bottom, from a fluorescence ratio of 0.25 to 4 (−2 to +2 in log-base 2 units). GC B and GC centroblasts indicate GC B cells and GC centroblasts from tonsil; FL, follicular lymphoma. Note that normal GC cells have very low levels of bcl-2 expression. All follicular lymphoma cases, the t(14;18)-positive SUDHL-6 cell line, and 6 of the 7 DLBCL cases with the t(14;18) translocation have overexpression of bcl-2 when assessed with clone 232714, which detects bcl-2 mRNA truncated at the 3′ end due to breakage at the mbr. The case DLCL-0018 has a breakpoint at the mcr by PCR assay and the intact message is detectable with all cDNA clones. The t(14;18)-negative DLBCLs that overexpress bcl-2 have bcl-2 mRNA detectable by clones other than 232714, indicating that the message is not truncated.
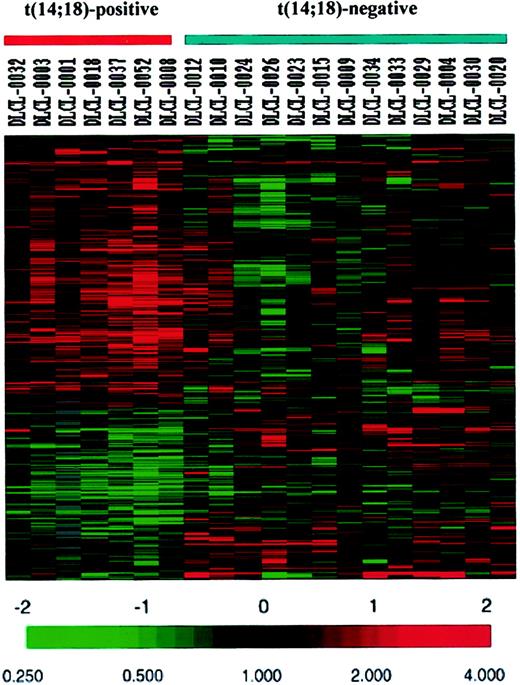
To search further for genes differentiating the t(14;18)-positive from t(14;18)-negative cases of GCBL-DLBCL, all genes on the Lymphochip (4026 clones) including GC B cell-associated genes were compared between the 7 cases with the t(14;18) and the 13 cases of GCBL-DLBCL without the t(14;18). We identified genes or expressed sequence tags (ESTs) represented by 403 cDNA clones showing significant differential expression (P < .05) using the 2-tailed Student t test. Genes or ESTs represented by 251 clones were expressed at higher levels in the t(14;18)-positive cases, whereas other genes or ESTs represented by 152 clones were expressed at lower levels in the t(14;18)-positive cases (Figure4). The 10 most differentially expressed genes are listed in Table 3.
Genes and ESTs represented by 403 clones showing differential expression between the t(14;18)-positive and t(14;18)-negative cases of GCBL-DLBCL.
Each horizontal row represents a separate cDNA clone on the microarray and each vertical column a separate case. The results are represented as the ratios of the hybridization signals on each spot on the microarray generated by the experimental mRNA samples versus the reference mRNA sample. These ratios are a measure of the relative gene expression of the experimental sample versus the reference standard and are depicted according to the color scale shown at the bottom. As indicated, the scale extends from a fluorescence ratio of 0.25 to 4 (−2 to +2 in log-base 2 units).
Genes and ESTs represented by 403 clones showing differential expression between the t(14;18)-positive and t(14;18)-negative cases of GCBL-DLBCL.
Each horizontal row represents a separate cDNA clone on the microarray and each vertical column a separate case. The results are represented as the ratios of the hybridization signals on each spot on the microarray generated by the experimental mRNA samples versus the reference mRNA sample. These ratios are a measure of the relative gene expression of the experimental sample versus the reference standard and are depicted according to the color scale shown at the bottom. As indicated, the scale extends from a fluorescence ratio of 0.25 to 4 (−2 to +2 in log-base 2 units).
The 10 genes most differentially expressed between the t(14; 18)-positive and t(14; 18)-negative cases of GCBL-DLBCL
| Name of gene . | Ratio3-150 . | P3-151 . |
|---|---|---|
| Similar to ribosomal protein S5; clone = 1241660 | 2.4 | .0001 |
| Protein phosphatase 2, regulatory subunit A (PR 65), beta isoform; clone = 626746 | 2.1 | .0001 |
| EWS = RNA binding protein; clone = 1192958 | 1.4 | .0001 |
| Unknown; clone = 1241826 | 1.8 | .0002 |
| Unknown; clone = 1241577 | 1.7 | .0002 |
| Lactate dehydrogenase B; clone = 624760 | 1.9 | .0002 |
| MP (inosine monophosphate) dehydrogenase 2; clone = 292008 | 2.2 | .0003 |
| Ribosomal protein L27; clone = 272185 | 1.9 | .0003 |
| Unknown; clone = 1340942 | 0.8 | .0004 |
| Unknown; clone = 1186027 | 0.7 | .0005 |
| Name of gene . | Ratio3-150 . | P3-151 . |
|---|---|---|
| Similar to ribosomal protein S5; clone = 1241660 | 2.4 | .0001 |
| Protein phosphatase 2, regulatory subunit A (PR 65), beta isoform; clone = 626746 | 2.1 | .0001 |
| EWS = RNA binding protein; clone = 1192958 | 1.4 | .0001 |
| Unknown; clone = 1241826 | 1.8 | .0002 |
| Unknown; clone = 1241577 | 1.7 | .0002 |
| Lactate dehydrogenase B; clone = 624760 | 1.9 | .0002 |
| MP (inosine monophosphate) dehydrogenase 2; clone = 292008 | 2.2 | .0003 |
| Ribosomal protein L27; clone = 272185 | 1.9 | .0003 |
| Unknown; clone = 1340942 | 0.8 | .0004 |
| Unknown; clone = 1186027 | 0.7 | .0005 |
Ratio of mean levels of mRNA for each cDNA clone between the t(14; 18)-positive and -negative cases.
Probability based on Student t test.
Discussion
The t(14;18)(q32;q21) translocation is a characteristic feature of follicular lymphoma and is thought to be the initiating event in lymphomagenesis.14,15 By analogy, DLBCL with the same translocation may be related to follicular lymphoma and the t(14;18) may also be important in its pathogenesis. In this study, we looked for the presence of the t(14;18) in a series of DLBCL using 2 techniques, FISH and PCR. The FISH assay has a higher sensitivity and detected the t(14;18) in 20% (7 of 35 cases) of DLBCL. Interestingly, these cases occurred exclusively in the GCBL subgroup, wherein the frequency was 35% (7 of 20 cases). Six of these 7 cases clustered closely with each other, and with normal GC cells and the t(14;18)-positive lymphoma cell line SUDHL-616 in the dendrogram, indicating very closely related gene expression profiles. The fact that these 6 cases shared a unique translocation and had gene expression profiles that were very similar to those of normal GC cells provides strong evidence that DLBCL with the t(14;18) is a distinctive subset within the GCBL subgroup. This observation also supports the validity of subdividing DLBCL into the GCBL and ABL subgroups, and indicates that tumors sharing an important genetic abnormality may also share a distinctive gene expression profile.
We also examined the messenger RNA (mRNA) and protein expression patterns of bcl-6 and CD10 because of their known association with normal and neoplastic GC B cells. The bcl-6 protein was expressed in all cases with the t(14;18), but it was also expressed in a high percentage of the remaining cases. CD10 was highly expressed at the mRNA level and frequently detected at the protein level in GCBL-DLBCL, but none of the cases of the ABL subgroup expressed CD10 by protein immunohistochemistry. These results suggest that CD10 protein expression is a specific, but not a very sensitive, marker for GCBL-DLBCL. Moreover, CD10 protein expression was demonstrated in a high percentage (86%) of the t(14;18)-positive cases.
Some of our t(14;18)-positive DLBCLs did not show detectable bcl-2 protein expression by immunohistochemistry, in keeping with previously published reports. Kramer et al17 reported that 12 of 25 cases of DLBCL with BCL2 gene rearrangement had low or absent expression of bcl-2 protein. In the study reported by Tang et al,18 only 2 of 15 cases of DLBCL with theBCL2 gene rearrangement were found to have a high level of bcl-2 protein expression. It is possible that bcl-2 expression is essential early in lymphomagenesis, but additional genetic abnormalities developing during tumor progression may abrogate the requirement for bcl-2. The mechanisms for dissociation betweenBCL2 gene rearrangement and bcl-2 protein expression are unclear. At the transcription level, bcl-2 expression was quite variable and showed no dependence on the presence of t(14;18). In t(14;18)-positive cases of DLBCL, bcl-2 transcripts were up-regulated in 6 of 7 cases with a probe that can detect bcl-2 mRNA in cases withBCL2 translocation at the mbr. Case DLCL-00003, which was negative for both bcl-2 and CD10 expression, showed a very low or undetectable level of bcl-2 transcripts with all cDNA clones. A group of t(14;18)-negative cases in both GCBL and ABL groups showed high levels of bcl-2 transcripts detected by all cDNA clones, indicating that alternative mechanisms up-regulating bcl-2 expression are operative in these cases. A set of genes differentially expressed between GCBL-DLBCL with and without the t(14;18) can be identified by statistical analysis (Figure 4 and Table 3). However, due to the small number of cases compared in our study, these results are considered preliminary and further analysis in a larger study is necessary to identify differentially expressed genes at a high level of confidence.
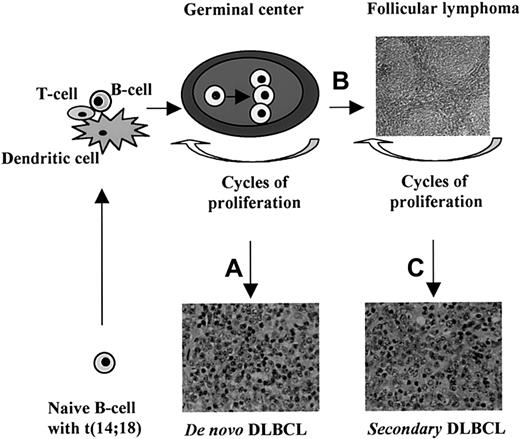
How is primary DLBCL with a t(14;18) different from follicular lymphoma that typically has a t(14;18)? In this study, the t(14;18)-positive cases of DLBCL were carefully reviewed and no follicular structures were seen morphologically or by immunostaining for follicular dendritic cells. Furthermore, none of these patients had a prior history of follicular lymphoma. Therefore, these cases are not follicular lymphomas misdiagnosed as DLBCL. The fact that primary DLBCLs with the t(14;18) have gene expression profiles similar to normal GC B cells suggests that the t(14;18) plays an important role in the pathogenesis of this subset of DLBCL, just as it does in follicular lymphoma. The t(14;18) is thought to occur as a low-frequency error duringIGH gene rearrangement in normal B-cell maturation, resulting in abnormal bcl-2 overexpression, which allows the cells to escape apoptosis if they become GC B cells. However, the t(14;18) is not sufficient for neoplastic transformation. In fact, rare cells with the t(14;18) are detectable in tonsils, lymph nodes with follicular hyperplasia, and blood lymphocytes from healthy individuals.19 20 These GC B cells harboring the t(14;18) are able to undergo repeated cycles of clonal expansion when the appropriate antigen is encountered. Extensive proliferation introduces additional genetic changes that lead to neoplastic transformation. The different genetic abnormalities occurring during this phase may lead to follicular lymphoma or primary DLBCL. We illustrate this concept in Figure 5.
Concept of the cellular origin of t(14;18)-positive DLBCL.
The t(14;18) occurs as a rare random event in naive B cells, probably during the D/J rearrangement of the IGH gene. Such a B cell may encounter the appropriate antigen in the context of dendritic cells and T cells in the paracortical region of a lymph node. The stimulated B cell then migrates to and proliferates in a GC. Overexpression of bcl-2 protein protects the cell and its progeny from apoptosis even when the affinity of the antibody is not very high. Repeated cycles of proliferation from subsequent antigenic encounters expand this clone and result in additional genetic abnormalities. Different secondary genetic alterations lead to the development of a primary DLBCL (A) or a follicular lymphoma (B). Further genetic changes occurring during the course of a follicular lymphoma will give rise to a secondary DLBCL in some cases (C). A number of genes may be able to initiate each of these pathways and the gene expression profile may be unique and distinguishable for each.
Concept of the cellular origin of t(14;18)-positive DLBCL.
The t(14;18) occurs as a rare random event in naive B cells, probably during the D/J rearrangement of the IGH gene. Such a B cell may encounter the appropriate antigen in the context of dendritic cells and T cells in the paracortical region of a lymph node. The stimulated B cell then migrates to and proliferates in a GC. Overexpression of bcl-2 protein protects the cell and its progeny from apoptosis even when the affinity of the antibody is not very high. Repeated cycles of proliferation from subsequent antigenic encounters expand this clone and result in additional genetic abnormalities. Different secondary genetic alterations lead to the development of a primary DLBCL (A) or a follicular lymphoma (B). Further genetic changes occurring during the course of a follicular lymphoma will give rise to a secondary DLBCL in some cases (C). A number of genes may be able to initiate each of these pathways and the gene expression profile may be unique and distinguishable for each.
It is likely that a number of different genetic lesions can result in the development of DLBCL from a t(14;18)-positive GC B cell, and each of these genetic alterations may be associated with a unique gene expression signature. One could speculate that the outlying case DLCL-0003 with the t(14;18) was associated with a different transformational event than the other 6 cases that clustered tightly together. We believe that genetic and gene expression analysis of additional cases will allow us to define the critical genetic events that lead to neoplastic transformation of GC B cells.
We thank Grant Wu and Greg Cochran for technical assistance.
Supported in part by grants U01-CA84967 and CA34233 from the National Cancer Institute and grant 6605-01 from the Leukemia and Lymphoma Society.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Wing C. Chan, Dept of Pathology and Microbiology, 983135 Nebraska Medical Center, Omaha, NE 68198; e-mail:jchan@unmc.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal