Abstract
Tumor necrosis factor–related apoptosis-inducing ligand (TRAIL, Apo2 ligand) effectively kills multiple myeloma (MM) cells in vitro irrespective of refractoriness to dexamethasone and chemotherapy. Because clinical trials with this anticancer agent are expected shortly, we investigated the signaling pathway of TRAIL-induced apoptosis in MM. We detected rapid cleavage of caspases-8, -9, -3, and -6, as well as the caspase substrates poly(ADP-ribose) polymerase (PARP) and DNA fragmentation factor-45 (DFF45), but not caspase-10, upon TRAIL treatment in sensitive MM cells, pointing to caspase-8 as the apical caspase of TRAIL signaling in MM cells. These phenomena were not observed or were significantly delayed in TRAIL-resistant MM cells, suggesting that resistance may arise from inhibition at the level of caspase-8 activation. Higher levels of expression for various apoptosis inhibitors, including FLICE-inhibitory protein (FLIP), and lower procaspase-8 levels were present in TRAIL-resistant cells and sensitivity was restored by the protein synthesis inhibitor cycloheximide (CHX) and the protein kinase C (PKC) inhibitor bisindolylmaleimide (BIM), which both lowered FLIP and cellular inhibitor of apoptosis protein-2 (cIAP-2) protein levels. Forced expression of procaspase-8 or FLIP antisense oligonucleotides also sensitized TRAIL-resistant cells to TRAIL. Moreover, the cell permeable nuclear factor (NF)–κB inhibitor SN50, which sensitizes TRAIL-resistant cells to TRAIL, also inhibited cIAP2 protein expression. Finally, CHX, BIM, and SN50 facilitated the cleavage and activation of procaspase-8 in TRAIL-resistant cells, confirming that inhibition of TRAIL-induced apoptosis occurs at this level and that these agents sensitize MM cells by relieving this block. Our data set a framework for the clinical use of approaches that sensitize MM cells to TRAIL by agents that inhibit FLIP and cIAP-2 expression or augment caspase-8 activity.
Introduction
Tumor necrosis factor (TNF)–related apoptosis-inducing ligand (TRAIL/Apo2L) is a member of the superfamily of cell death ligands, which also includes TNF-α and Fas ligand (FasL or CD95L).1,2 TRAIL induces apoptosis in tumor cells of diverse origins3-5 but not in normal cells in vitro1,2 or in experimental animals,6,7 even though the messenger RNAs for TRAIL and its receptors are expressed in a wide range of normal tissues.8 TRAIL induces apoptosis by cross-linking 1 of 2 apoptosis-signaling receptors, DR4 (or TRAIL-R1) and DR5 (or TRAIL-R2/KILLER/TRICK2). DR4 and DR5 receptors transduce the death signal via their 80-amino acid intracellular death domains (DDs), which recruit adaptor protein(s). The latter facilitate the binding and autocatalytic activation of caspases, generating a cascade of proteolytic activation of downstream caspases that culminates in irreversible commitment of cells to undergo apoptosis.9 Both caspase-8 and caspase-10 have been reported in different models to be the apical caspases of the TRAIL signaling pathway3,4,10,11 and in the case of caspase-8, Fas-associated death domain protein (FADD) was identified as the adaptor molecule, similarly to their interaction observed during Fas signaling. Two additional TRAIL receptors, TRAIL-R3/TRID/DcR1 and TRAIL-R4/TRUNDD/DcR2, lack a functional DD and cannot transduce a proapoptotic signal. They are therefore considered “decoy receptors,” competing with DR4 and DR5 for TRAIL binding. This markedly complex system of TRAIL receptors, and in particular the expression of decoy receptors, was initially postulated to provide the explanation for the specificity of the proapoptotic action of TRAIL against cancer cells,12 but not their normal cell counterparts.
Recently, we showed that TRAIL induces apoptosis of human multiple myeloma (MM) cell lines,13 as well as freshly obtained samples from patients with MM, and exhibits in vivo antimyeloma activity in nude mice xenografted with human plasmacytomas.14 TRAIL spares normal peripheral blood B cells and bone marrow mononuclear cells and imposes no significant adverse effect on hematopoietic progenitors.14Furthermore, TRAIL is active against dexamethasone- and cytotoxic drug- resistant MM cells.14 Importantly, doxorubicin, peptide inhibitors of nuclear translocation of nuclear factor (NF)–κB (SN50), and proteasome inhibitors can selectively sensitize MM cells, but not normal B cells, to TRAIL-induced apoptosis.14These data set the framework for the clinical use of TRAIL/Apo2L against MM.
In the present study, we investigated the intracellular signaling of TRAIL/Apo2L-induced apoptosis in MM cell lines and the mechanism(s) of resistance. We found that TRAIL activates caspase-8 and caspase-6, but not caspase-10, in sensitive MM cells. Caspase-9 and caspase-3 are also activated, but are not necessary for the execution of apoptosis. Resistance to TRAIL is due to lack of activation of caspase-8 and is associated with a low procaspase-8/FLICE-inhibitory protein (FLIP) ratio. Consistent with this, down-regulation of FLIP by antisense oligonucleotides or transfection of procaspase-8 sensitized resistant MM cells to TRAIL. The protein synthesis inhibitor cycloheximide (CHX) and the protein kinase C (PKC) inhibitor bisindolylmaleimide (BIM) down-regulated FLIP and cIAP-2 levels and sensitized MM cells to TRAIL. The cell-permeable NF-κB inhibitor SN50, which reverses TRAIL resistance of MM cells also down-regulated cellular inhibitor of apoptosis protein-2 (cIAP-2). These data delineate the intracellular signaling pathway of TRAIL/Apo2L-induced apoptosis in MM and suggest approaches to overcome potential clinical resistance.
Materials and methods
MM cell lines
Dexamethasone-sensitive MM-1S and dexamethasone-resistant MM-1R human MM cell lines were kindly provided by Dr Steven Rosen (Northwestern University, Chicago, IL). RPMI-8226/S cells and its doxorubicin (Dox40)–resistant sublines were a kind gift from Dr William Dalton (Lee Moffit Cancer Center, Tampa, FL). ARP-1 cells were kindly provided by Dr O. Barchinova (Boston University, Boston, MA). ARH-77, HS-Sultan, and IM-9 human cell lines were obtained from the American Type Culture Collection (Manassas, VA).
As we have previously shown,14 MM-1S, MM-1R, RPMI-8226/S, RPMI-Dox40, and ARP-1 cells are very sensitive to TRAIL/Apo2L-induced apoptosis (300 ng/mL for 18 hours induces > 60% apoptosis), whereas ARH-77, IM-9, and HS-Sultan cells exhibit only 15%, 3%, and 3% cell death, respectively, under the same conditions. Therefore, ARH-77 cells are deemed to be moderately and IM-9 and HS-Sultan cells highly resistant to TRAIL, and as such these cell lines were used in this study as a model of resistance to TRAIL/Apo2L. It should be noted, however, that this resistance is not complete, because prolonged exposure to higher concentrations of TRAIL leads to a modest loss of viability, even in IM-9 and HS-Sultan cells.
All cell lines were cultured in RPMI 1640 medium (Gibco Laboratories, Grand Island, NY) supplemented with 10% charcoal dextran-treated fetal bovine serum (FBS; Hyclone, Logan, UT) as well asl-glutamine, penicillin, and streptomycin (Gibco).
Materials
Recombinant human TRAIL was obtained from Immunex Corporation (Seattle, WA), in a leucine zipper (LZ) form that promotes and stabilizes the formation of trimers, as previously reported.6 Goat polyclonal antibodies for human DR4, DR5, DcR1, caspase-3, as well as mouse monoclonal antibodies for Bcl2, BclxL, and tubulin were obtained from Santa Cruz Biotechnology (Santa Cruz, CA); anti-FADD monoclonal antibody and polyclonal antisera against cIAP-1, cIAP-2, and XIAP from R & D Systems (Minneapolis, MN); polyclonal antiserum against survivin from Oncogene Research (Boston, MA); monoclonal antibody for caspase-8 and rabbit polyclonal antisera for caspase-10, caspase-6, and FLIP from Upstate Biotechnology (Lake Placid, NY); rabbit polyclonal antibody for caspase-10 from Research Diagnostics (Flanders, NJ); mouse MsIgG1 and MsIgG2b isotype controls from Beckman Coulter/Immunotech (Miami, FL); antihuman DcR2 rabbit polyclonal antibody from Imgenex (San Diego, CA); goat antimouse IgG fluorescein isothiocyanate (FITC)–conjugated F(ab′)2 fragment, donkey antigoat IgG FITC-conjugated F(ab′)2 fragment, and donkey antirabbit IgG FITC-conjugated F(ab′)2 fragment from Jackson Immunoresearch Laboratories (West Grove, PA); 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT), CHX, and BIM III from Sigma Chemical (St Louis, MO); the inhibitory peptide SN50 from Biomol (Plymouth Meeting, PA); complete-TM mixture of proteinase inhibitors, Ig-free normal horse serum, and sodium dodecyl sulfate (SDS) from Life Technologies (Gaithersburg, MD); and the enhanced chemiluminescence (ECL) kit, which includes the peroxidase-labeled antimouse and antirabbit secondary antibodies, from Amersham (Arlington Heights, IL).
Immunoblot analysis
Immunoblot analysis was performed as previously described.15 Briefly, cells were lysed for 30 minutes on ice in lysis buffer (50 mM Tris-HCl pH 8, with 120 mM NaCl and 1% NP-40) supplemented with the complete-TM mixture of proteinase inhibitors. The samples were cleared by microcentrifugation (14 000 rpm, 30 minutes, 4°C) and assessed for protein concentration. Thirty micrograms of the protein/sample were electrophoresed in a 12% SDS-polyacrylamide gel (SDS-PAGE), and electroblotted onto nitrocellulose membranes. After 1 hour incubation in blocking solution (20% IgG-free normal horse serum, in phosphate-buffered saline, PBS), the membranes were exposed overnight at 4°C to the primary antibody. Following washing in PBS, the respective secondary peroxidase-labeled antibody was applied at 1:10 000 dilution for 1 hour at room temperature. The proteins were visualized with the ECL technique.
Cleavage of caspases, poly(ADP-ribose) polymerase, and DNA fragmentation factor-45
The involvement of caspases in TRAIL-induced apoptosis in MM cells was studied by evaluating the levels of procaspases-8, -10, -3, -6, and -9 as well as the emergence of their cleaved active forms by immunoblotting in lysates of cells treated with 300 ng/mL TRAIL for 0.5, 1, and 5 hours. Also, the cleavage of 2 known targets of caspase activity, poly(ADP-ribose) polymerase (PARP) and DNA fragmentation factor-45 (DFF45), were also studied in the same lysates.
Caspase-3 activity assays
The activity of caspase-3 was monitored in MM cells following treatment with 300 ng/mL TRAIL for 5 hours with the Caspase-3 Intracellular Activity Assay kit (Calbiochem, La Jolla, CA), according to the manufacturer's instructions. The assay depends on the detection of the cleavage of the peptide PhiPhiLux, which is mediated by caspase-3. When the peptide is cleaved, it provides an intense fluorescent signal.
MTT colorimetric survival assays
The effect of TRAIL on MM cell survival was examined by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) colorimetric assay, as previously described.15 Cells were plated in 48-well plates at 70% to 80% confluence and then incubated for 18 hours with LZ-TRAIL (100-300 ng/mL), in 2% bovine serum–supplemented RPMI medium at 37°C. Prior to exposure to TRAIL, MM cells were preincubated with caspase inhibitors (pancaspase inhibitor ZVAD-FMK, caspase-8 inhibitor IETD-FMK, caspase-3 inhibitor DEVD-FMK, caspase-9 inhibitor LEHD-FMK, and caspase-6 inhibitor VEID-FMK; all purchased from Calbiochem, and used at 20 μM) for 1 hour, or, in other experiments, with cycloheximide (0.1 μg/mL) or BIM (20 μM) for 6 hours. All experiments were repeated at least 3 times and each experimental condition was repeated at least in quadruplicate wells in each experiment. At the end of each treatment, cells were incubated with 1 mg/mL MTT in fresh media for 4 hours at 37°C, and then a mixture of isopropanol and 1N HCl (23:2, vol/vol) was added under vigorous pipetting to dissolve the formazan crystals. Dye absorbance in viable cells was measured at 570 nm, with 630 nm as a reference wavelength. Cell survival was estimated as a percentage of the value of untreated controls.
Cloning and transfection of procaspase-8 into myeloma cells
The full-length human procaspase-8 sequence was amplified by polymerase chain reaction using an HT29 colon carcinoma expression library cloned into the pCDM8 vector and appropriate synthetic oligonucleotide primers, corresponding to sequences encoding the amino- and carboxy-terminal extremities of the precursor protein, and cloned into the mammalian expression vector pcDNA3.1-TOPO-V5-His (Invitrogen, Carlsbad, CA). The correct orientation and sequence were verified by restriction analysis and dideoxy sequencing, respectively. IM-9 cells were transfected with the procaspase-8 construct or the empty vector with the help of Superfect (Qiagen, Valencia, CA) or left untreated. Forty-eight hours later, the cells were treated overnight with either TRAIL (200 ng/mL) or vehicle. Cell survival was quantified with the MTT assay.
Transfection of FLIP antisense and control oligonucleotides
To delineate the role of FLIP as a negative regulator of TRAIL-induced apoptosis, we transfected the TRAIL-resistant line IM-9, which expresses high levels of FLIP with fully phosphorothioated single-stranded ASO directed against the human FLIP translation initiation codon (sequence, 5′-GACTTCAGCAGACATCCTAC-3′) or control phosphorothioate oligodeoxynucleotides (sequence, 5′-TGGATCCGACATGTCAGAGA-3′), as previously described by Perlman et al.16 IM-9 cells were plated in 24-well plates and transfected with the help of Oligofectamine (Gibco BRL, Gaithersburg, MD). Forty-eight hours later, TRAIL (200 ng/mL) was added to appropriate wells and the cells were incubated for an additional 18 hours. Cell death was quantified by MTT as described above.
Caspase-8 activity assay
The IM-9 cells were pretreated with CHX (0.1 μg/mL), BIM (20 μM), or SN50 (30 μg/mL) for 6 hours, and then stimulated with TRAIL (300 ng/mL) for 30 more minutes. The enzymatic activity of caspase-8 was measured with the ApoAlert caspase-8 colorimetric kit (Clontech, Palo Alto, CA), according to the manufacturer's instructions. Each experimental condition was performed both in the presence and absence of the caspase-8 inhibitor IETD-FMK (20 μM). The value obtained in the presence of IETD-FMK was subtracted from that obtained in its absence, to yield the caspase-8–specific activity of the sample.
Flow cytometric analysis for TRAIL receptor expression
The MM cell lines were characterized for their surface expression of TRAIL receptors by flow cytometry. Staining was performed as previously described.17 Briefly, for each cell line, 106 cells were incubated with the appropriate anti-TRAIL receptor antibody or a respective control (5.0 μg) for 45 minutes. Specifically, DR4 and DR5 expression was assessed with anti-DR4 and anti-DR5 monoclonal antibodies (mouse IgG1 and IgG2b as respective controls) and confirmed with goat antihuman DR4 and DR5 polyclonal antibodies, using goat IgG for control stainings. DcR1 and DcR2 expression was assessed with goat anti-DcR1 (or rabbit anti-DcR1) and goat anti-DcR2 polyclonal antibodies. Cells were then washed with PBS and incubated for 45 minutes with 2.0 μg goat antimouse IgG FITC-conjugated F(ab′)2 fragment for anti-DR4 and anti-DR5 monoclonal antibody phenotypic analyses or with a donkey antigoat IgG FITC-conjugated F(ab′)2 fragment for the DcR1, DcR2, DR4, and DR5 analyses with the goat polyclonal antibodies. Cells were then washed, fixed with 1% formaldehyde PBS, and analyzed on a EPICS-XL-MCL flow cytometer (Coulter, Hialeah, FL).
Statistical analysis
Statistical significance was examined by analysis of variance, followed by the Duncan posthoc test. P < .05 was considered consistent with statistical significance, in all analyses.
Results
Immunoblot analysis for activation of caspases and cleavage of PARP and DFF45 in myeloma cells exhibiting differential sensitivities to TRAIL
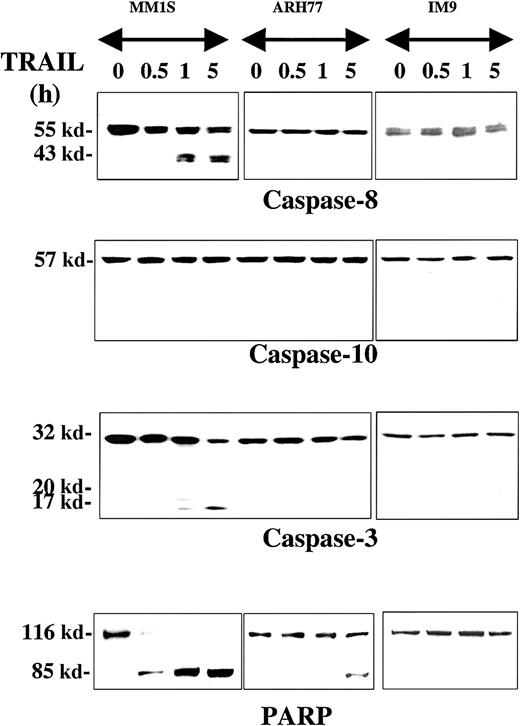
The proteolytic cleavage of caspase-8, -10, -9, and -3 and their substrates PARP and DFF45 were studied in TRAIL-sensitive (MM.1S), moderately resistant (ARH-77), and highly resistant (IM-9) cells that were treated with TRAIL (300 ng/mL) for 0.5, 1, and 5 hours. Rapid cleavage of procaspase-8 was detected in MM.1S cells (Figure1), but not in ARH-77 or IM-9 cells. Interestingly, procaspase-10 was not cleaved in any cell line throughout the duration of the experiment, contrary to our previous finding of caspase-10 cleavage during TRAIL-induced apoptosis of thyroid carcinoma cells3 using the same antibody from Upstate Biotechnology. To confirm this finding, we used another antibody against a different epitope of caspase-10, which was obtained from Research Diagnostics, and compared our findings in TRAIL-treated MM and thyroid carcinoma cells (SW579 line). Once again, caspase-10 cleavage was detected with the Research Diagnostics antibody in TRAIL-treated thyroid carcinoma cells, but not in MM.1S cells (data not shown), thus confirming the specificity of our findings. These results, therefore, suggest that caspase-8, but not caspase-10, mediates TRAIL signaling in MM cells, and that there are tissue-dependent differences in the TRAIL signaling pathway.
Immunoblot analysis for caspases and PARP in myeloma cells exhibiting differential sensitivities to TRAIL.
MM.1S, ARH-77, and IM-9 cells were treated with TRAIL (300 ng/mL) for 0.5, 1, and 5 hours. Rapid cleavage of caspase-8, caspase-3, and PARP was observed in the TRAIL-sensitive MM.1S cells. Due to lower caspase-8 protein levels in TRAIL-resistant cells, longer exposures were used to ensure adequate visualization of the bands. Caspase-10 was not cleaved, excluding it as an apical caspase in the TRAIL pathway for MM. In the moderately TRAIL-resistant ARH-77 cells, cleavage of caspases and PARP was significantly inhibited, though eventually low-level cleavage of caspase-3 (on longer exposure of the film) and PARP was seen at 5 hours of treatment. No cleavage of any caspases or PARP was detected in highly TRAIL-resistant IM-9 cells.
Immunoblot analysis for caspases and PARP in myeloma cells exhibiting differential sensitivities to TRAIL.
MM.1S, ARH-77, and IM-9 cells were treated with TRAIL (300 ng/mL) for 0.5, 1, and 5 hours. Rapid cleavage of caspase-8, caspase-3, and PARP was observed in the TRAIL-sensitive MM.1S cells. Due to lower caspase-8 protein levels in TRAIL-resistant cells, longer exposures were used to ensure adequate visualization of the bands. Caspase-10 was not cleaved, excluding it as an apical caspase in the TRAIL pathway for MM. In the moderately TRAIL-resistant ARH-77 cells, cleavage of caspases and PARP was significantly inhibited, though eventually low-level cleavage of caspase-3 (on longer exposure of the film) and PARP was seen at 5 hours of treatment. No cleavage of any caspases or PARP was detected in highly TRAIL-resistant IM-9 cells.
Caspase-3 was also cleaved in TRAIL-treated MM.1S cells, but not to any significant degree in ARH-77 and IM-9 cells. A weak band of cleaved caspase-3 in ARH-77 was, however, detected at 5 hours after treatment with TRAIL only after prolonged film exposure. The same pattern was found for the caspase substrate PARP, with rapid complete cleavage in TRAIL-sensitive MM.1S cells, weak delayed cleavage in ARH-77, and lack of cleavage in IM-9 cells (Figure 1).
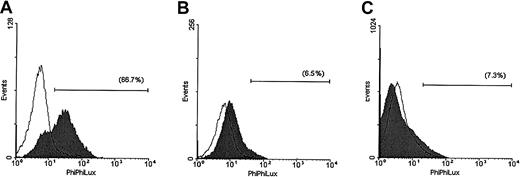
The enzymatic activity of caspase-3 was also evaluated in the same cell lines after 5 hours of TRAIL treatment (Figure2). These studies demonstrated that TRAIL induced a strong increase in the number of MM.1S cells that were positive for caspase-3 activity and still retained intact cellular membrane as evidenced by lack of propidium iodide uptake. In contrast, only minor responses were detected in TRAIL-treated ARH-77 and IM-9 cells.
Assessment of caspase-3 enzymatic activity in TRAIL-sensitive and -resistant cells.
Caspase-3 enzymatic activity was measured with a caspase-3 intracellular activity assay kit in TRAIL-sensitive (MM.1S, panel A), moderately resistant (ARH-77, panel B), and highly resistant (IM-9, panel C) cells treated with (shaded curves) or without (unshaded curves) TRAIL (300 ng/mL) for 5 hours. A large population of cells was found to be positive for caspase-3 activity in MM.1S, but not ARH-77 or IM-9 cells treated with TRAIL.
Assessment of caspase-3 enzymatic activity in TRAIL-sensitive and -resistant cells.
Caspase-3 enzymatic activity was measured with a caspase-3 intracellular activity assay kit in TRAIL-sensitive (MM.1S, panel A), moderately resistant (ARH-77, panel B), and highly resistant (IM-9, panel C) cells treated with (shaded curves) or without (unshaded curves) TRAIL (300 ng/mL) for 5 hours. A large population of cells was found to be positive for caspase-3 activity in MM.1S, but not ARH-77 or IM-9 cells treated with TRAIL.
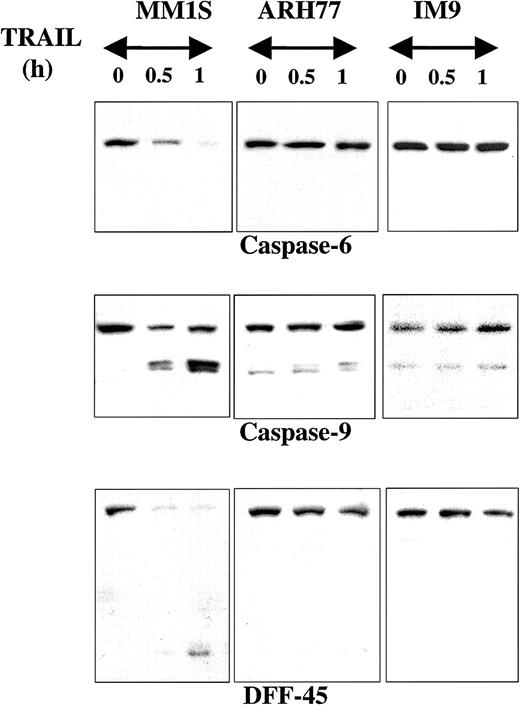
Caspase-6, another executioner caspase, was also rapidly cleaved in MM1.S but not in ARH-77 or IM-9 cells treated with TRAIL (Figure3). Caspase-9 similarly underwent rapid cleavage in MM.1S cells (Figure 3). This effect was not detected in ARH-77 or IM-9 cells, but both cell lines contained low but detectable levels of cleaved caspase-9. It is possible that these cells maintain their viability by quenching the activity of caspase-9 with the help of apoptosis inhibitors, potentially XIAP, which binds and inactivates caspase-9.18 DFF45, another caspase substrate, was also rapidly cleaved in MM.1S cells but not in ARH-77 or IM-9 cells (Figure3).
Immunoblot analysis for caspase-6, caspase-9, and DFF45 in myeloma cells exhibiting differential sensitivities to TRAIL.
Immunoblot analysis for caspase-6, caspase-9, and DFF45 was performed for TRAIL-sensitive (MM.1S), moderately resistant (ARH-77), and highly resistant (IM-9) cells treated with TRAIL (300 ng/mL) for 0.5 and 1 hour. Rapid cleavage of caspase-6, caspase-9, and DFF45 was observed in MM.1S, but not ARH-77 or IM-9 cells. A band corresponding to cleaved caspase-9 was constitutively present in ARH-77 or IM-9 cells, but remains unchanged after TRAIL treatment.
Immunoblot analysis for caspase-6, caspase-9, and DFF45 in myeloma cells exhibiting differential sensitivities to TRAIL.
Immunoblot analysis for caspase-6, caspase-9, and DFF45 was performed for TRAIL-sensitive (MM.1S), moderately resistant (ARH-77), and highly resistant (IM-9) cells treated with TRAIL (300 ng/mL) for 0.5 and 1 hour. Rapid cleavage of caspase-6, caspase-9, and DFF45 was observed in MM.1S, but not ARH-77 or IM-9 cells. A band corresponding to cleaved caspase-9 was constitutively present in ARH-77 or IM-9 cells, but remains unchanged after TRAIL treatment.
These data therefore suggest that in TRAIL-sensitive MM cells a rapid activation of the caspase cascade occurs following TRAIL treatment, and that caspase-8 likely functions as the apical caspase in TRAIL-induced signaling in MM cells. In contrast, caspase-8 cleavage and ensuing downstream events are significantly delayed or absent in TRAIL-resistant cells, thereby suggesting that a potential antiapoptotic block takes place at the level of caspase-8 activation.
Modulation of TRAIL-induced cytotoxicity in MM.1S myeloma cells by selective inhibition of caspases
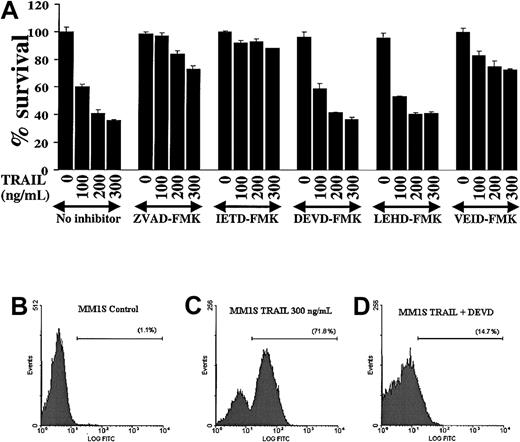
Because caspases are proteolytically cleaved and activated during TRAIL-induced apoptosis in MM cells, we next attempted to discern which caspases were indispensable for apoptotic signaling in MM. We therefore evaluated the effect of various caspase inhibitors (the pancaspase inhibitor ZVAD-FMK, the caspase-8 inhibitor IETD-FMK, the caspase-3/-7 inhibitor DEVD-FMK, the caspase-9 inhibitor LEHD-FMK, and the caspase-6 inhibitor VEID-FMK19) on TRAIL-induced apoptosis in MM.1S cells (Figure 4A). These studies showed that the pancaspase inhibitor ZVAD-FMK protected MM.1S cells from TRAIL-induced apoptosis, confirming that caspase activity was indispensable in TRAIL-induced apoptosis. Moreover, the caspase-8 inhibitor IETD-FMK also had a protective effect, confirming that caspase-8 activation is an integral part of this apoptotic pathway. In contrast, the caspase -3 and -7 inhibitor DEVD-FMK did not protect MM.1S cells from TRAIL. This suggests that the activation of the executioner caspases-3 and -7 is not critical for TRAIL-induced apoptosis in MM cells. The executioner caspase in this pathway appears to be caspase-6, because the caspase-6 inhibitor VEID-FMK also protected MM.1S cells from TRAIL-induced apoptosis. Interestingly, the function of caspase-9 appeared to be redundant because the caspase-9 inhibitor LEHD-FMK did not protect MM.1S cells from TRAIL-mediated apoptosis.
Modulation of TRAIL-induced cytotoxicity in MM.1S myeloma cells by selective inhibition of caspases.
(A) TRAIL-sensitive MM.1S myeloma cells were pretreated 1 hour before overnight incubation with TRAIL (100, 200, and 300 ng/mL) with or without the pancaspase inhibitor ZVAD-FMK, the caspase-8 inhibitor IETD-FMK, the caspase-3 and -7 inhibitor DEVD-FMK, the caspase-9 inhibitor LEHD-FMK, or the caspase-6 inhibitor VEID-FMK. Cell survival was determined using the MTT colorimetric assay. Bars represent mean ± SD values. (B-D) Caspase-3 enzymatic activity was measured by a caspase-3 intracellular activity assay in TRAIL-sensitive MM.1S cells as in Figure 2. (B) Control MM.1S cells. (C) MM.1S cells were treated with TRAIL (300 ng/mL) for 5 hours. (D) MM.1S cells were treated with TRAIL in the presence of DEVD-FMK (1 hour before treatment). DEVD-FMK inhibits caspase-3 activity, confirming that it is indeed internalized by MM cells and is functional.
Modulation of TRAIL-induced cytotoxicity in MM.1S myeloma cells by selective inhibition of caspases.
(A) TRAIL-sensitive MM.1S myeloma cells were pretreated 1 hour before overnight incubation with TRAIL (100, 200, and 300 ng/mL) with or without the pancaspase inhibitor ZVAD-FMK, the caspase-8 inhibitor IETD-FMK, the caspase-3 and -7 inhibitor DEVD-FMK, the caspase-9 inhibitor LEHD-FMK, or the caspase-6 inhibitor VEID-FMK. Cell survival was determined using the MTT colorimetric assay. Bars represent mean ± SD values. (B-D) Caspase-3 enzymatic activity was measured by a caspase-3 intracellular activity assay in TRAIL-sensitive MM.1S cells as in Figure 2. (B) Control MM.1S cells. (C) MM.1S cells were treated with TRAIL (300 ng/mL) for 5 hours. (D) MM.1S cells were treated with TRAIL in the presence of DEVD-FMK (1 hour before treatment). DEVD-FMK inhibits caspase-3 activity, confirming that it is indeed internalized by MM cells and is functional.
The finding that the caspase-3/-7 inhibitor DEVD-FMK did not protect MM.1S cells from TRAIL-induced apoptosis agrees with previous data in another model.20 Still, we wanted to confirm that this inhibitor is actually internalized by MM cells and does block caspase-3 activity. Therefore, we assessed caspase-3 activity in MM.1S cells, as previously, and found that DEVD-FMK indeed blocks caspase-3 activity in TRAIL-treated MM.1S cells (Figure 4B-D) and thus the negative data obtained with this compound are valid.
Expression of intracellular regulators of apoptosis in TRAIL-sensitive and -resistant MM cells
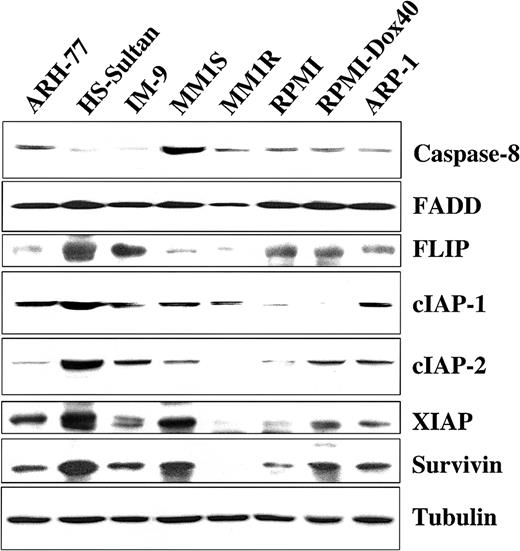
We next examined TRAIL-sensitive and -resistant MM cells for protein expression of several important proapoptotic and antiapoptotic regulators to discern differences that might account for variations in TRAIL sensitivity. Cell extracts from TRAIL-sensitive MM-1S, MM-1R, RPMI-8226/S, RPMI-Dox40, and ARP-1 cells; moderately TRAIL-resistant ARH-77 cells; and highly TRAIL-resistant IM-9 and HS-Sultan cells14 were examined by Western blot analysis for the presence of the proapoptotic molecules caspase-8 and FADD along with the intracellular apoptosis inhibitors FLIP, cIAP1, cIAP2, XIAP, and survivin. Although no differences in the expression of the proapoptotic protein FADD were observed among TRAIL-sensitive and -resistant cells, highly TRAIL-resistant HS-Sultan and IM-9 cells exhibited very low levels of the proapoptotic caspase-8 protein in comparison to TRAIL-sensitive and moderately TRAIL-resistant (ARH-77) cell lines. In addition, these studies showed that no single apoptosis inhibitor was per se correlated with resistance to TRAIL/Apo2L, though in general, higher levels of expression for the various apoptotic inhibitor proteins that we examined were present in TRAIL-resistant cells (Figure5).
Expression of intracellular regulators of apoptosis in TRAIL-sensitive and TRAIL-resistant myeloma cells.
Immunoblot analysis was used to examine cell lysates from myeloma cells that are moderately resistant (ARH-77), highly resistant (HS-Sultan, IM-9), or sensitive (MM.1S, MM.1R, RPMI 8226, RPMI-Dox 40, ARP-1) to TRAIL for expression of apoptosis-inducing (procaspase-8 and FADD) or apoptosis-blocking (FLIP, cIAP1, cIAP2, XIAP, and survivin) proteins. Equal protein loading was ensured by demonstration of uniform tubulin expression.
Expression of intracellular regulators of apoptosis in TRAIL-sensitive and TRAIL-resistant myeloma cells.
Immunoblot analysis was used to examine cell lysates from myeloma cells that are moderately resistant (ARH-77), highly resistant (HS-Sultan, IM-9), or sensitive (MM.1S, MM.1R, RPMI 8226, RPMI-Dox 40, ARP-1) to TRAIL for expression of apoptosis-inducing (procaspase-8 and FADD) or apoptosis-blocking (FLIP, cIAP1, cIAP2, XIAP, and survivin) proteins. Equal protein loading was ensured by demonstration of uniform tubulin expression.
Increased expression of procaspase-8 sensitizes TRAIL-resistant MM cells to TRAIL
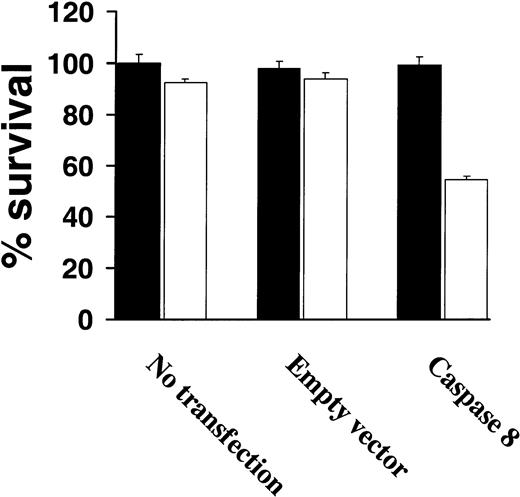
Because very low levels of procaspase-8 were observed among highly TRAIL-resistant cells correlated with resistance to TRAIL, we next attempted to restore TRAIL sensitivity by overexpressing procaspase-8 in TRAIL-resistant IM-9 cells. As shown in Figure6, transient expression of procaspase-8 restored TRAIL sensitivity of IM-9 cells, whereas untransfected or control vector–transfected IM-9 cells remained TRAIL resistant (Figure 6).
Increased procaspase-8 expression sensitizes TRAIL-resistant myeloma cells to TRAIL.
Cell survival of highly TRAIL-resistant IM-9 cells that were either untransfected or transfected with an empty vector or a procaspase-8 complementary DNA (cDNA)–containing vector, were treated with or without TRAIL (white bars) or vehicle (black bars) for 18 hours and assessed for survival using the MTT assay. Bars represent mean ± SD values.
Increased procaspase-8 expression sensitizes TRAIL-resistant myeloma cells to TRAIL.
Cell survival of highly TRAIL-resistant IM-9 cells that were either untransfected or transfected with an empty vector or a procaspase-8 complementary DNA (cDNA)–containing vector, were treated with or without TRAIL (white bars) or vehicle (black bars) for 18 hours and assessed for survival using the MTT assay. Bars represent mean ± SD values.
CHX and BIM down-regulate FLIP and cIAP2 expression and overcome TRAIL resistance in MM cells
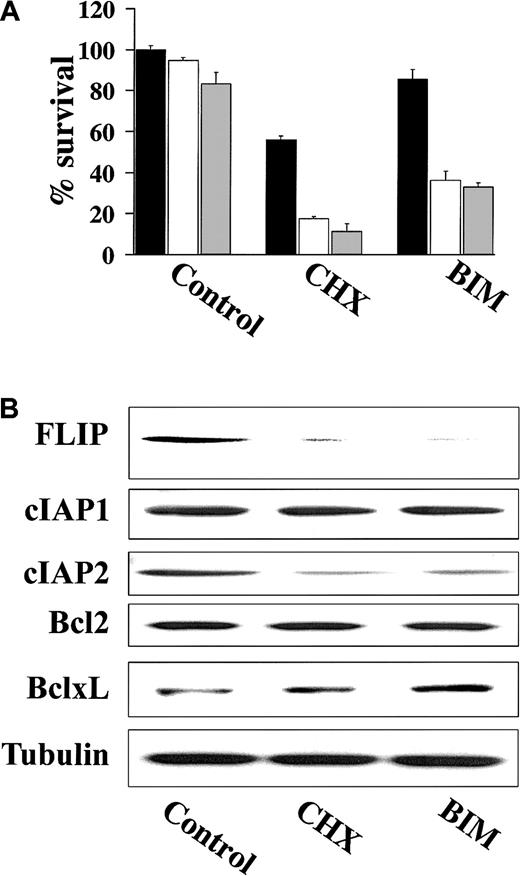
Previous studies have demonstrated that the activity of various proapoptotic agents can be enhanced following treatment with CHX, a protein synthesis inhibitor that can block expression of short-lived proteins including the antiapoptotic FLIP.21,22 As such, we examined whether treatment of TRAIL-resistant cells with CHX resulted in TRAIL-induced apoptosis. These studies showed that CHX did in fact sensitize highly TRAIL-resistant IM-9 cells to TRAIL (Figure7A). Because highly TRAIL-resistant HS-Sultan and IM-9 cells expressed high FLIP protein levels, which mediate TRAIL resistance in mature dendritic cells,23 we next examined whether BIM, an agent known to down-regulate FLIP,22 similarly restored TRAIL sensitivity in highly TRAIL-resistant IM-9 cells. BIM, analogous to CHX, also restored TRAIL sensitivity to IM-9 cells (Figure 7A). Moreover, immunoblot analysis showed that both BIM and CHX down-regulated the protein levels of FLIP as well as cIAP2, but not those of Bcl-2 or Bcl-xL in IM-9 cells (Figure 7B).
CHX and BIM down-regulate FLIP and cIAP2 expression and overcome TRAIL resistance.
(A) Cell survival of IM-9 cells pretreated with CHX (0.1 μg/mL) or BIM (20 μM) for 6 hours and then treated with 0 ng/mL (black bars), 100 ng/mL (white bars), or 200 ng/mL (gray bars) TRAIL for an additional 18 hours. Cell survival was determined using the MTT colorimetric assay. Bars represent mean ± SD values. (B) Immunoblotting for FLIP, cIAP1, cIAP2, Bcl-2, and Bcl-xL of cell lysates from IM-9 cells treated with CHX (0.1 μg/mL) or BIM (20 μM) for 6 hours. Levels of tubulin are shown for confirmation of equal loading.
CHX and BIM down-regulate FLIP and cIAP2 expression and overcome TRAIL resistance.
(A) Cell survival of IM-9 cells pretreated with CHX (0.1 μg/mL) or BIM (20 μM) for 6 hours and then treated with 0 ng/mL (black bars), 100 ng/mL (white bars), or 200 ng/mL (gray bars) TRAIL for an additional 18 hours. Cell survival was determined using the MTT colorimetric assay. Bars represent mean ± SD values. (B) Immunoblotting for FLIP, cIAP1, cIAP2, Bcl-2, and Bcl-xL of cell lysates from IM-9 cells treated with CHX (0.1 μg/mL) or BIM (20 μM) for 6 hours. Levels of tubulin are shown for confirmation of equal loading.
Inhibition of FLIP expression with antisense oligonucleotides sensitizes TRAIL-resistant MM cells to TRAIL
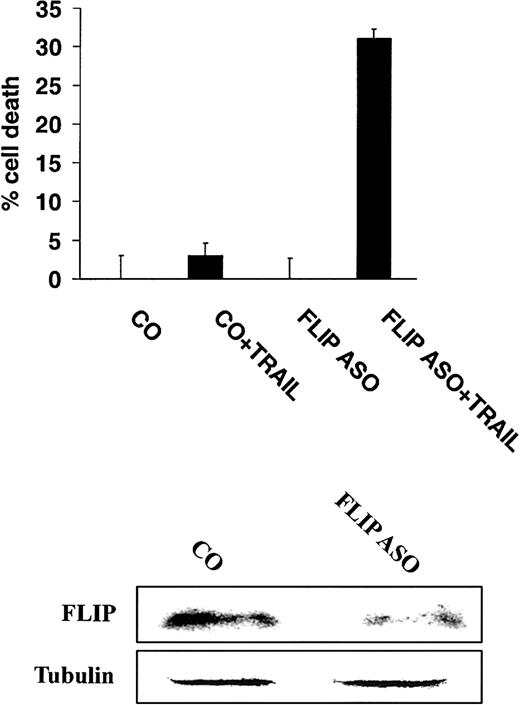
To more directly demonstrate a role for FLIP in mediating TRAIL resistance, highly TRAIL-resistant IM-9 cells were incubated with either control or antisense oligonucleotides directed at FLIP,16 and subsequently treated with TRAIL (200 ng/mL). These studies showed that incubation of cells with antisense oligonucleotides to FLIP sensitized IM-9 cells to TRAIL (Figure8).
Inhibition of FLIP expression with antisense oligonucleotides sensitizes TRAIL-resistant MM cells to TRAIL.
Cell death of highly TRAIL-resistant IM-9 cells transfected with either FLIP antisense (ASO) or control (CO) oligonucleotides, and then treated with or without TRAIL (200 ng/mL) for 18 hours. FLIP and tubulin protein levels are shown for comparison.
Inhibition of FLIP expression with antisense oligonucleotides sensitizes TRAIL-resistant MM cells to TRAIL.
Cell death of highly TRAIL-resistant IM-9 cells transfected with either FLIP antisense (ASO) or control (CO) oligonucleotides, and then treated with or without TRAIL (200 ng/mL) for 18 hours. FLIP and tubulin protein levels are shown for comparison.
Modulation of apoptosis regulatory proteins by the NF-κB inhibitor SN50
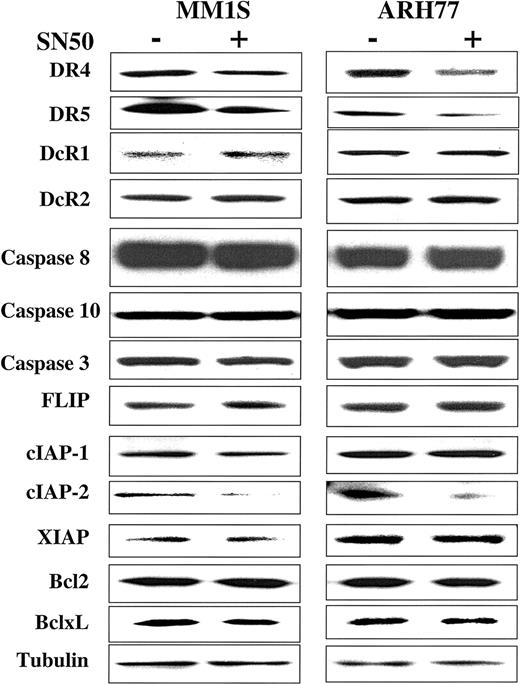
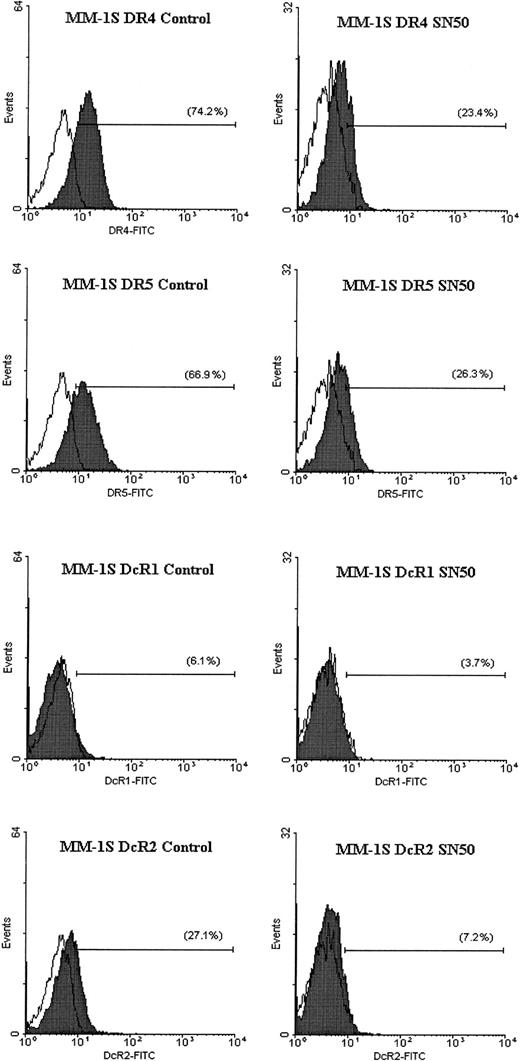
In previous work, we showed that the cell-permeable NF-κB inhibitor SN50 increased the sensitivity of TRAIL-sensitive MM cells (ie, MM.1S) to TRAIL, and restored TRAIL sensitivity to TRAIL-resistant cells (ARH-77, IM-9).14 As an extension of this work, we next sought to identify the molecular target of SN50 in the apoptotic machinery. Cells (MM.1S, ARH-77, and IM-9) were treated for 6 hours with the SN50 peptide (30 μg/mL), and protein levels of DR4, DR5, DcR1, DcR2, procaspase-8, procaspase-10, procaspase-3, FLIP, cIAP1, cIAP2, XIAP, survivin, Bcl-2, and Bcl-xL were then examined by immunoblotting. As shown in Figure 9, the increased response to TRAIL could not be attributed to changes in levels of TRAIL receptor expression. In fact, a moderate decrease of DR4/DR5 was found in all cell lines examined (Figures 9 and10), in agreement with a recent report in another model,24 suggesting therefore that sensitivity to TRAIL can be restored even in the presence of weak(er) DR4/DR5 receptor expression and that low-level expression of the TRAIL death receptors does not necessarily account for lack of TRAIL activity in certain tumor cells.
Expression of TRAIL receptors and intracellular apoptosis regulator proteins in myeloma cells treated with the NF-κB inhibitor SN50.
Immunoblot analysis for TRAIL receptors (DR4, DR5, DcR1, DcR2), procaspase-8, procaspase-10, procaspase-3, FLIP, cIAP1, cIAP2, XIAP, survivin, Bcl2, and Bcl-xL in TRAIL-sensitive (MM.1S) and moderately resistant (ARH-77) cells treated with and without the NF-κB inhibitor SN50 (30 μg/mL) for 6 hours. Levels of tubulin are shown for confirmation of equal protein loading.
Expression of TRAIL receptors and intracellular apoptosis regulator proteins in myeloma cells treated with the NF-κB inhibitor SN50.
Immunoblot analysis for TRAIL receptors (DR4, DR5, DcR1, DcR2), procaspase-8, procaspase-10, procaspase-3, FLIP, cIAP1, cIAP2, XIAP, survivin, Bcl2, and Bcl-xL in TRAIL-sensitive (MM.1S) and moderately resistant (ARH-77) cells treated with and without the NF-κB inhibitor SN50 (30 μg/mL) for 6 hours. Levels of tubulin are shown for confirmation of equal protein loading.
Flow cytometric analysis for TRAIL receptor expression on myeloma cells treated with the NF-κB inhibitor SN50.
Flow cytometric analysis was performed to determine the cell surface expression of TRAIL receptors DR4, DR5, DcR1, and DcR2 (shaded peaks) on MM.1S cells treated with and without the NF-κB inhibitor SN50 (30 μg/mL) for 6 hours. Isotype control antibody staining appears as unshaded peaks.
Flow cytometric analysis for TRAIL receptor expression on myeloma cells treated with the NF-κB inhibitor SN50.
Flow cytometric analysis was performed to determine the cell surface expression of TRAIL receptors DR4, DR5, DcR1, and DcR2 (shaded peaks) on MM.1S cells treated with and without the NF-κB inhibitor SN50 (30 μg/mL) for 6 hours. Isotype control antibody staining appears as unshaded peaks.
Among the other proapoptotic and antiapoptotic proteins studied, only cIAP2 protein expression was significantly decreased after treatment with the NF-κB inhibitor SN50 (Figure 9), suggesting that cIAP2 has a role in mediating TRAIL resistance and is also a target for TRAIL-related sensitization strategies.
Caspase-8 activation in TRAIL-resistant myeloma cells preincubated with CHX, BIM, or SN50 prior to TRAIL treatment
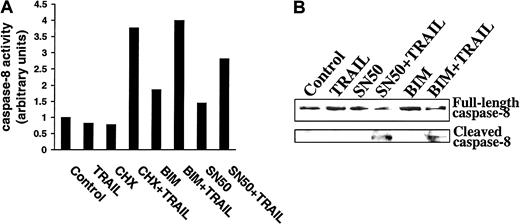
Because CHX, BIM, and SN50 sensitized MM cells to TRAIL, and because these agents also down-regulated FLIP and cIAP2, we next determined whether these agents relieved inhibitors of caspase-8 processing and activation. As shown in Figure11A, increased caspase-8 activity using the ApoAlert caspase-8 colorimetric kit was observed in IM-9 cells only when preincubated with CHX, BIM, or SN50 prior to TRAIL exposure. Moreover, caspase-8 cleavage was not induced by TRAIL treatment of control IM-9 cells, whereas caspase-8 cleavage was readily induced by TRAIL in IM-9 cells pretreated with CHX (not shown), BIM, or SN50 (Figure 11B). These studies therefore confirm the proapoptotic effect of CHX, BIM, and SN50, BIM at the level of caspase-8 activation, and further suggest that inhibition of procaspase-8 cleavage is a major mechanism of resistance to TRAIL in MM cells.
Demonstration of caspase-8 activation in TRAIL-resistant myeloma cells preincubated with CHX, BIM, or SN50 before TRAIL treatment.
(A) Caspase-8 activity was determined by use of the ApoAlert caspase-8 colorimetric kit in highly TRAIL-resistant IM-9 cells that were incubated with CHX (0.1 μg/mL), BIM (20 μM), or SN50 (30 μg/mL) for 6 hours before treatment with TRAIL (300 ng/mL) for 30 minutes. (B) Immunoblot analysis for caspase-8 cleavage in IM-9 cells preincubated with BIM (20 μM) or SN50 (30 μg/mL) for 6 hours before treatment with TRAIL (300 ng/mL) for 1 hour.
Demonstration of caspase-8 activation in TRAIL-resistant myeloma cells preincubated with CHX, BIM, or SN50 before TRAIL treatment.
(A) Caspase-8 activity was determined by use of the ApoAlert caspase-8 colorimetric kit in highly TRAIL-resistant IM-9 cells that were incubated with CHX (0.1 μg/mL), BIM (20 μM), or SN50 (30 μg/mL) for 6 hours before treatment with TRAIL (300 ng/mL) for 30 minutes. (B) Immunoblot analysis for caspase-8 cleavage in IM-9 cells preincubated with BIM (20 μM) or SN50 (30 μg/mL) for 6 hours before treatment with TRAIL (300 ng/mL) for 1 hour.
Discussion
In the present study, we investigated the mechanism of TRAIL-induced apoptosis in MM. We found that cell death triggered by TRAIL is mediated by the activation of caspases-8 and -6. Caspases-3 and -9 were also cleaved and activated, yet their role appears to be dispensable. Resistance to TRAIL was associated with lack of cleavage of caspase-8 and downstream caspases, but could not be attributed solely to the presence of any single apoptosis inhibitor protein. However, diminished expression of the proapoptotic protein caspase-8 and expression of the apoptosis inhibitor proteins FLIP and cIAP2 were associated with decreased TRAIL sensitivity because overexpression of caspase-8 or inhibition of FLIP or cIAP2 restored sensitivity to TRAIL. Interestingly, the sensitization of resistant cells to TRAIL by either CHX, BIM, or SN50 resulted in activation of caspase-8 after TRAIL stimulation, thus confirming that inhibition of the apoptotic cascade in TRAIL-resistant cells rested at the level of caspase-8 cleavage.
TRAIL is a member of the superfamily of TNF-related ligands that potently induces apoptosis of a wide range of cancer cells, while sparing normal cells.1-7,12,25-27 The temporary concerns raised by a report suggesting that recombinant TRAIL/Apo2L killed normal human hepatocytes in vitro28 were addressed by subsequent studies that attributed it to nonoptimized recombinant ligand preparations.29 Therefore, there is strong interest for the upcoming phase I clinical trials of this anticancer agent. We and others13,14,30-32 have demonstrated that TRAIL induces apoptosis of MM cell lines and patient tumor cells in vitro. Importantly, TRAIL sensitivity was noted even in MM cells resistant to dexamethasone, melphalan, and adriamycin, suggesting that TRAIL may be clinically active even in the setting of MM refractory to conventional therapies. Five cell lines (ARH-77, HS-Sultan, IM-9 and MM-AS, MM-SV), which are all CD20+ and therefore likely to represent an earlier plasmacytic differentiation stage, showed significantly lower apoptotic responses to TRAIL than MM cell lines that were CD20− (eg, MM.1S, MM.1R, RPMI-8226/S, RPMI-Dox40).14 Consistent with these findings, we recently showed that TRAIL sensitivity in B cells may be differentiation-stage specific: preplasmacytic CD20+ B-cell neoplastic cells (5 diffuse large B-cell lymphoma cell lines, a Burkitt lymphoma cell line, as well as tumor cells from 4 patients with Waldenstrom macroglobulinemia) were resistant to TRAIL, whereas 2 pre–B-cell tumor cell lines were TRAIL sensitive.33 Our ongoing studies suggest that expression of FLIP, along with other caspase-regulating intracellular proteins, rather than TRAIL receptor expression, regulates this differentiation stage sensitivity to TRAIL.
Apoptosis induced by cell surface death receptors (eg, TNF-R1, Fas, DR4, DR5) is mediated by the recruitment and activation of an apical caspase (either caspase-8 or caspase-10) to the receptor, which subsequently activates downstream executioner caspases (caspase-3, -7, or -6). The mechanism of TRAIL-triggered signaling, as well as the identity of the apical caspase in the TRAIL pathway, are controversial. In contrast to Fas signaling where caspase-8 is unequivocally recruited to the receptor signaling complex and is the first caspase to be activated, both caspase-8 and caspase-10 have been implicated as the apical caspases of TRAIL-induced apoptosis. Specifically, interaction of caspase-10 with TRAIL receptors has been shown in cells cotransfected with plasmids encoding the respective genes,34 and we have recently shown recruitment of endogenous caspase-10 to the TRAIL receptor signaling complex in thyroid carcinomas3 and Ewing sarcomas.4 In contrast, studies in human lymphoid lines10 11 have shown recruitment of endogenous caspase-8 to the TRAIL-activated death receptors DR4 and DR5. In agreement with the latter studies on hematopoietic malignancies, we found that caspase-8, but not caspase-10, is proteolytically cleaved and the initiator caspase of the TRAIL pathway in MM. Therefore, it appears that the initial step of the apoptotic cascade triggered by TRAIL is tissue dependent.
Previous studies on Fas-mediated apoptosis have shown that the death receptor–mediated autocatalytic cleavage of procaspase-8 can signal apoptosis via direct cleavage of caspase-3 and other executioner caspases (caspase-7, caspase-6).35 Additionally, caspase-8 can cleave Bid, a cytoplasmic member of the bcl-2 family, which induces the release of cytochrome c from mitochondria.36,37 Cytochrome c in complex with apoptotic protease-activating factor 1 (Apaf-1) activates caspase-9, which in turn activates the executioner caspases.38 This mitochondria-dependent pathway can be inhibited by bcl-2.39,40 Our data suggest that TRAIL-induced apoptosis in sensitive MM cells proceeds rapidly through cleavage of downstream caspases by caspase-8. The pivotal role of caspases is evidenced by the inhibitory effect of a pancaspase inhibitor, as well as by specific inhibitors for caspase-8 and caspase-6. Furthermore, the well-known caspase substrates PARP and DFF45 were rapidly cleaved in MM.1S cells on TRAIL treatment. PARP is the most widely used marker for the detection of caspase activity.41 DFF45 is the 45-kd inhibitory subunit of the heterodimeric DNA fragmentation factor (DFF), which is cleaved and inactivated by executioner caspases, thus allowing the enzymatically active 40-kd subunit, otherwise known as caspase-activated DNase (CAD), to translocate to the nucleus and mediate the degradation of genomic DNA.42 43
Activation of the mitochondrial pathway may also occur during TRAIL-induced apoptosis in MM cells, as evidenced by the cleavage of caspase-9. However, it appears that this pathway is redundant, because a caspase-9 inhibitor did not protect MM.1S cells from TRAIL-induced cell death. Moreover, in another study transfection of bcl-2 into TRAIL-sensitive RPMI 8226 cells, which blocks the release of cytochromec from mitochondria, failed to protect against TRAIL.30 Caspase-3 activation also appears to be dispensable, because TRAIL-induced apoptosis was not attenuated by the caspase-3 inhibitor DEVD-FMK. Therefore, caspase-3 activation is a marker that correlates with cell sensitivity to TRAIL, but may also be functionally redundant. This supports previous data in other models of overlapping activation of alternative caspases.20 44Moreover, caspase-7 is also inhibited by DEVD-FMK, excluding it as an executioner caspase in TRAIL-induced apoptosis in MM. In contrast, caspase-6 is a candidate executioner caspase, because a caspase-6–specific inhibitor protected MM cells from TRAIL.
Procaspase-8 cleavage, as well as cleavage of all other downstream caspases and caspase substrates, was significantly inhibited or even absent in TRAIL-resistant cells. This provided the first piece of evidence for the existence of an antiapoptotic block at the level of caspase-8 activation. To identify possible reasons for the inefficient activation of caspase-8, a panel of cell lines with high (MM-1S, MM-1R, RPMI-8226/S, RPMI-Dox40, and ARP-1), low (ARH-77), and no (IM-9, HS-Sultan) sensitivity to TRAIL were examined by immunoblotting for the presence of the apoptosis inhibitors FLIP, cIAP1, cIAP2, XIAP, and survivin, as well as for caspase-8 and the adaptor molecule FADD. Although expression of no single apoptosis inhibitor per se correlated with resistance to TRAIL/Apo2L, there was a tendency for higher levels of the various apoptotic inhibitors, in particular FLIP, and lower levels of procaspase-8 in the cell lines with lower response to TRAIL. Moreover, restoration of high protein levels of procaspase-8 by forced expression sensitized IM-9 cell to TRAIL.
The identification of low procaspase-8 levels along with FLIP expression in highly TRAIL-resistant cells led us to study the effect of 2 more compounds on TRAIL-induced apoptosis of MM cells. CHX and BIM have been previously shown to overcome resistance to Fas-induced apoptosis,3,21,22,45 likely due to down-regulation of FLIP protein levels.21,22 In this study, we extended their use in TRAIL signaling and found that both can overcome the resistance of IM-9 cells to TRAIL. Importantly, both CHX and BIM down-regulated FLIP and cIAP2 protein levels, but not cIAP1, Bcl-2 or Bcl-xL. Using an antisense oligonucleotide, we then specifically lowered FLIP protein expression in IM-9 cells, which sensitized these cells to TRAIL, thereby confirming the protective role of FLIP against TRAIL-induced apoptosis. Although the inhibitory role of FLIP in Fas signaling is established, its role in TRAIL-induced apoptosis has been controversial, because elevated FLIP levels have been correlated with susceptibility to TRAIL in some,12,46 but not all, studies.47 Because the antiapoptotic role of FLIP depends on its recruitment to receptor complexes through FADD and procaspase-8 binding, which occurs in some but not all malignancies,3,4,34,46,48-51 it is possible that FLIP may exert an inhibitory role on TRAIL signaling in some, but not all, tissues. Alternatively, equal levels of FLIP may be inhibitory only in tumors with low death receptor and procaspase-8 levels, as demonstrated previously in another model39 and shown for IM-9 and HS-Sultan cells in our study.
We have previously shown that SN50, a cell permeable inhibitor of the nuclear translocation and transcriptional activity of NF-κB, can increase the sensitivity of TRAIL-sensitive MM cells (ie, MM.1S) as well as overcome resistance in TRAIL-resistant cells (ARH-77, IM-9).14 We now attempted to identify the molecular mediator of SN50 on apoptosis. We found that sensitization to TRAIL by SN50 was associated with down-regulation of cIAP2, which occurred in all cell lines tested irrespective of the baseline sensitivity to TRAIL. cIAP2 is a member of the IAP family of antiapoptotic proteins, which is recruited to the signaling complex of TNF receptor I via its interaction with TRAF1 and TRAF2 and exerts an inhibitory function on caspase-8 activation.52 cIAP2 expression was found to be regulated by NF-κB in human leukemic cells.53 Thus, cIAP2 appears to modulate TRAIL-induced apoptosis in MM cells and mediate the prosurvival effect of NF-κB.
Finally, to confirm that the inhibitory activity of FLIP and cIAP2 on TRAIL-induced apoptosis is exerted at the level of procaspase-8 cleavage, we pretreated TRAIL-resistant cells with CHX, BIM, or SN50, and then stimulated them with TRAIL. We found that cleavage and enzymatic activation of procaspase-8 on stimulation with TRAIL occurred only in cells that had been pretreated with CHX, BIM, or SN50. This proves that the observed sensitization to TRAIL by these compounds is due to their effect on the processing of procaspase-8 and, furthermore, that the inhibition of caspase-8 activation is a major mechanism of TRAIL-resistance in MM cells.
In summary, the potent antimyeloma activity of TRAIL/Apo2L is mediated by the rapid activation of caspase-8, followed by caspase-6. Low levels of procaspase-8, as well as the antiapoptotic function of FLIP and cIAP2, may hinder caspase-8 activation, and thereby confer resistance to TRAIL/Apo2L. Pharmacologic agents that down-regulate FLIP and cIAP2, such as inhibitors of NF-κB and PKC, may be useful clinically as adjunctive agents in the treatment of MM.
Supported by the Laurie Strauss Leukemia Foundation (N.M., C.S.M.), the Multiple Myeloma Research Foundation (N.M., C.S.M., S.P.T.), the Bailey Family Research Fund (N.M., C.S.M.), a National Institutes of Health Career Development Award (S.P.T.), and the Doris Duke Distinguished Clinical Research Scientist Award (K.C.A.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Steven P. Treon, Department of Adult Oncology, Dana Farber Cancer Institute, 44 Binney St, Mayer Bldg, Rm M557, Boston MA 02115; e-mail: steven_treon@dfci.harvard.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal