Abstract
Plasminogen activator inhibitor–1 (PAI-1) plays a key role in control of coagulation and tissue remodeling and has been shown to be regulated by a number of cell stimuli, among those hypoxia. In this study we characterize the hypoxia-mediated induction of PAI-1 in human hepatoma cell line HepG2. We found that PAI-1 is tightly regulated in a narrow oxygen gradient. After incubation at oxygen concentrations of 1% to 2%, a 60-fold increase in PAI-1 messenger RNA levels was observed, whereas mild hypoxic conditions of more than 3.5% did not appear to induce transcription. Moreover, increased levels of PAI-1 protein were observed after incubation at low oxygen tensions. Through sequence analysis, several putative hypoxia-response elements (HREs 1-5) were identified in the human PAI-I promoter. Reporter gene assays showed that the HRE-2 (−194 to −187) was necessary and sufficient for the hypoxia-mediated response. By electrophoretic mobility assay we observed hypoxia-dependent binding of a protein complex to the HRE-2 motif. Further analysis demonstrated that HRE-2 was specifically recognized by the hypoxia-inducible transcription factor 1α–arylhydrocarbon nuclear translocator complex. Taken together, our data demonstrate that hypoxia-induced transcription is mediated through HIF-1 interaction with the HRE-2 site of the human PAI-1 promoter.
Introduction
Plasminogen activator inhibitor–1 (PAI-1) plays a central role in the control of physiologically important mechanisms involved in the homeostasis of blood coagulation and remodeling of extracellular matrix (reviewed by Booth1). The effect of PAI-1 is mediated through inhibition of urokinase and tissue type plasminogen activators. The importance of PAI-1 in the regulation of fibrinolytic activity is highlighted by several studies documenting an association between increased levels of PAI-1 and the risk of developing a cardiovascular disease.2-4 Furthermore, numerous clinical studies of different types of cancer have identified high levels of plasma PAI-1 as a strong prognostic factor for more metastatic forms of cancer, concomitant with a poorer clinical outcome (reviewed by Harbeck et al5).
PAI-1 is produced by a variety of cell types in vitro, such as hepatocytes,6 platelets,7 smooth muscle cells,8 and endothelial cells.9 The sources of PAI-1 in vivo have not as yet been identified. However, studies in rabbits indicate that the liver and endothelial cells are the most important producers.10 Several agents induce PAI-1 at the transcriptional level, including phorbol esters,11inflammatory cytokines,12 transforming growth factor β,13 and hypoxia.14 For most of these stimuli, the signal transduction pathways have been identified, including target transcription factors and the corresponding specific transcriptional control elements within the regulated target genes.15,16 Recently, it has been found that a 300–base pair stretch in the promoter region of the humanPAI-1 gene is necessary for the responses to hypoxia and that these responses were mediated by the hypoxia-inducible transcription factor, HIF-1.17 18 There is, however, no information available about molecular mechanisms involved in the control of transcriptional activation of PAI-1 by hypoxia, nor has the degree of hypoxia necessary for induction been determined.
We have previously found that PAI-1 was up-regulated in 4 different human liver cell lines on incubation in 1% oxygen.19 In the present study, we have characterized hypoxia-induced expression of PAI-1 in the human hepatoma cell line HepG2 cultured in a controlled atmosphere with oxygen concentrations ranging from 1% to 8%. Moreover, we have identified an element in the human PAI-1 promoter mediating hypoxic responses and demonstrated that this element binds HIF-1.
Materials and methods
Cell cultures
The human hepatoma cell line, HepG2 (ATCC HB-8065), was obtained through American Type Culture Collection (ATCC; Manassas, VA). The cells were cultured in Eagle minimal essential medium supplemented with 10% fetal bovine serum, nonessential amino acids, 100 IU/mL penicillin, and 100 μg/mL streptomycin.
Hypoxic treatment
Before exposure to hypoxia, HepG2 cells were seeded in 2 mL medium in 6-well plates (Costar, Acton, MA) to achieve a final concentration of 0.5 × 106 cells/well and incubated for 24 hours in a humidified ambient atmosphere with 5% CO2 at 37°C. For the hypoxic treatment, the cells were transferred to a combined workbench/incubator20 with a humidified atmosphere of 1%, 2%, 3.5%, 5%, 6.5%, and 8% oxygen and 5% CO2balanced with N2 and incubated at 37°C for up to 48 hours. After 1, 2, 4, 8, 12, 16, 24, 32, 40, and 48 hours, the cell cultures were processed directly in the incubator. The control cultures maintained in 21% oxygen and 5% CO2 were processed every 8 hours for the duration of the experiment. For each oxygen tension, the experiment was carried out on 2 separate occasions.
Real time reverse transcriptase–polymerase chain reaction
For preparation of RNA, the GenElute messenger RNA (mRNA) kit (Sigma, St Louis, MO) was used according to the manufacturer's instructions. Briefly, the medium was withdrawn from one well, and the cells were lysed by the addition of 300 μL lysis buffer. The lysate was collected and stored at −80°C until completion of the experiment, when all samples were processed for RNA extraction simultaneously. After isolation, the RNA was DnaseI treated and used for complementary DNA (cDNA) synthesis. The cDNA was prepared from approximately 2 μg RNA by using the M-MLV reverse transcriptase (Sigma) with random decamer primers. The levels of PAI-1 transcript were determined by real-time reverse transcriptase–polymerase chain reaction (RT-PCR) as described.19 To normalize for input load of cDNA between the samples, 18S rRNA was used as an external endogenous standard. The forward, 5′-AGGACCGCGGTTCTATTTTGTTGG-3′, and reverse, 5′-CCCCCGGCCGTCCCTCTTA-3′, primers for 18S rRNA were designed by using the PrimerSelect program of the Lasergene software package (DNASTAR, Madison, WI) and were used at a concentration of 1 pmol per reaction. The amplification was performed in an iCycler (Bio-Rad, Hercules, CA), using a 2-temperature cycling, consisting of a denaturation step for 15 seconds at 95°C and an annealing/extension step for 30 seconds at 68°C. For the detection of the PCR products in real-time, the SYBR Green I fluorophore (Molecular Probes, Leiden, The Netherlands) was used in a final 22 000-fold dilution from the stock supplied by the manufacturer. For quantitative analysis of the PAI-1 transcripts and 18S rRNA, the cDNA from each sample was analyzed in duplicate and on 2 separate occasions.
Quantitation of cell- and matrix-associated and soluble PAI-1
For immunoblotting, the cells were lysed in SDS sample buffer containing 1 mM phenylmethylsulfonyl fluoride and protease inhibitors (Complete; Roche, Mannheim, DE), and the protein concentration was determined by NanoOrange (Molecular Probes). Protein (20 μg) was loaded onto a 10% SDS-polyacrylamide gel and after electrophoresis was blotted onto a polyvinylidene diflouride membrane (Millipore, Bedford, MA). Proteins were stained with Sypro Ruby (Bio-Rad), detected in a Flour-S MultiImager (Bio-Rad), and analyzed by using the TotalLab software package (Phoretix, Newcastle, United Kingdom) to normalize for differences between samples in protein load and transfer. The detection of PAI-1 was carried out by using a 1000-fold dilution of primary rabbit antibody against human PAI-1 (Santa Cruz Biotechnology, Santa Cruz, CA) and a 50 000-fold dilution of biotinylated secondary antibody (goat antirabbit immunoglobulin G; DAKO, Copenhagen, Denmark). Finally, 60 000-fold diluted horseradish peroxidase–conjugated streptavidin (Amersham, Uppsala, Sweden) was used, and, for visualization, the SuperSignal West Femto substrate (Pierce, Rockford, IL) was used. The detection was performed in the Fluor-S MultiImager, and the signals were analyzed by using the TotalLab software package. For each sample the Western blotting procedure was performed at least twice. The concentration of secreted PAI-1 in the medium was determined by enzyme-linked immunosorbent assay (Imulyse PAI-1, Biopool, Ventura, CA), and each sample was analyzed in duplicate.
Plasmid constructs, transfection procedure, and reporter assay
The human PAI-1 promotor region, extending from positions −806 to +19 was amplified from a pEMBL8cat plasmid containing the PAI-promotor (a kind gift from P. Andreasen, University of Aarhus, Denmark) with the aid of an upstream and downstream primer incorporating restriction sites for MluI andBglII, respectively. The sequence of the upstream primer was 5′-TGAACGCGTAAGCTTTTACCATGGTAACCCCT-3′ and that of the downstream primer was 5′-TGAAGATCTGCAGCCAAACACAGCTGTGCT-3′. After restriction, the PCR product was ligated into the luciferase reporter plasmid, pGL3basic (Promega, Madison, WI), yielding a pGL3-PAI-wt12 345 construct containing the 5 putative hypoxia-response element (HRE) motifs (Figure3A). The individual HRE motifs were mutated by using a QuickChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA), and the mutation primers are listed in Table 1. The generated plasmids were designated pGL3-PAI-wtx, where x denotes the presence of wild-type putative HREs.
Oligonucleotides used for mutagenesis and electrophoretic mobility shift assay
| Name . | Sequence . | Position . |
|---|---|---|
| wt1 | 5′-gcacacacacacacacacacatgcctcagcaagtccc-3′ | −175/−139 |
| mut1 | 5′-gcacacacacacacacacttaatcctcagcaagtccc-3′ | |
| wt2 | 5′-cctgaatgctcttacacacgtacacacacagagcagc-3′ | −210/−174 |
| mut2 | 5′-cctgaatgctcttacacttaatcacacacagagcagc-3′ | |
| wt3 | 5′-gccctgggggaaaacttccacgttttgatggaggttatc-3′ | −471/−433 |
| mut3 | 5′-gccctgggggaaaacttccttaatttgatggaggttatc-3′ | |
| wt4 | 5′-cagacaaaacctagacaatcacgtggctggctgcattgccc-3′ | −585/−545 |
| mut4 | 5′-cagacaaaacctagacaatcttaatgctggctgcattgccc-3′ | |
| wt5 | 5′-ggggcacagagagagtctggacacgtggggagtcagccg-3′ | −703/−664 |
| mut5 | 5′-ggggcacagagagagtctggacttaatgggagtcagccg-3′ |
| Name . | Sequence . | Position . |
|---|---|---|
| wt1 | 5′-gcacacacacacacacacacatgcctcagcaagtccc-3′ | −175/−139 |
| mut1 | 5′-gcacacacacacacacacttaatcctcagcaagtccc-3′ | |
| wt2 | 5′-cctgaatgctcttacacacgtacacacacagagcagc-3′ | −210/−174 |
| mut2 | 5′-cctgaatgctcttacacttaatcacacacagagcagc-3′ | |
| wt3 | 5′-gccctgggggaaaacttccacgttttgatggaggttatc-3′ | −471/−433 |
| mut3 | 5′-gccctgggggaaaacttccttaatttgatggaggttatc-3′ | |
| wt4 | 5′-cagacaaaacctagacaatcacgtggctggctgcattgccc-3′ | −585/−545 |
| mut4 | 5′-cagacaaaacctagacaatcttaatgctggctgcattgccc-3′ | |
| wt5 | 5′-ggggcacagagagagtctggacacgtggggagtcagccg-3′ | −703/−664 |
| mut5 | 5′-ggggcacagagagagtctggacttaatgggagtcagccg-3′ |
Only the sequences of the sense oligonucleotides are shown. For mutagenesis and electrophoretic mobility shift assay, the complementary probes were used. The putative hypoxia-response elements are shown in boldface type, and the mutations are underlined.
HepG2 cells were transfected with the pGL3-PAI-wtx constructs. Briefly, 1.5 × 106 cells in 400 μL growth medium were mixed with 10 μg plasmid DNA and electroporated (220 V, 800 μF) by using the Gene Pulser II system (Bio-Rad). The cells were then seeded in 200 μL medium in 96-well plates in sixplicate at a concentration of 50 × 103 cells/well and incubated for 3 hours at 21% O2, after which half of the plates were transferred to the hypoxic chamber and incubated for 24 hours before analysis. The luciferase activity was determined by using the Steady-Glo luciferase assay (Promega) according to the manufacturer's instructions.
Preparation of nuclear extracts
For nuclear extract preparation, HepG2 cells in 10-cm dishes were cultured at 1% or 21% oxygen for 6 hours. The cells were then washed twice with ice-cold phosphate-buffered saline (PBS), scraped into 10 mL PBS and pelleted by centrifugation at 2000 rpm for 5 minutes at 4°C. The cell pellet was resuspended in hypotonic buffer HB (10 mM Tris-HCl [pH 7.3], 10 mM KCl, 1.5 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol), incubated on ice for 10 minutes and pelleted by centrifugation at 1500 rpm for 5 minutes at 4°C. The cell pellet was resuspended in lysis buffer (HB with 0.4% NP-40) and incubated on ice for 10 minutes. The nuclei were pelleted by centrifugation at 1500 rpm for 5 minutes at 4°C and washed once in HB. The nuclear proteins were extracted by incubation in high-salt buffer (20 mM Tris-HCl [pH 7.3], 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.42 M KCl, 20 μg/mL leupeptin, 1 μg/mL pepstatin) for 20 minutes on ice. The nuclear debris was pelleted by centrifugation at 14 000 rpm for 20 minutes at 4°C. The total protein concentration was determined by the Bradford method.
Alignment of human and rat promoter
Alignment of the promoters was performed by using the Megalign program (DNASTAR).
Electrophoretic mobility shift assay
Sequences of probes used for electrophoretic mobility shift assay (EMSA) encompassing the putative HRE motifs and mutant probes are shown in Table 1. The probes were end-labeled with T4 polynucleotide kinase and γ-32P]-ATP and purified on G25 Microspin columns (Pharmacia). DNA binding reactions were carried out with 12 μg of proteins, 6-fmol labeled probe in 20 μL reaction volume with final concentrations of 10 mM Hepes, 2.8 mM MgCl2, 16% glycerol, 0.15 mM EDTA, 0.25 mM dithiothreitol, 0.1 M KCl, 5 mM Tris-HCl [pH 7.3], 1 μg poly(dI-dC), and 1 μg poly(dC). The reactions were incubated on ice for 30 minutes. In inhibition experiments cold competitor probe was added at 5-, 50-, and 500-fold molar excess. In control experiments, HIF-1α and ARNT proteins were expressed by in vitro translation in rabbit reticulocyte lysate (Promega) by using plasmids pSP72/HIF-1α21 and pGEM7/Arnt22 for HIF-1α and ARNT, respectively. For supershift experiments, polyclonal antibodies against HIF-1α and its partner factor ARNT were added after an initial 30-minute incubation of the probe with nuclear extract and incubated for 1 hour at 4°C. The complexes were resolved on a 4% polyacrylamide gel in a Tris-glycine-EDTA buffer at 30 mA at 4°C.
Results
Hypoxia-induced expression of PAI-1
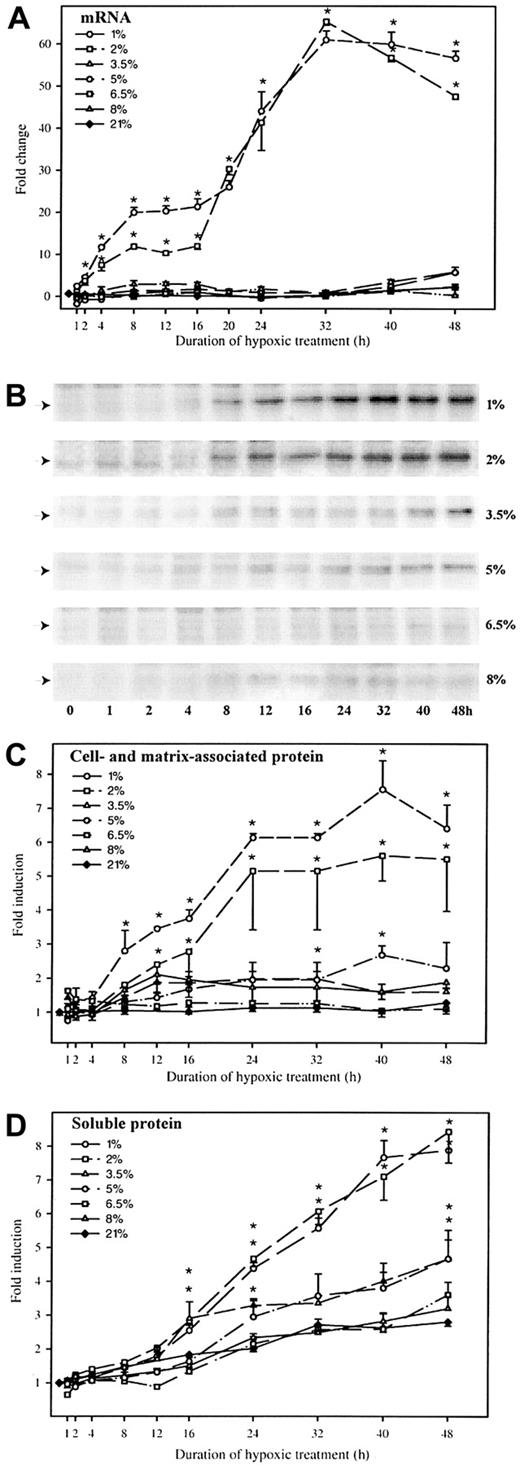
HepG2 cells were cultured for up to 48 hours under different oxygen concentrations, ranging from 1% to 8%, and at ambient air conditions. As shown in Figure 1A, oxygen concentrations of 1% and 2% rapidly induced PAI-1 mRNA expression leading to statistically significant (P < .05) increases after only 2 hours and reaching maximal levels of a 60-fold induction response after 32 hours of hypoxic treatment. At oxygen concentrations from 3.5% to 5%, a scant 5-fold induction of PAI-1 mRNA expression was observed after 40 hours of hypoxic treatment, whereas no changes in PAI-1 mRNA levels could be observed at oxygen tensions above 6.5%, relative to the levels observed in the ambient air control samples.
Kinetics of hypoxia-induced PAI-1 mRNA and protein.
HepG2 cells were cultured at oxygen concentrations of 1%, 2%, 3.5%, 5%, 6.5%, 8%, and 21% for up to 48 hours. At the indicated intervals, RNA, protein, and medium were harvested. (A) Levels of PAI-1 mRNA were determined by real-time RT-PCR. The levels of PAI-1 mRNA were normalized to 18S rRNA within each sample and to the starting values, which were set to 1.0. Each graph represents the average of 2 separate experiments, each analyzed in duplicate on 2 separate occasions (n = 8). Error bars represent SEM; *P < .001 versus respective ambient controls. (B) For each oxygen tension one representative Western blot of PAI-1 is shown. Autoradiographic signals were obtained by chemiluminescence and detected in a multi-imager. (C) Quantitative measurements of the PAI-1 protein detected by the Western blots. The signals were normalized to the total amount of protein transferred to the membranes. Each of the graphs represents the average of 2 separate experiments, each analyzed in duplicate (n = 4). Error bars represent SEM; *P < .005 versus respective ambient controls. (D) Determination of soluble PAI-1 protein in the cell culture medium was performed by enzyme-linked immunosorbent assay. Each of the graphs represents values from 2 separate experiments, each determined in triplicate (n = 6). Error bars represent SEM; *P < .001 versus ambient controls.
Kinetics of hypoxia-induced PAI-1 mRNA and protein.
HepG2 cells were cultured at oxygen concentrations of 1%, 2%, 3.5%, 5%, 6.5%, 8%, and 21% for up to 48 hours. At the indicated intervals, RNA, protein, and medium were harvested. (A) Levels of PAI-1 mRNA were determined by real-time RT-PCR. The levels of PAI-1 mRNA were normalized to 18S rRNA within each sample and to the starting values, which were set to 1.0. Each graph represents the average of 2 separate experiments, each analyzed in duplicate on 2 separate occasions (n = 8). Error bars represent SEM; *P < .001 versus respective ambient controls. (B) For each oxygen tension one representative Western blot of PAI-1 is shown. Autoradiographic signals were obtained by chemiluminescence and detected in a multi-imager. (C) Quantitative measurements of the PAI-1 protein detected by the Western blots. The signals were normalized to the total amount of protein transferred to the membranes. Each of the graphs represents the average of 2 separate experiments, each analyzed in duplicate (n = 4). Error bars represent SEM; *P < .005 versus respective ambient controls. (D) Determination of soluble PAI-1 protein in the cell culture medium was performed by enzyme-linked immunosorbent assay. Each of the graphs represents values from 2 separate experiments, each determined in triplicate (n = 6). Error bars represent SEM; *P < .001 versus ambient controls.
To establish whether the increased mRNA levels resulted in any increase in PAI-I protein levels, the intracellular/matrix-associated and soluble levels of PAI-1 protein were determined by immunoblotting (Figures 1B,C) and enzyme-linked immunosorbent assay (Figure 1D), respectively. In the case of cells cultured at 1% and 2% O2, an increase in intracellular and matrix-associated PAI-1 content was evident after 8 hours (Figure 1B), and the levels reached a maximum of an approximately 7-fold increase over the levels detected at ambient air controls after 24 hours of incubation (Figure1C). In the case of cells cultured at oxygen concentrations ranging from 3.5% to 8%, only moderate increases (approximately 2-fold) over to ambient air controls were observed.
In addition, the levels of soluble PAI-1 protein were determined (Figure 1D). The increase in intracellular and matrix-bound PAI-1 protein levels preceded that of soluble PAI-1 by 8 hours. PAI-1 was secreted continuously for the duration of the experiment. After 48 hours of hypoxic treatment at 1% and 2% oxygen, the secreted levels of PAI-1 were approximately 5-fold higher than those for the ambient controls. The cells cultured at 3.5% to 5% secreted twice as much as controls, whereas the secretion of PAI-1 by cells cultured at 6.5% oxygen or more was comparable to that of the control cells.
Identification of the hypoxia-responsive element in the human PAI-1 promotor
Sequence analysis of the human PAI-1 promotor revealed 5 putative HRE motifs, showing homology with the HIF-1 binding consensus sequence BACGTSSK (B = G/C/T, S = G/C, and K = G/T).23 The first potential HIF-1 binding site, HRE-1, at positions −158 to −151, the second site, HRE-2 (positions −194 to −187), and the third site, HRE-3 (positions −453 to −446), shared homology with the consensus sequence in 6 of 8 bases, the fourth site, HRE-4 (positions −566 to −559), in 7 of 8 bases, and, finally, the fifth site, HRE-5 (positions −681 to −674), in all 8 bases (Figure2). Alignment with the rat PAI-1 promoter showed that the HRE-1 site corresponding to the HIF-1 binding site of the rat promoter24 deviated in position 4 from the consensus sequence and that there was complete conservation between the HRE-2 site and the site identified in the rat promoter as an upstream stimulatory factor 2a binding site (Figure 2).25 None of the other motifs shared complete conservation with the rat promoter, however. In fact, motifs 4 and 5 contained the E box sequence CACGTG.
Alignment of the promotor regions of human and rat PAI-1.
The promoter region of human PAI-1, stretching from position −806 to +19 relative to transcription start, was aligned with the homologous sequence from rat. The 5 putative HREs are highlighted in gray, and uppercase letters indicate homology with the canonical HRE sequence BACGTSSK, B = B = G/C/T, S = C/G, and K = G/T.23The rat HIF-1 and USF-2a binding sites are highlighted in black. Finally, the CACAG element that is important for the functionality of various HREs is boxed.29 In the rat sequence * denotes identical sequences, letters indicate substitutions, and deletions are represented by dashes.
Alignment of the promotor regions of human and rat PAI-1.
The promoter region of human PAI-1, stretching from position −806 to +19 relative to transcription start, was aligned with the homologous sequence from rat. The 5 putative HREs are highlighted in gray, and uppercase letters indicate homology with the canonical HRE sequence BACGTSSK, B = B = G/C/T, S = C/G, and K = G/T.23The rat HIF-1 and USF-2a binding sites are highlighted in black. Finally, the CACAG element that is important for the functionality of various HREs is boxed.29 In the rat sequence * denotes identical sequences, letters indicate substitutions, and deletions are represented by dashes.
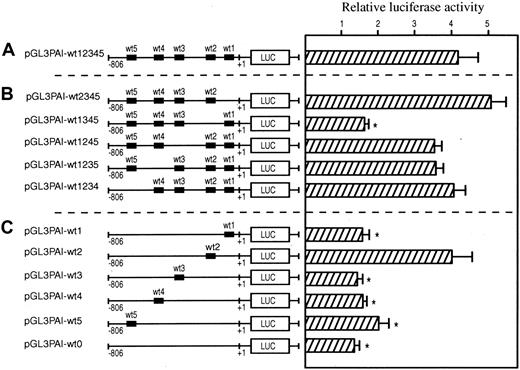
Transfection experiments with the wild-type promoter (Figure3A) and 5 mutant constructs in which the putative HRE motifs were individually mutated revealed that the HRE-2 was necessary for hypoxia-dependent activation (Figure 3B). Mutations of the HRE-1 and -3 motifs did not affect the luciferase activity. Finally, when the HRE-4 or -5 motifs were mutated, the basal levels of luciferase activity were decreased (data not shown). However, this decrease did not affect hypoxia-dependent activation of promoter activity. Transfection experiments in which the mutant constructs contained only one wild-type putative HRE demonstrated that the HRE-2 motif was sufficient to mediate the maximal response (Figure 3C). The transfection experiments with the construct in which all putative HRE motifs were mutated showed no hypoxia-dependent activation response, demonstrating that no other functional HRE was present in the promoter.
Transcriptional activation of human PAI-1 promotor by hypoxia.
The transcriptional activation was measured by a luciferase reporter assay. HepG2 cells were transiently transfected with wild-type and mutated promotor constructs. For each construct wild-type HRE1-5 are indicated. The sequence of the mutations and HREs can be found in Table1. After transfection, the cells were cultured at either 21% or 1% oxygen for 24 hours before the assessment of luciferase activity. In each experiment, luciferase activity of cells cultured in hypoxia was determined relative to that of ambient control cells. Each experiment was performed in triplicate on at least 5 separate occasions. Error bars denote SEM; *P < .05 relative to pGL3PAI-wt12 345. (A) Enhancement of luciferase activity in wild-type promotor construct after incubation at 1% oxygen. (B) Effect of mutations of single HREs on the hypoxia-mediated induction. (C) Capacity of individual wild-type HREs to drive transcriptional expression from the PAI-1 promotor after hypoxic treatment.
Transcriptional activation of human PAI-1 promotor by hypoxia.
The transcriptional activation was measured by a luciferase reporter assay. HepG2 cells were transiently transfected with wild-type and mutated promotor constructs. For each construct wild-type HRE1-5 are indicated. The sequence of the mutations and HREs can be found in Table1. After transfection, the cells were cultured at either 21% or 1% oxygen for 24 hours before the assessment of luciferase activity. In each experiment, luciferase activity of cells cultured in hypoxia was determined relative to that of ambient control cells. Each experiment was performed in triplicate on at least 5 separate occasions. Error bars denote SEM; *P < .05 relative to pGL3PAI-wt12 345. (A) Enhancement of luciferase activity in wild-type promotor construct after incubation at 1% oxygen. (B) Effect of mutations of single HREs on the hypoxia-mediated induction. (C) Capacity of individual wild-type HREs to drive transcriptional expression from the PAI-1 promotor after hypoxic treatment.
Analysis of DNA binding activity
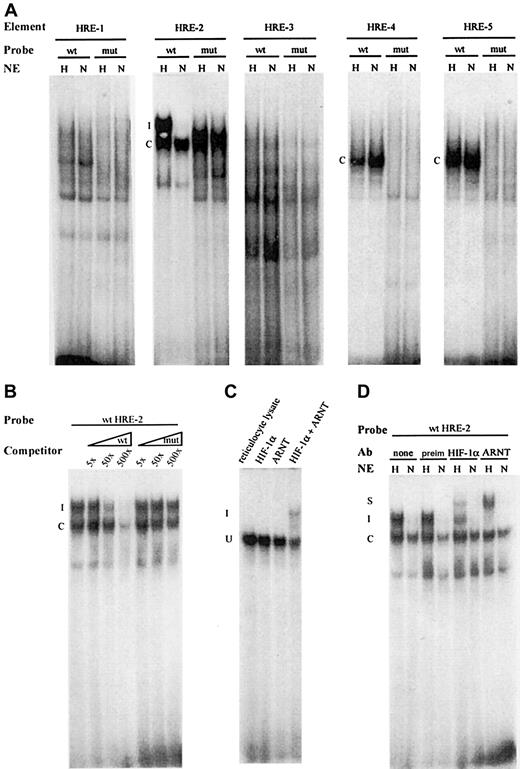
We next performed protein-DNA interaction studies with the sequences spanning the individual HRE motifs. To this end protein-DNA interaction was assessed by EMSA using nuclear extracts obtained from cells incubated at ambient air or under hypoxic conditions (1% O2) for 6 hours. In the case of probes containing the HRE-1, -4, and -5 motifs, a distinctive constitutive, hypoxia-independent complex was formed. This complex was not formed with any of the mutated probes (Figure4A). When the probe containing HRE-2 was incubated with nuclear extract from hypoxic cells, 2 distinct complexes were formed, a constitutive and a hypoxia-inducible one. The hypoxia-dependent complex was not detected on mutation of the core sequence 5′-CACGTACA-3′ to 5′-CTTAATCA-3′, demonstrating that the HRE consensus sequence was essential for hypoxia-induced complex formation. Finally, we observed no complex formation with the probes containing either the wild-type or mutated HRE-3 motifs (Figure 4A). The specificity of the binding was verified by adding increasing amounts of cold probe, which competed for binding of the complex, whereas addition of mutated probe did not influence the formation of the hypoxia-inducible complex (Figure 4B). As the HRE-2 differs from the consensus HIF-1 binding sequence with respect to 2 nucleotides, gel shift experiments were performed by using in vitro–translated HIF-1α and ARNT and wild-type HRE-2 probe. As shown in Figure 4C, when either HIF-1α or ARNT were incubated with the probe, as expected no specific complexes were formed. However, when HIF-1α and ARNT were allowed to dimerize, a specific complex was created. This finding indicated that the HRE-2 sequence was capable of binding the HIF-1α–ARNT heterodimer. Finally, the identity of the hypoxia-dependent DNA-protein complex generated with nuclear extract was confirmed in a supershift analysis (Figure 4D). The addition of antibodies against HIF-1α or ARNT to the binding reaction led to the formation of supershifted complexes, the specificity of which was confirmed when the preimmune serum was used. In conclusion, these results demonstrate that the HRE-2 motif was recognized by the HIF-1α–ARNT complex, in excellent agreement with the ability of the HRE-2 motif to mediate hypoxia-inducible promoter activation in functional assays.
EMSA with the HREs of the human PAI-1 promotor.
Oligonucleotides corresponding to wild-type (wt) or mutated (mut) HREs (sequences are listed in Table 1) were incubated with either nuclear extracts (NE) from normoxic cells (N) or hypoxic cells (H) or from in vitro–translated proteins. (A) NE from H and N cells was incubated with wt and mut probes corresponding to all 5 putative HRE elements of human PAI-1. The constitutive complexes are marked with a C, and the hypoxia-induced complex with an I. (B) For competition assay, radiolabeled wt HRE-2 probe was incubated with NE from hypoxic cells with cold wt or mut probe in 5-, 50-, or 500-fold molar excess, as indicated. (C) Radiolabeled wt HRE-2 probe was incubated with either in vitro–translated HIF-1, ARNT, or both. Rabbit reticulocyte lysate was used as control. (D) For supershift analysis, the wt HRE-2 probe was incubated with NE from H or N cells, as indicated. After initial binding, the preimmune serum, anti–HIF-1 antibody, or anti-ARNT antibody was added. The constitutive complexes are marked C, the inducible I, and the supershifted S.
EMSA with the HREs of the human PAI-1 promotor.
Oligonucleotides corresponding to wild-type (wt) or mutated (mut) HREs (sequences are listed in Table 1) were incubated with either nuclear extracts (NE) from normoxic cells (N) or hypoxic cells (H) or from in vitro–translated proteins. (A) NE from H and N cells was incubated with wt and mut probes corresponding to all 5 putative HRE elements of human PAI-1. The constitutive complexes are marked with a C, and the hypoxia-induced complex with an I. (B) For competition assay, radiolabeled wt HRE-2 probe was incubated with NE from hypoxic cells with cold wt or mut probe in 5-, 50-, or 500-fold molar excess, as indicated. (C) Radiolabeled wt HRE-2 probe was incubated with either in vitro–translated HIF-1, ARNT, or both. Rabbit reticulocyte lysate was used as control. (D) For supershift analysis, the wt HRE-2 probe was incubated with NE from H or N cells, as indicated. After initial binding, the preimmune serum, anti–HIF-1 antibody, or anti-ARNT antibody was added. The constitutive complexes are marked C, the inducible I, and the supershifted S.
Discussion
In the present report we have characterized the mechanism of induction of PAI-1 mRNA and protein expression by hypoxia. We observed that substantial up-regulation of PAI-1 mRNA and protein levels in human liver cells occurs mainly at oxygen concentrations below 2%. Interestingly, the PAI-1 gene was transcriptionally activated shortly after the onset of hypoxia, but the protein synthesis lagged behind the transcriptional up-regulation by several hours. Thus, our data demonstrate high responsiveness of PAI-1 in vitro in a narrow range of oxygen tensions and are in line with the observations in vivo in which the pathophysiologically low oxygen partial tensions found in tumors, during wound healing, and in the ischemic heart disease are accompanied by increased PAI-1 gene expression levels.26-28
To elucidate the molecular mechanisms involved in the hypoxic induction, we analyzed the upstream promoter region of thePAI-I gene. We have identified a functional hypoxia-responsive element, HRE-2, located at positions −194 to −187 upstream of the transcriptional start site. This element was necessary, as well as sufficient, for hypoxia-mediated activation of the PAI-1 promoter. Subsequent analysis by EMSA revealed that a hypoxia-inducible protein complex containing HIF-1α and ARNT interacted with the HRE-2 element. Previously, hypoxia-dependent regulation of expression ofPAI-1 gene has been analyzed in rat hepatocytes. Intriguingly, even mild hypoxia of 8% was capable to induce PAI-1 transcription in rat.24 This finding is discordant with the present study in which more severe hypoxia, less than 2% oxygen, was necessary to induce human PAI-1 expression levels. We have compared the sequences of human and rat PAI-1 promoters (GenBank accession No.X13323 and J05206, respectively) to identify possible factors underlying such a difference. Remarkably, a single substitution, A for G, in the HRE-1 site of human PAI-1 promoter became apparent when compared with the cognate site that binds HIF-1 in rat. Further experiments demonstrated the inability of the HRE-1 element to drive hypoxia-dependent transcriptional activation and to bind any specific hypoxia-inducible protein complex. Thus, a point mutation of the canonical E box motif 5′-CACGTG-3′ in rat to 5′-CACATG-3′ in humans renders the site nonfunctional.
With regard to the human HRE-2 motif, our analysis revealed that this 8 base–long recognition motif 5′-CACGTACA-3′ was identical with the corresponding site present in the rat PAI-I promoter. Yet, the strong hypoxia-dependent induction of transcription and binding of HIF-1 that was observed with human HRE-2 motif is in striking contrast with the only weak hypoxic responses that were described in rat.24In this context, it is noteworthy that there is adjacent, downstream to the human HRE-2, a 5′-CACAG-3′ motif that matches a functionally essential sequence found downstream of the HIF-1-binding site in the human erythropoietin promoter.29 This motif, however, is absent from the rat promoter. Thus, the features associated specifically with the sequence immediately downstream from these particular HREs may provide a basis for the discrepancies in functional properties of the 2 elements from the 2 species.
The HRE-4 and -5 sites were found to bind protein complexes independently of the oxygen partial tension. Consequently, the participation of HIF-1 in formation of these complexes can be excluded. However, each of the sites contained an E box sequence, which has been shown to bind a number of basic helix-loop-helix transcription factors,30 including the ARNT homodimer,31 or the Clock-BMAL heterodimer, that is involved in the control of the circadian expression of the PAI-1 gene.32 Given the fact that the levels of PAI-1 peak in early morning,33concomitantly with low levels of oxygen in the circulation,34 it is plausible that HIF-1 is also involved in the control of the circadian oscillations of PAI-1. Further studies are thus required to elucidate the relative contributions of CLOCK and HIF-1 to this phenomenon.
We thank Professor P. Andreasen, University of Aarhus, Denmark, for the kind gift of the pEMBL8cat plasmid. The technical assistance of Lisbeth Rasmussen, Mette Bøgh, and Hanne Møller is highly appreciated.
Supported by Danish Medical Research council grant no. 9802548 and Danish Cancer Society grant no. 9821560.
P.E. is co-owner of the company that produced the prototype combined workbench-incubator sold to the host institute and used in the present work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Trine Fink, Danish Cancer Society, Department of Virus and Cancer, Gustav Wieds Vej 10, 8000 Århus C, Denmark; e-mail:trine@virus.au.dk.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal