The BCR-ABL fusion, the molecular equivalent of the Philadelphia translocation, gains importance for treatment stratification in adult acute lymphoblastic leukemia (ALL). In this prospective study, samples from 478 patients with CD10+ B-cell precursor ALL (c-ALL and pre-B ALL) underwent BCR-ABL reverse transcription–polymerase chain reaction (RT-PCR) analysis with double testing of positive samples. Patients were stratified according to the PCR result and treated in 2 German Multicenter Trials of Adult ALL. The outcome was followed and the prognostic impact of BCR-ABL was compared to clinical risk features. Of the 478 samples, 432 had an evaluable BCR-ABL result. Thirty-seven percent of the c-ALL and pre-B ALL patients were BCR-ABL+ (p190, 77%; p210, 20%; simultaneous p190/p210, 3%). BCR-ABL positivity was associated with the high-risk features of older age (45 years versus 30 years median age; P = .0001) and higher white blood cell counts (23 500/μL versus 11 550/μL; P = .0001). Univariate and multivariate analyses revealed BCR-ABL as the leading factor for a poor prognosis (P = .0001) in comparison to clinical risk criteria. Irrespective of the breakpoint, presence of any BCR-ABL transcript predicted a lower chance of initial treatment response (68.4% versus 84.6%; P = .001) and a lower probability of disease-free survival at 3 years (0.13 versus 0.47;P = .0001). This bad outcome was not influenced by postinduction high-dose treatment stratifications. The results show a high prevalence of BCR-ABL fusion transcripts with predominance of p190. BCR-ABL RT-PCR is confirmed as a sensitive, rapid method to diagnose t(9;22), and p190 and p210 are unequivocally demonstrated as the most important predictors of poor long-term survival despite intensified chemotherapy.
Introduction
The Philadelphia (Ph) chromosome is the most frequent cytogenetic abnormality known in human leukemias and can be detected in more than 95% of patients with chronic myeloid leukemia (CML), in a range of 20% to 40% of adults with acute lymphoblastic leukemia (ALL), 2% to 5% of children with ALL, and in rare cases of acute myelogenous leukemia.1-6 This t(9;22) translocation leads to a head-to-tail fusion of the abl proto-oncogene from chromosome 9 with the 5′ half of the breakpoint cluster region (bcr) sequences on chromosome 22.7,8 Transcription of bcr-abl results either in a 8.5-kilobase (kb) messenger RNA (mRNA) that codes for a 210-kd protein or in a 7.5-kb mRNA encoding a 190-kd protein.8,9 For the p210 protein, exon b2 or exon b3 of the BCR gene (M-bcr region) is coupled toABL exon 2 (b2a2 or b3a2 junction), whereas the p190-kd protein results from a break within the first intron of BCR(m-bcr region) splicing the first exon of the BCR gene to the second exon of the ABL gene (e1a2).1 Other fusion products are observed at much lower frequencies. BCR-ABL proteins demonstrate enhanced tyrosinase kinase activity compared to the normal 145-kd ABL gene product. Moreover, p190 exhibits a higher transforming potential than p210 in transfection assays and transgenic mouse models.10,11 BCR-ABL expression in hemopoietic cells is known to induce resistance to apoptosis, growth factor independence, as well as alterations in cell-cell and cell-matrix interactions.12-15
The p210 product is the hallmark of CML, whereas p190 and the p210 subtype occur in B-lineage ALL.1,2,5,8,9 Studies on childhood ALL demonstrated that most children carry p190 rather than p210, and that the Ph chromosome is an important negative prognostic factor.3,4 In adult ALL, the occurrence of BCR-ABL positivity is confined to CD10+ B-cell precursor ALL (c-ALL and pre-B ALL precursor).5 The prevalence and the distribution of the various BCR-ABL translocations is less clear in adults, although a correlation with a poor outcome and long-term survival of less than 10% is evident. 2,5,6,16
Whereas mature B-ALL, T-ALL, and pro-B ALL achieved a significant increase in survival, prognosis of overall adult ALL stagnated in the 1990s mainly due to the only slight improvements in c-ALL and pre-B ALLs. Despite Ph+ patients representing the subgroup with the worst prognosis, no prospective evaluation of this high-risk feature has been reported within CD10+ ALL with polymerase chain reaction (PCR) methods.
The goal of this prospective trial was to clarify the prevalence of the BCR-ABL recombination subtypes in c-ALL and pre-B ALL by reverse transcription–PCR (RT-PCR). One further aim was to increase the reliability of PCR by simultaneously evaluating positive probes in a second laboratory. This study within the German Multicenter Trials of Adult ALL (GMALL) also investigated the prognostic significance of the bcr breakpoints and offers the first data on the validity of risk criteria in this subset of ALL patients.
Patients, materials, and methods
Patients and diagnosis
Pretreatment specimens were obtained from 875 adults entering the GMALL trials 04/89 and 05/93 between January 1992 and July 1999. The following analyses were all performed in prospective, controlled procedures and were essential for treatment stratification.
Before start of treatment, the diagnosis of ALL was confirmed at the central cytologic laboratory (H. Loeffler, Kiel, Germany) by evaluating bone marrow and peripheral blood smears according to the French-American-British criteria.17 In addition, immunophenotyping was performed at the GMALL central reference laboratory (E. Thiel, Berlin, Germany); details of the methods used for immunophenotyping and classification have been published elsewhere.18 19 Special attention was paid to CD10-positivity within the B-lineage marker-positive (CD19+, cytoplasmic CD22+) cases, which was controlled by a second antibody in ambiguous cases (5%-20% CD10 positivity). Immunophenotypic subgroups of B-cell precursor ALL were defined as follows: pro-B ALL, TdT+ CD19+CD10− cytoplasmatic (cy) IgM− surface (S) Ig−, c-ALL, TdT+ CD19+CD10+cyIgM− SIg−, pre-B ALL, TdT+ CD19+ CD10+cyIgM+SIg−.
Molecular detection of the BCR-ABL fusion
Prospective PCR testing was performed on cell samples from 478 c-ALL or pre-B ALL patients at diagnosis (04/89, n = 61; 05/93, n = 417). Because only 2 of 96 pro-B ALLs were BCR-ABL+, pro-B ALLs were excluded for clinical follow-up to achieve a homogeneous patient population. After negative results in 30 T-ALL blast cell samples, BCR-ABL PCR was omitted in the remaining cases with T-ALL immunophenotype (n = 215). Immunophenotypically unclassified leukemias (n = 17) and B-ALL (n = 69) were also excluded from BCR-ABL evaluation.
Peripheral blood was accepted for analysis in rare cases in an overt leukemic state. At least 2 samples containing 5 × 106 cells each were used for PCR, which was performed at 2 institutions (Department of Hematology, Oncology, and Transfusion Medicine, Department of Hematology, Berlin, and Institute of Human Genetics, Heidelberg). Total RNA was isolated from fresh or cryopreserved leukemia cells by standard methods. Amplification of either ABL-ABL or BCR-ABL fusion products was performed on complementary DNA (cDNA) reaction mixtures in an automatic thermocycler (Perkin Elmer Biosystems, Foster City, CA) using a nested primer approach (25 cycles each). The 2 laboratories used different PCR protocols2 and oligonucleotide primers (Table1). PCR products were visualized by electrophoresis in 2% agarose gels stained with ethidium bromide.
Primers used for PCR analysis
| Primer . | Sequence . |
|---|---|
| PCR Berlin | |
| RT-PCR primer | 5′-TGATTATAGCCTAAGACCCGGA-3′ (2o, ABL II) |
| 1. PCR round | 5′-TGATTATAGCCTAAGACCCGGA-3′ (2o, ABL II) |
| 5′-ATCTGCCTGAAGCTGGTGGGCT-3′ (1o1, ABL Ia) | |
| 5′-GCAGCAGCCTGGAAAAGTACTT-3′ (1o2, ABL Ib) | |
| 5′-GAAGTGTTTCAGAAGCTTCTCC-3′ (5o, BCR 2) | |
| 5′-ACCATCGTGGGCGTCCGCAAGA-3′ (7o, BCR I) | |
| 2. PCR round | 5′-ACTGAAGCCGCTCGTTGGAACTCCAA-3′ (1i, ABL II) |
| 5′-ATCTCCAGTGGCCAGAAAATCATACA-3′ (2i, ABL II) | |
| 5′-TGGAGCTGCAGATGCTGACCAACTCG-3′ (5i, BCR 2) | |
| 5′-AGATCTGGCCCAACGATGGCGAGGGC-3′ (7i, BCR I) | |
| PCR Heidelberg | |
| RT-PCR | Random hexamers |
| 1. PCR round | 5′-CAGCGGCCAGTAGCATCTGACTTTG-3′ (3o, ABL II) |
| 5′-CCATTTTTGGTTTGGGCTTCACACCATTCC-3′ (4o, ABL III) | |
| 5′-GACGCGTGCAGAGTGGAGGGAGAACATCCGG-3′ (6o, BCR 1) | |
| 5′-ATGGCGAGGGCGCCTTCCAT-3′ (8o/i, BCR I) | |
| 2. PCR round | 5′-GCCTCAGGGTCTGAGTGAAGCCGCTCGTTG-3′ (3i, ABL II) |
| 5′-TGTGATTATAGCCTAAGACCCGGAGCTTTTC-3′ (4i, ABL III) | |
| 5′-GAAGAAGTGTTTCAGAAGCTTCTCC-3′ (6i, BCR 2) | |
| 5′-ATGGCGAGGGCGCCTTCCAT-3′ (8o/i, BCR I) |
| Primer . | Sequence . |
|---|---|
| PCR Berlin | |
| RT-PCR primer | 5′-TGATTATAGCCTAAGACCCGGA-3′ (2o, ABL II) |
| 1. PCR round | 5′-TGATTATAGCCTAAGACCCGGA-3′ (2o, ABL II) |
| 5′-ATCTGCCTGAAGCTGGTGGGCT-3′ (1o1, ABL Ia) | |
| 5′-GCAGCAGCCTGGAAAAGTACTT-3′ (1o2, ABL Ib) | |
| 5′-GAAGTGTTTCAGAAGCTTCTCC-3′ (5o, BCR 2) | |
| 5′-ACCATCGTGGGCGTCCGCAAGA-3′ (7o, BCR I) | |
| 2. PCR round | 5′-ACTGAAGCCGCTCGTTGGAACTCCAA-3′ (1i, ABL II) |
| 5′-ATCTCCAGTGGCCAGAAAATCATACA-3′ (2i, ABL II) | |
| 5′-TGGAGCTGCAGATGCTGACCAACTCG-3′ (5i, BCR 2) | |
| 5′-AGATCTGGCCCAACGATGGCGAGGGC-3′ (7i, BCR I) | |
| PCR Heidelberg | |
| RT-PCR | Random hexamers |
| 1. PCR round | 5′-CAGCGGCCAGTAGCATCTGACTTTG-3′ (3o, ABL II) |
| 5′-CCATTTTTGGTTTGGGCTTCACACCATTCC-3′ (4o, ABL III) | |
| 5′-GACGCGTGCAGAGTGGAGGGAGAACATCCGG-3′ (6o, BCR 1) | |
| 5′-ATGGCGAGGGCGCCTTCCAT-3′ (8o/i, BCR I) | |
| 2. PCR round | 5′-GCCTCAGGGTCTGAGTGAAGCCGCTCGTTG-3′ (3i, ABL II) |
| 5′-TGTGATTATAGCCTAAGACCCGGAGCTTTTC-3′ (4i, ABL III) | |
| 5′-GAAGAAGTGTTTCAGAAGCTTCTCC-3′ (6i, BCR 2) | |
| 5′-ATGGCGAGGGCGCCTTCCAT-3′ (8o/i, BCR I) |
o indicates outer primer; i, inner primer.
All recommended precautions were taken to avoid contaminations. A negative (sterile water) and a positive control were included in each amplification experiment. Also the following precautions were undertaken to ensure the accuracy of results: (1) No cells, no RNA, and no amplified samples were allowed in the room where PCR mixtures were prepared. (2) No amplified samples were brought back into the room where RNA processing was performed. (3) The thermal cyclers were kept in a separate laboratory, away from the room where cell collection and RNA processing were performed.
In the case of a positive BCR-ABL result in the initial PCR, the other laboratory performed a control PCR on a second fraction of the patient's cell probe. BCR-ABL–negative samples were controlled in 120 cases by a second PCR.
Concordant results between 2 different PCR assays were classified as BCR-ABL+. If the laboratories obtained divergent results, PCR assays were repeated and if consistently divergent results were obtained, this PCR result was finally classified as ambiguous. Samples containing no amplifiable RNA were evaluated as insufficient.
Cytogenetic analysis and fluorescence in situ hybridization
Cytogenetic analysis and fluorescence in situ hybridization (FISH) were performed prospectively on 212 cell samples, which were prepared for chromosome analysis and G-banded as described elsewhere.20 Chromosome abnormalities were identified and classified according to the International System for Human Cytogenetic Nomenclature.21,22 FISH was carried out according to a method described elsewhere23 and was applied in part retrospectively on 17 cell samples.
Treatment schedules
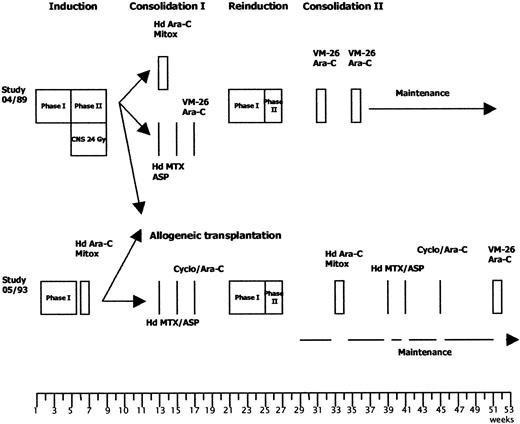
Treatment comprised induction, consolidation I, reinduction, consolidation II, maintenance, and central nervous system (CNS) prophylaxis and has been published elsewhere for the 04/89 study.19,24 Protocols were adapted to risk groups as reviewed by Hoelzer and colleagues25 and Ph+/BCR-ABL+ patients were allocated to the high-risk arm (Figure 1 and Table2).
Treatment strategies for high-risk patients in the GMALL studies 04/89 and 05/93.
The therapy protocols are given in detail in Table 2.
Treatment strategies for high-risk patients in the GMALL studies 04/89 and 05/93.
The therapy protocols are given in detail in Table 2.
Treatment elements for high-risk ALL patients in the GMALL studies 04/89 and 05/93
| Therapy . | Dose . | Route . | Days . |
|---|---|---|---|
| Induction | |||
| Phase I | |||
| Prednisolone | 3 × 20 mg/m2 | PO | 1-28 |
| Vincristine | 2 mg | IV | 1, 8, 15, 22 |
| Daunorubicin | 45 mg/m2 | IV | 1, 8, 15, 22 |
| L-asparaginase | 5000 E/m2 | IV | 15-28 |
| Methotrexate | 15 mg | IT | 1 |
| Phase II | |||
| 04/89 and 05/93 low-risk patients and high-risk patients older than 50 y | |||
| Cyclophosphamide | 650 mg/m2* | IV | 29, 43, 57 |
| Cytarabine | 75 mg/m2 | IV | 31-34, 38-41, 45-48, 52-55 |
| 6-Mercaptopurine | 60 mg/m2 | PO | 29-57 |
| Methotrexate | 15 mg | IT | 31, 38, 45, 52 |
| 05/93 high-risk patients younger than 50 y (wk 6) | |||
| Cytarabine | 3 g/m2 | IV | 1-4 (12 hourly) |
| Mitoxantrone | 10 mg/m2 | IV | 3-5 |
| Reinduction | |||
| Phase I | |||
| Prednisone | 3 × 20 mg/m2 | PO | 1-28 |
| Vincristine | 2 mg | IV | 1, 8, 15, 22 |
| Doxorubicin | 25 mg/m2 | IV | 1, 8, 15, 22 |
| Methotrexate | 15 mg | IT | 1 |
| Cytarabine | 40 mg | IT | 1 |
| Dexamethasone | 4 mg | IT | 1 |
| Phase II | |||
| Cyclophosphamide | 650 mg/m2* | IV | 29 |
| Cytarabine | 75 mg/m2 | IV | 31-34, 38-41 |
| Thioguanine | 60 mg/m2 | PO | 29-42 |
| Methotrexate | 15 mg | IT | 29 |
| Cytarabine | 40 mg | IT | 29 |
| Dexamethasone | 4 mg | IT | 29 |
| Consolidation | |||
| HD cytarabine/mitoxantrone (wk 13 of 04/89; wk 33 of 05/93) | |||
| Cytarabine | 1 g/m2 | IV | 1-4 (12 hourly) |
| Mitoxantrone | 10 mg/m2 | IV | 2-5 |
| HD methotrexate/L-asparaginase (wk 13, 15, 17 of 04/89, wk 13, 15, 39, 41 of 05/93) | |||
| Methotrexate | 1500 mg/m2 | IV | 1 |
| L-asparaginase | 10 000 IU/m2 | IV | 2 |
| 6-Mercaptopurine (wk 13, 15 of 05/93) | |||
| 6-Mercaptopurine | 25 mg/m2 | PO | 1-5 |
| Cyclophosphamide/Ara-C (wk 17, 45 of 05/93) | |||
| Cyclophosphamide | 1000 mg/m2 | IV | 1 |
| Cytarabine | 500 mg/m2 | IV | 1 (24-h infusion) |
| Teniposide/cytarabine (wk 31, 35 of 04/89, wk 51 of 05/93) | |||
| Teniposide | 60 mg/m2 | IV | 1-5 |
| Cytarabine | 75 mg/m2 | IV | Day 1-5 |
| Therapy . | Dose . | Route . | Days . |
|---|---|---|---|
| Induction | |||
| Phase I | |||
| Prednisolone | 3 × 20 mg/m2 | PO | 1-28 |
| Vincristine | 2 mg | IV | 1, 8, 15, 22 |
| Daunorubicin | 45 mg/m2 | IV | 1, 8, 15, 22 |
| L-asparaginase | 5000 E/m2 | IV | 15-28 |
| Methotrexate | 15 mg | IT | 1 |
| Phase II | |||
| 04/89 and 05/93 low-risk patients and high-risk patients older than 50 y | |||
| Cyclophosphamide | 650 mg/m2* | IV | 29, 43, 57 |
| Cytarabine | 75 mg/m2 | IV | 31-34, 38-41, 45-48, 52-55 |
| 6-Mercaptopurine | 60 mg/m2 | PO | 29-57 |
| Methotrexate | 15 mg | IT | 31, 38, 45, 52 |
| 05/93 high-risk patients younger than 50 y (wk 6) | |||
| Cytarabine | 3 g/m2 | IV | 1-4 (12 hourly) |
| Mitoxantrone | 10 mg/m2 | IV | 3-5 |
| Reinduction | |||
| Phase I | |||
| Prednisone | 3 × 20 mg/m2 | PO | 1-28 |
| Vincristine | 2 mg | IV | 1, 8, 15, 22 |
| Doxorubicin | 25 mg/m2 | IV | 1, 8, 15, 22 |
| Methotrexate | 15 mg | IT | 1 |
| Cytarabine | 40 mg | IT | 1 |
| Dexamethasone | 4 mg | IT | 1 |
| Phase II | |||
| Cyclophosphamide | 650 mg/m2* | IV | 29 |
| Cytarabine | 75 mg/m2 | IV | 31-34, 38-41 |
| Thioguanine | 60 mg/m2 | PO | 29-42 |
| Methotrexate | 15 mg | IT | 29 |
| Cytarabine | 40 mg | IT | 29 |
| Dexamethasone | 4 mg | IT | 29 |
| Consolidation | |||
| HD cytarabine/mitoxantrone (wk 13 of 04/89; wk 33 of 05/93) | |||
| Cytarabine | 1 g/m2 | IV | 1-4 (12 hourly) |
| Mitoxantrone | 10 mg/m2 | IV | 2-5 |
| HD methotrexate/L-asparaginase (wk 13, 15, 17 of 04/89, wk 13, 15, 39, 41 of 05/93) | |||
| Methotrexate | 1500 mg/m2 | IV | 1 |
| L-asparaginase | 10 000 IU/m2 | IV | 2 |
| 6-Mercaptopurine (wk 13, 15 of 05/93) | |||
| 6-Mercaptopurine | 25 mg/m2 | PO | 1-5 |
| Cyclophosphamide/Ara-C (wk 17, 45 of 05/93) | |||
| Cyclophosphamide | 1000 mg/m2 | IV | 1 |
| Cytarabine | 500 mg/m2 | IV | 1 (24-h infusion) |
| Teniposide/cytarabine (wk 31, 35 of 04/89, wk 51 of 05/93) | |||
| Teniposide | 60 mg/m2 | IV | 1-5 |
| Cytarabine | 75 mg/m2 | IV | Day 1-5 |
1000 mg/m2 in 05/93.
Induction consisted of two 4-week protocols. Induction phase I included prednisone, vincristine, daunorubicin, and l-asparaginase and was similar for all patients. Induction phase II of the 04/89 study was composed of cyclophosphamide, 6-mercaptopurine, and cytosine arabinoside (ara-C); high-risk patients younger than 50 years from the 05/93 study received high-dose (HD) ara-C (3 g/m2intravenously [IV] every 12 hours, days 1-4) and mitoxantrone (10 mg/m2 IV, days 3-5). In the 04/89 study, high-risk patients were randomized to receive either HD ara-C/mitoxantrone or HD methotrexate (MTX) and asparaginase as consolidation therapy. In the 05/93 study, consolidation combined HD MTX, asparaginase, 6-mercaptopurine (25 mg/m2 orally [PO], days 1-5 and 15-19), ara-C (500 mg/m2 IV, day 29), and cyclophosphamide (1000 mg/m2 IV, day 29). If an HLA-compatible sibling donor was available, high-risk patients were treated with allogenic bone marrow transplantation (BMT) in the first complete remission (CR). Reinduction phase I was performed with prednisone, vincristine, and adriamycin; cyclophosphamide, thioguanine, and ara-C were given during phase II reinduction. Consolidation II treatment in the 04/89 study was based on ara-C and VM-26. In the 05/93 trial, these agents were supplemented for high-risk patients by alternating cycles of HD ara-C/mitoxantrone, HD MTX/asparaginase, and cyclophosphamide/ara-C. Maintenance therapy combined 6-mercaptopurine and MTX starting at week 39 (04/89 study) or week 29 (05/93 study). In the 05/93 study, maintenance therapy was stopped during the consolidation cycles. All patients except the high-risk group in the 05/93 study received 5 intrathecal (IT) injections of MTX and cranial irradiation during induction therapy as well as repeated applications of triple therapy (MTX, dexamethasone, ara-C) during postremission and maintenance therapy.
Statistical analysis
The database was set up at the data center of the GMALL study group (Gesellschaft für Informationsverarbeitung in der Medizin, Munich, Germany) and the statistical analysis was performed at the GMALL study center (Frankfurt, Germany). The median follow-up was 39 months (range, 1-94 months) for surviving patients. The data of the 05/93 study were updated in October 2000. The χ2 test was used to compare the clinical parameters between BCR-ABL+and BCR-ABL− patients and between the different BCR-ABL subtypes. The 2-sided Wilcoxon-Mann-Whitney test was applied for comparison of median values. Survival was defined as the time from the start of therapy to the last follow-up or death. Disease-free survival (DFS) was the time from achievement of CR to relapse or death in remission (event) or date of last follow-up (censored). DFS and survival time were estimated by the Kaplan-Meier method and compared by the log-rank test.26,27 To identify factors of independent value, multivariate analysis of potential prognostic factors was performed using the Cox proportional hazards model.28 The statistical analysis was performed with the SAS program (SAS-PC, Version 6.12; SAS Institute, Cary, NC).
Results
Prevalence of BCR-ABL fusion
Ninety-one percent of the samples (432 of 478) could be analyzed successfully by BCR-ABL RT-PCR with high concordance of the results (4% discordance). BCR-ABL positivity was recorded by both laboratories in 39% of c-ALL (150 of 383) and in 26% of pre-B ALL (25 of 95) leading to a prevalence of 37% BCR-ABL transcripts in the whole group of the CD10+ B-lineage precursor ALL (Table3). For patients with successful central BCR-ABL PCR diagnostics, a prevalence of 41% BCR-ABL+ patients was evident.
Results of prospective BCR-ABL PCR analysis in pre-B ALL and c-ALL patients of the GMALL studies 04/89 and 05/93
| PCR result . | BCR-ABL transcript . | c-ALL n = 383 (%) . | Pre-B ALL n = 95 (%) . | Total N = 478 (%) . |
|---|---|---|---|---|
| BCR-ABL− | 198 (52) | 59 (63) | 257 (54) | |
| BCR-ABL+ | 150 (39) | 25 (26) | 175 (37) | |
| p190 | 116 (77) | 19 (76) | 135 (77) | |
| p210 | 30 (20) | 4 (16) | 34 (20) | |
| p190/p210 | 4 (3) | 2 (8) | 6 (3) | |
| Discordant results | 16 (4) | 2 (2) | 18 (4) | |
| PCR in one laboratory only | 5 (1) | 2 (2) | 7 (1) | |
| Insufficient material for PCR | 14 (4) | 7 (7) | 21 (4) |
| PCR result . | BCR-ABL transcript . | c-ALL n = 383 (%) . | Pre-B ALL n = 95 (%) . | Total N = 478 (%) . |
|---|---|---|---|---|
| BCR-ABL− | 198 (52) | 59 (63) | 257 (54) | |
| BCR-ABL+ | 150 (39) | 25 (26) | 175 (37) | |
| p190 | 116 (77) | 19 (76) | 135 (77) | |
| p210 | 30 (20) | 4 (16) | 34 (20) | |
| p190/p210 | 4 (3) | 2 (8) | 6 (3) | |
| Discordant results | 16 (4) | 2 (2) | 18 (4) | |
| PCR in one laboratory only | 5 (1) | 2 (2) | 7 (1) | |
| Insufficient material for PCR | 14 (4) | 7 (7) | 21 (4) |
Concerning treatment protocols, in the 04/89 study 30 of 61 (49%) patients were diagnosed BCR-ABL+. In the trial 05/93, 145 of 417 (35%) patients yielded a BCR-ABL fusion transcript.
Prevalence of the different bcr breakpoints
Both subtypes of BCR-ABL transcripts (m-bcr/p190 and M-bcr/p210) were similarly distributed in c-ALL and pre-B ALL (Table4). The p190 variant was present in 77% (135 of 175) and p210 products in 20% (34 of 175) of the BCR-ABL+ patients. Simultaneous occurrence of p190 and p210 was determined in 3% (6 of 175) of the patients. Seventy-one percent of the patients with p210 (M-bcr) expressed b2a2 (n = 24), whereas 26% had b3a2 (n = 8) transcripts. PCR revealed simultaneous amplification of b2a2 and b3a2 in only one sample.
Comparison of PCR results, cytogenetic, as well as FISH analysis
| Cytogenetics . | PCR BCR-ABL+ . | PCR BCR-ABL− . | PCR ambiguous . | PCR ne . |
|---|---|---|---|---|
| Ph+ | 51 | 3 | 0 | 3 |
| Ph− | 16 | 98 | 6 | 3 |
| Ph ne | 8 | 20 | 0 | 4 |
| Cytogenetics . | PCR BCR-ABL+ . | PCR BCR-ABL− . | PCR ambiguous . | PCR ne . |
|---|---|---|---|---|
| Ph+ | 51 | 3 | 0 | 3 |
| Ph− | 16 | 98 | 6 | 3 |
| Ph ne | 8 | 20 | 0 | 4 |
In each lane the number of patients is given.
Ph ne indicates t(9;22) not evaluable; PCR ne, insufficient material.
In the different treatment trials (04/89; 05/93), p190 and p210 fusion transcripts occurred at a similar prevalence as in the total study population.
Cytogenetic detection of the Ph translocation and comparison to the PCR result
Concurrent BCR-ABL PCR and cytogenetic analyses were performed on 212 patients' cell samples (Table 4). PCR failed to obtain an evaluable result in 12 (6%) patients (6 ambiguous, 6 insufficient PCR results), cytogenetics in 28 (13%), and both methods in 4 patients (2%). In the remaining 168 patients, 89% concordant results were obtained by cytogenetics and PCR (n = 149).
Nineteen specimens remained discordant after PCR and karyotyping analysis. Sixteen positive PCR amplificates could not be confirmed by a t(9;22) karyotype. FISH was performed in 6 of these samples and detected a translocation in all cases. One further patient exhibited a t(9;22) at relapse, suggesting the presence of a BCR-ABL fusion also at diagnosis. PCR missed a t(9;22) in 3 patients. This includes one patient with a variant t(9;20;22) and another one in whom retrospectively repeated PCR reactions identified a false-negative initial PCR result.
Cytogenetic evaluation was performed on 6 of 17 cases with an ambiguous PCR result and a t(9;22) was evident in none of these patients. FISH was carried out in one additional, ambiguous ALL cell sample and did not yield a BCR-ABL fusion.
Prevalence of BCR-ABL fusion transcripts in patients without sufficient central BCR-ABL PCR data
Twenty-one ALL cell samples contained fewer than 2 × 105 cells and were considered insufficient for double PCR (Table 3). Seven samples did not yield evaluable RNA in both laboratories and were classified as one laboratory only (Table 3). In 19 of these 28 patients, BCR-ABL PCR was performed at local study centers and central cytogenetic analysis was done in 11 patients. These additional analyses in 6 of 19 evaluable patients yielded a BCR-ABL fusion transcript or a t(9;22) or both. Thus, a prevalence of 32% t(9;22) is evident even in those patients without sufficient cell counts for double PCR.
Clinical characteristics
Clinical characteristics before treatment were available in 432 patients with c-ALL or pre-B ALL with successful BCR-ABL analysis (256 males, 176 females) and are summarized in Table5. The number of patients within 3 age groups (< 20 years, 20-50 years, > 50 years) differed significantly between BCR-ABL+ and BCR-ABL−patients (P = .001) and the median age was significantly higher in the BCR-ABL+ group (45 versus 30 years;P = .0001). BCR-ABL+ patients were also characterized by higher median white blood cell (WBC) counts (23 500/μL versus 11 550/ı̀L; P = .0001), higher median neutrophil counts (2219/μL versus 1080/μL;P = .0001), and hemoglobin values (10.2 g/dL versus 9.2 g/dL; P = .004) although platelet counts and blast infiltration were similar in both BCR-ABL+ and BCR-ABL− patients. Pretreatment complications like bleeding and infection occurred at similar rates in the BCR-ABL+ and BCR-ABL− groups. Concerning the immunophenotype, BCR-ABL+ ALL blasts expressed at a higher level CD34 (94% versus 66%; P = .001) and myeloid antigens (CD13, 27% versus 14%, P = .001; CD33, 27% versus 18%, P = .018). Organomegaly was more often observed in the BCR-ABL− patients (hepatomegaly, 43% versus 34%; splenomegaly, 54% versus 45%), but significance was not reached. Patients with p190 or a p210 fusion subtype showed no difference in clinical presenting features apart from a statistically significant higher median age in the p210+group (51 versus 42 years; P = .01).
Comparison of the clinical and immunologic characteristics of the BCR-ABL+ and BCR-ABL− patients
| Characteristics . | BCR-ABL−5-150 n = 257 (%) . | BCR-ABL+5-150 n = 175 (%) . | P . |
|---|---|---|---|
| Age (y) | |||
| 15-20 | 64 (25) | 8 (5) | .0015-151 |
| 20-50 | 141 (55) | 98 (56) | |
| 50-65 | 52 (20) | 69 (39) | |
| Median | 30 (15-64) | 45 (15-65) | .00015-152 |
| WBC (× 109/L)5-153 | |||
| < 30 000 | 185 (72) | 97 (55) | .0015-155 |
| > 30 000 | 71 (28) | 78 (45) | |
| Median | 11 550 (22-720 000) | 23 500 (1500-3 531 000) | .00015-152 |
| Granulocytes (× 109/L) | |||
| Median | 1080 (0-28 400) | 2219 (0-494 340) | .00015-152 |
| Hemoglobin (g/dL) | |||
| Median | 9.2 (2.5-16) | 10.2 (3-16.3) | .0045-152 |
| Immunophenotype | |||
| CD34 > 20% | 169 (66) | 164 (94) | .0015-155 |
| CD13 > 20%5-154 | 36 (14) | 46 (27) | .0015-155 |
| CD33 > 20%5-159 | 45 (18) | 126 (27) | .0015-155 |
| Characteristics . | BCR-ABL−5-150 n = 257 (%) . | BCR-ABL+5-150 n = 175 (%) . | P . |
|---|---|---|---|
| Age (y) | |||
| 15-20 | 64 (25) | 8 (5) | .0015-151 |
| 20-50 | 141 (55) | 98 (56) | |
| 50-65 | 52 (20) | 69 (39) | |
| Median | 30 (15-64) | 45 (15-65) | .00015-152 |
| WBC (× 109/L)5-153 | |||
| < 30 000 | 185 (72) | 97 (55) | .0015-155 |
| > 30 000 | 71 (28) | 78 (45) | |
| Median | 11 550 (22-720 000) | 23 500 (1500-3 531 000) | .00015-152 |
| Granulocytes (× 109/L) | |||
| Median | 1080 (0-28 400) | 2219 (0-494 340) | .00015-152 |
| Hemoglobin (g/dL) | |||
| Median | 9.2 (2.5-16) | 10.2 (3-16.3) | .0045-152 |
| Immunophenotype | |||
| CD34 > 20% | 169 (66) | 164 (94) | .0015-155 |
| CD13 > 20%5-154 | 36 (14) | 46 (27) | .0015-155 |
| CD33 > 20%5-159 | 45 (18) | 126 (27) | .0015-155 |
In each column is given the number (percent) of patients with a certain clinical feature such as age or WBC within the group of all BCR-ABL− or BCR-ABL+ patients. Prefers to the comparison of each clinical parameter in BCR-ABL− versus BCR-ABL+ patients.
c-ALL and pre-B ALL only.
P by Fisher exact text.
P by Wilcoxon 2-sample test.
Data for 1 patient missing.
P by χ2 test.
Data for 5 patients are missing.
Data for 3 patients are missing.
Treatment response
Of the total, 432 patients were evaluable for treatment response and outcome. A significant difference in the initial response rate according to BCR-ABL status was evident. A complete remission (CR) was achieved in 217 of 257 (84.6%) BCR-ABL− patients and 120 of 175 (68.4%) of BCR-ABL+ patients (P = .001; Table 6); 19 (7%) BCR-ABL− and 32 (18%) of the BCR-ABL+ patients did not respond to induction therapy. One hundred and nine (50%) of the BCR-ABL− and only 20 (17%) of the BCR-ABL+ patients maintained a CR.
Results of induction therapy and outcome of BCR-ABL positive and negative c-ALL and pre-B ALL patients
| . | BCR-ABL− patients n = 257 (%) . | BCR-ABL+ patients n = 175 (%) . | P χ2 . |
|---|---|---|---|
| Results of induction therapy | |||
| CR | 217 (84.6) | 120 (68.4) | .001 |
| Failure | 19 (7) | 32 (18) | |
| Aplasia | 2 (1) | 3 (2) | |
| Early death | 14 (5) | 19 (11) | |
| Withdrawal | 4 (2) | 0 | |
| Not evaluable | 1 (0.4) | 1 (0.6) | |
| Outcome of CR patients | N = 217 (%) | N = 120 (%) | |
| CCR | 109 (50) | 20 (17) | |
| Death in CR | 18 (8) | 26 (21) | |
| Relapse | 90 (42) | 74 (62) |
| . | BCR-ABL− patients n = 257 (%) . | BCR-ABL+ patients n = 175 (%) . | P χ2 . |
|---|---|---|---|
| Results of induction therapy | |||
| CR | 217 (84.6) | 120 (68.4) | .001 |
| Failure | 19 (7) | 32 (18) | |
| Aplasia | 2 (1) | 3 (2) | |
| Early death | 14 (5) | 19 (11) | |
| Withdrawal | 4 (2) | 0 | |
| Not evaluable | 1 (0.4) | 1 (0.6) | |
| Outcome of CR patients | N = 217 (%) | N = 120 (%) | |
| CCR | 109 (50) | 20 (17) | |
| Death in CR | 18 (8) | 26 (21) | |
| Relapse | 90 (42) | 74 (62) |
Early death is death in less than 56 days.
N indicates number of evaluable patients; CCR, continuous CR.
Irrespective of the therapeutic protocol, in both treatment trials the presence of a BCR-ABL fusion predicted (P = .0001) a lower survival (04/89 study, median survival 330 days; 05/93 study, median survival 334 days) in comparison to BCR-ABL− patients (04/89 study, median survival 907 days; 05/93 study, median survival 873 days). In both treatment trials the DFS of BCR-ABL+patients was similar with a median of 245 days (04/89 study) and 255 days (05/93 study). Both studies were therefore combined for further analyses.
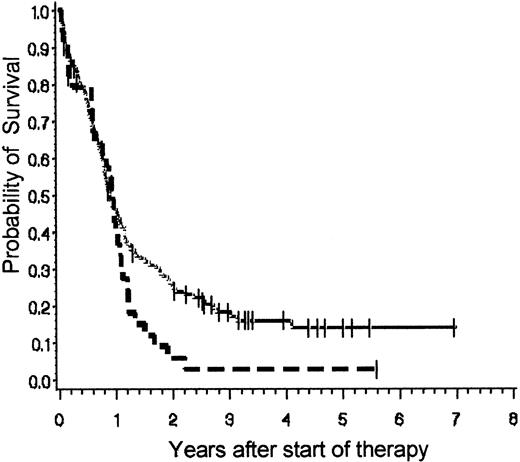
The probabilities of survival and DFS for the whole group of BCR-ABL+ and BCR-ABL− patients are demonstrated in Figure 2A and 2B, respectively. Figure 2C illustrates the DFS excluding patients who underwent BMT. The probability of survival at 3 years after diagnosis accounts for 0.47 (± 0.03 SE) in BCR-ABL− (n = 257; median survival, 905 days) versus 0.15 (± 0.03 SE) in BCR-ABL+ patients (n = 175; median survival, 334 days;P = .0001). Even BCR-ABL+ patients who reached CR (n = 120) have only a 0.19 (± 0.04 SE) survival probability at 3 years in comparison to 0.55 (± 0.04 SE) in BCR-ABL−cases (n = 217; P = .0001) with a median survival of 394 days in BCR-ABL+ and 1585 days in BCR-ABL−patients. The DFS of BCR-ABL+ patients remains markedly low (without BMT at 3 years, 0.06 ± 0.03 SE; with BMT at 3 years, 0.13 ± 0.03 SE), despite a higher percentage of BCR-ABL+CR patients having stem cell transplantation procedures (n = 17 BCR-ABL−; n = 57 BCR-ABL+). However, although this treatment intensification accounted for a higher mortality for BCR-ABL+ patients in remission, there were no long-term survivors after chemotherapy alone in the BCR-ABL+ patients.
Probabilities of overall survival and DFS.
(A) Probability of overall survival of BCR-ABL+patients versus BCR-ABL− patients. Probability of overall survival at 3 years of 175 BCR-ABL+ patients (0.15 ± 0.03 SE) and 257 BCR-ABL− patients (0.47 ± 0.03 SE) treated in the GMALL studies 04/89 and 05/93. Log-rank P = .0001. (B) Probability of DFS of BCR-ABL+ patients versus BCR-ABL− patients including those undergoing BMT. Probability of DFS at 3 years for 120 BCR-ABL+ patients (0.13 ± 0.03 SE) and 217 BCR-ABL− patients (0.47 ± 0.04 SE) treated in the GMALL studies 04/89 and 05/93. Log-rank P = .0001. (C) Probability of DFS survival of BCR-ABL+ patients versus BCR-ABL− patients without including the patients undergoing BMT. Probability of DFS at 3 years for 62 BCR-ABL+ patients (0.06 ± 0.03 SE) and 200 BCR-ABL− patients (0.49 ± 0.04 SE) treated in the GMALL studies 04/89 and 05/93. Log-rank P = .0001. Solid line (top line in each graph) indicates BCR-ABL+; broken line (bottom line in each graph), BCR-ABL−.
Probabilities of overall survival and DFS.
(A) Probability of overall survival of BCR-ABL+patients versus BCR-ABL− patients. Probability of overall survival at 3 years of 175 BCR-ABL+ patients (0.15 ± 0.03 SE) and 257 BCR-ABL− patients (0.47 ± 0.03 SE) treated in the GMALL studies 04/89 and 05/93. Log-rank P = .0001. (B) Probability of DFS of BCR-ABL+ patients versus BCR-ABL− patients including those undergoing BMT. Probability of DFS at 3 years for 120 BCR-ABL+ patients (0.13 ± 0.03 SE) and 217 BCR-ABL− patients (0.47 ± 0.04 SE) treated in the GMALL studies 04/89 and 05/93. Log-rank P = .0001. (C) Probability of DFS survival of BCR-ABL+ patients versus BCR-ABL− patients without including the patients undergoing BMT. Probability of DFS at 3 years for 62 BCR-ABL+ patients (0.06 ± 0.03 SE) and 200 BCR-ABL− patients (0.49 ± 0.04 SE) treated in the GMALL studies 04/89 and 05/93. Log-rank P = .0001. Solid line (top line in each graph) indicates BCR-ABL+; broken line (bottom line in each graph), BCR-ABL−.
Looking at the different BCR-ABL subtypes with regard to prognostic relevance, no clear statistically significant difference for p190 and p210 transcripts was observed, although p210+ patients tended to do worse. CR rates after induction therapy were 67% for p190 (n = 91) versus 74% for p210 (n = 25) and the probability of survival after 3 years was 0.19 (± 0.04 SE) for p190 (n = 135) and 0.03 (± 0.03 SE) for p210 (n = 34; P = .07; Figure3). Although p190+patients in CR (n = 91) had survival estimates after 3 years of 0.23 (± 0.05 SE) compared to 0.04 (± 0.04 SE; P = .07) for p210 transcripts (n = 25), the probability of DFS after 3 years was 0.15 (± 0.04 SE) for p190 (n = 89) and 0.04 (± 0.04 SE) for p210 (n = 24; P = .23) with a median remission duration of 8.3 months for p190 and 7.6 months for p210.
Probability of overall survival of p190+patients versus p210+ patients.
Probability of overall survival at 3 years of 135 p190+, indicated by the top solid line, (patients (0.19 ± 0.04 SE) and 34 p210+ patients, indicated by the bottom broken line, (0.03 ± 0.03 SE) treated in the GMALL studies 04/89 and 05/93. Log-rank P = .07. Median follow-up of 328 days for m-BCR or p190 versus 340 days for M-bcr or p210.
Probability of overall survival of p190+patients versus p210+ patients.
Probability of overall survival at 3 years of 135 p190+, indicated by the top solid line, (patients (0.19 ± 0.04 SE) and 34 p210+ patients, indicated by the bottom broken line, (0.03 ± 0.03 SE) treated in the GMALL studies 04/89 and 05/93. Log-rank P = .07. Median follow-up of 328 days for m-BCR or p190 versus 340 days for M-bcr or p210.
Multivariate analysis
Based on former prognostic factors of the GMALL studies for survival of CR patients in the univariate analysis, that is, time to CR (< or > 4 weeks), WBC count (< or > 30 000/μL), age (< or > 50 years) and BCR-ABL (positive or negative), were selected for multivariate analysis. This evaluation, based on 334 cases, confirmed only the WBC count and especially the BCR-ABL PCR result as independent prognostic factors (Table7).
Univariate and multivariate analysis of the risk factors for the survival of CR patients (n = 334) by the Cox proportional hazard model
| Variable . | Subgroup . | Survival of CR patients . | Univariate analysis P . | Multivariate analysisP . | Multivariate analysis risk ratio . |
|---|---|---|---|---|---|
| BCR-ABL | Negative vs positive | 0.44 vs 0.14 | .0001 | .0001 | 2.766 |
| WBC | < vs > 30/nL | 0.38 vs 0.22 | .006 | .0025 | 1.587 |
| Age | < vs > 50 y | 0.34 vs 0.27 | .01 | .40 | 1.155 |
| CR | < vs > 4 wk | 0.34 vs 0.20 | .04 | .22 | 1.325 |
| Variable . | Subgroup . | Survival of CR patients . | Univariate analysis P . | Multivariate analysisP . | Multivariate analysis risk ratio . |
|---|---|---|---|---|---|
| BCR-ABL | Negative vs positive | 0.44 vs 0.14 | .0001 | .0001 | 2.766 |
| WBC | < vs > 30/nL | 0.38 vs 0.22 | .006 | .0025 | 1.587 |
| Age | < vs > 50 y | 0.34 vs 0.27 | .01 | .40 | 1.155 |
| CR | < vs > 4 wk | 0.34 vs 0.20 | .04 | .22 | 1.325 |
< vs > indicates less versus more than.
Discussion
One aim of this study was to evaluate the prevalence of BCR-ABL positivity prospectively in a large series of adult c-ALL and pre-B ALL patients. Thirty-seven percent of these ALLs were identified as BCR-ABL+ in comparison to our previously published retrospective analysis.2 Thus, we record a similar frequency of BCR-ABL positivity as given by other authors for adult CD-10+ ALL.5 Five percent of the c-ALL and pre-B ALL specimens failed to obtain sufficient blast cell counts, but even in those patients a 30% prevalence of BCR-ABL transcripts or t(9;22) translocations was evident. Because only 2% of the pro-B ALL patients were diagnosed BCR-ABL+ and the vast majority if not all T-ALL and L3-type B-ALL may be assumed to be BCR-ABL−, a calculated frequency of 20% BCR-ABL+ ALL samples can be estimated for the whole adult ALL series and a similar result has been published recently.29 As in childhood ALL,4,30 our data demonstrate the predominance of p190 products (77%) compared to p210 amplificates (20%) as already published elsewhere,2,5although trials with smaller patient cohorts detected a higher prevalence of p210 transcripts.31,32 Looking at the p210+ group, b2a2 transcripts were more frequent than b3a2. Coexpression of p190/p210 was a rare event in our study (1%), but some authors reported a high rate of very low-level p190 in p210+ leukemias by competitive PCR.33 PCR and karyotype analysis agree in 90% of the cases,5,23 34-36which could be confirmed in our study. With the application of double testing, consistently false-positive PCR results were rare and mainly caused by contamination. Despite only very few patients having false-negative PCR results as revealed by cytogenetics and confirmed PCR, ongoing trials continue to apply double testing in all cell samples to achieve the best accuracy.
Analysis by FISH and standard cytogenetics may provide additional information, but karyotype analysis may lead to false-negative results and FISH is hampered by the possibility of false-positive results. Thus, in the context of therapeutic treatment stratification, PCR is the preferred method of BCR-ABL detection and offers higher sensitivity than karyotyping as well as the opportunity to differentiate p190 and p210.
The demonstration of discrete oncogenic potential of the different BCR-ABL transcripts10,11 led to the speculation that the breakpoint region has an impact on clone biology and clinical manifestations. Relating to the breakpoint, there are no differences in presenting features beside the higher age of p210+patients. Despite a trend, statistical significant differences in remission achievement or DFS have not been reached.31Because in our study population an age bias in the p210+patients is not completely ruled out, the evaluation of larger study cohorts will be necessary to clarify prognostic differences of BCR breakpoint regions.
The molecular and clinical findings underscore the similarities of adult and childhood Ph+ ALL and support the view that BCR-ABL+ ALL is a single clinical entity with a wide age range. Our data confirm the hypothesis that BCR-ABL+ ALL in comparison to BCR-ABL− disease represents a subgroup with a worse prognosis within the CD10+ B-lineage ALL. One main difference is the frequency of patients exhibiting a BCR-ABL translocation (37% compared to 3% by children).4Furthermore, in contrast to childhood ALL,4 our data indicate a higher degree of immaturity of BCR-ABL+ blasts as evidenced by the coexpression of CD34 and myeloid antigens. On clinical grounds, in children, prednisone response, age, and leukocyte count at the time of diagnosis predict outcome of BCR-ABL+ALL, but the estimates of DFS for the children with the worst prognosis still exceeds the outcome of BCR-ABL+ adult ALL by far.4
Despite the fact that HD chemotherapy with ara-C and MTX was part of the protocol and a substantial proportion of patients underwent BMT, the overall survival of our BCR-ABL+ adult patients remained markedly low, which is in keeping with recent results.37,38 Our data demonstrate that BCR-ABL positivity is associated with a worse prognosis resulting from a lower initial CR rate and earlier occurrence of relapse. Analysis of prognostic criteria confirmed in our patients the well-established clinical high-risk factors in univariate analyses,25 that is, high WBC count, age above 50 years, and time to achievement of CR. However, the highest level of significance was reached for the BCR-ABL rearrangement and remarkably the multivariate analysis revealed the presence of BCR-ABL as the leading risk factor. Similar to other trials,37 age at disease onset, time to reach CR, and in part WBC count lost their relevance and may be influenced in earlier trials by their correlation to the Ph translocation.25
Biologic and clinical investigations have revealed that ALL is not a uniform disease but is comprised of immunophenotypical and genetical subentities, which differ in their natural history, clinical presentation, and prognosis. Biologic characterization of certain ALL subtypes is one of the major advances in adult ALL. BCR-ABL is revealed as an outstanding poor prognostic factor, which occurs at a high prevalence in c-ALL and pre-B ALL patients and may explain in part the poor outcome of B-lineage ALL in adults.
Thus, in all patients with these immunophenotypes BCR-ABL RT-PCR should be performed to rapidly identify low-risk patients. For BCR-ABL+ patients, so far no improvement of outcome has been achieved even with intensive multiagent chemotherapy including HD ara-C and HD MTX. BMT in first CR from a matched related or unrelated donor may offer the only chance of cure.40,41 Because of the older age of most BCR-ABL+ adults, the option of BMT is not available for substantial numbers of these patients. Thus, new therapeutic approaches are urgently required. Specific inhibitors (STI-571) of BCR-ABL tyrosine kinase activity are now being tested in clinical trials42,43 and may soon be included in front-line therapy of adult Ph+ ALL. Usage of designer ribozymes targeted against BCR-ABL sequences constitutes another promising tool, which is in preclinical evaluation.44 Application of these drugs may hopefully induce higher and longer lasting CR rates and supports the perspective for subtype-adjusted therapy in adult ALL.
We thank C. Bloomfield for kindly reviewing the paper. The excellent technical assistance of A. Sindram, P. Haveman, D. Erz, K. Weidenauer, R. Bielen, E. Strohmaier, A. Wunderlich, U. Spadinger, and H. Kessler is gratefully acknowledged.
Participating institutions include: Aachen: RWTH Klinik (Floege, S. Handt; R. Osieka, U. Fabry); Augsburg: Zentralklinik (D. Renner, G. Schlimock); Bad Saarow: Humaine Klinikum (W. Schultzke, H. Fuss); Berlin: UK Benjamin Franklin (E. Thiel, W.-U. Knauf); Krankenhaus Moabit (K.-P. Hellriegel, H.-H. Fülle); Krankenhaus Neukölln (A. Grüneisen); Campus Virchow-Charité (D. Huhn, Strohscheer); Campus Buch-Charité (B. Dörken, W.-D. Ludwig); Campus Charité (K. Possinger, R. Arnold); St-Hedwig-Krankenhaus (C. Boewer, B. Oldenkott); Bielefeld: Franziskus-Hospital (H.J. Weh, A. Zumsprekel); Bochum: Universitätsklinik (W. Schmiegel, Petrasch); Bonn: Universitätsklinik (T. Sauerbruch, A. Glasmacher); Braunschweig: Klinikum (B. Wörmann, Pies); Bremen: Zentralkrankenhaus (H. Rasche, A. Peyn); Evangelische Diakonissenanstalt (K.-H. Pflüger, T. Wolff); Bremerhaven: St Joseph Hospital (H.-H. Heidtmann, A. Pott); Chemnitz: Klinikum (F. Fiedler, K. Troll); Cottbus: Carl-Thiem-Klinikum (H.B. Steinhauer, C. Rudolph); Dessau: Klinikum (W. Rosahl, B. Seidel); Dortmund: Klinik Mitte (T.U. Hausamen, W. Freund); St Johannes-Hospital (H.-J. Pielken, K. Wietholt); Dresden: Universitätsklinik (G. Ehninger, R. Naumann); Krankenhaus Dresden-Friedrichstadt (H. Nüsslein, J. Steglich); Duisburg: St Johannes-Hospital (C. Aul, J. Anhuf); Johanniter-Krankenhaus (K. Ziegert, R. Lang); Städtische Kliniken (H.Gerhartz, P. Ritter); Düren: Krankenhaus Düren (J. Karow, M. Engels); Düsseldorf: Universitätsklinik (R. Haas, A. Heyll); Erlangen: Universitätsklinik (J. R. Kalden, M. Gramatzki); Eschweiler: St-Antonius-Hospital (R. Fuchs, Thomalla); Essen: Klinik der GHS (U. Dührsen, C. Kasper); Universitätsklinik (S. Seeber, M. R. Nowrousian); Essen-Werder: Evangelisches Krankenhaus (W. Heit, W. Langer); Frankfurt: Universitätsklinik (D. Hoelzer, B. Wassmann, O.G. Ottmann); Krankenhaus Nordwest (A. Knuth, E. Jäger); Freiburg: Universitätsklinik (R. Mertelsmann, W. Digel); Giessen: Universitätsklinik ZIM (H. Pralle, A. Matzdorff); Göttingen: Universitätsklinikum (Brittinger, Martin); Graz: Universitätsklinik (W. Linkesch, F. Bauer); Greifswald: Ernst-Moritz-Arndt-Universität (G. Dölken, M. Schwenke); Gütersloh: Krankenhaus (C. Gropp, R. Depenbusch); Hagen: Katholisches Krankenhaus (H. Eimermacher, W. Kalitzschke); Halle/Saale: Martin-Luther-Universität (H.-J. Schmoll, C. Schöber); Hamburg: Krankenhaus St Georg (R. Kuse, T. Faak); Krankenhaus Altona (D. Braumann, P. Hoelzer), Universitätsklinik Eppendorf (D.K. Hossfeld, M. de Wit); Hameln: Krankenhaus (H. Schmidt); Hamm: Evangelische Krankenhaus (L. Balleisen; A. Grote-Metke); Hannover: Medizinische Hochschule (A. Ganser, H. Diedrich); Krankenhaus Siloah (H. Kirchner, A. Renzelmann); Heidelberg: Universitätsklinik (A. Ho, A. Viardot); Homburg/Saar: Universitätsklinik (M. Pfreundschuh, F. Hartmann); Idar-Oberstein: Klinik für Knochenmarktransplantation (A. A .Fauser, F.-W. Blau); Jena: Universitätsklinik (K. Höffken, H.-J. Fricke); Kaiserslautern: Westpfalz Klinikum (Link, F.-G. Hagmann); Karlsruhe: Klinikum (J. T. Fischer, S. Wilhelm); Kassel: Klinikum (W.-D. Hirschmann, B. Eggeling); Kiel: Universitätsklinik (K. Kneba, C. Pott; U. R. Fölsch, F. Gieseler, Meyer-Alber); Koblenz: Stift St Martin (H.-H. Dormeyer, M. Hoffknecht); Köln: Universitätsklinik (V. Diehl, D. Voliotis); Krefeld: Krankenanstalten (K. Becker, M. Planker); Leipzig: Universitätsklinik (D. Niederwieser, T. Skibbe); Leverkusen: Klinikum (N. Niederle, M. Kress); Lübeck: Krankenhaus Süd (H. Bartels, E.Maass); Universitätsklinik (T. Wagner, F. Heitz); Lüdenscheid: Kreiskrankenhaus (D. Kämpfe, H. Misko); Ludwigshafen am Rhein: Klinikum (M. Uppenkamp, M. Baldus); Magdeburg: Otto-von-Guericke-Universität (A. Franke, K. Jentsch-Ulrich); Mainz: Universitätsklinik (C. Huber, J. Beck); Mannheim: Klinikum (R. Hehlmann, A. Weiss); Marburg: Klinikum (A. Neubauer, B. Reckzeh); Minden: Klinikum (H. Bodenstein, H. Lampe); Mönchengladbach: Krankenhaus Maria Hilf II (H.E. Reis, G. Trenn); München: Krankenhaus München-Schwabing (C. Nerl, T. Lipp); Klinikum Rechts der Isar (C. Peschel, F. Schneller); Universitätsklinik Innenstadt (B. Emmerich, H. Buhmann); Universitätsklinik Groβhadern (R. Hiddemann; W. Kern); Krankenhaus München-Harlaching (R. Hartenstein, N. Brack); Städtisches Krankenhaus München-Neuperlach (M. Garbrecht); Münster: Universitätsklinik (W. E. Berdel, M. Stelljes); Neubrandenburg: Klinikum (H. Rühle, N. Grobe); Offenburg: Klinikum (F. Hirsch, G. Köchling); Oldenburg: Klinikum (H. J. Illiger, B Metzner); Osnabrück: Städtische Kliniken (J. Hartlapp, T. Hegge); Potsdam: St Josefs-Krankenhaus (G. Kautzsch, A. Ruppert); Klinikum Ernst von Bergmann (R. Pasold, R. Rothmann); Regensburg: Universitätsklinik (R. Andreesen, A. Reichle); Rostock: Universitätsklinik (M. Freund, J. Caspar); Saarbrücken: Klinik St Theresia (J. Preiss, P. Schmidt); Sande: Nordwest-Krankenhaus (H. Schönborn, F. K. Natt); Schwäbisch-Hall: Diakonie-Krankenhaus (H. Heissmeyer, T. Geer); Schwerin: Klinikum (R. Subert, D. Hähling); Siegen: St Marienkrankenhaus (W. Gassmann, M. Winkemann); Stralsund: Klinikum (T. H. Ittel); Stuttgart: Robert Bosch-Krankenhaus (W. Aulitzky, B. Löffler); Diakonissenkrankenhaus (E. Heidemann, R. Mück); Bürgerhospital (H.C. Benöhr, W. Grimminger); Katharinenhospital (H.-G. Mergenthaler, H.-M. Reinhold); Trier: Krankenhaus Barmherzige Brüder (C. B. Kölbel, W. Weber); Mutterhaus der Borromäerinnen (M. Clemens, R. Mahlberg); Tübingen: Universitätsklinik (Kanz, M. Mohren); Ulm: Universitätsklinik (H. Döhner, G. Munzert); Wiesbaden: Klinik der Landeshauptstadt (N. Frickhofen, H.-G.Fuhr); Deutsche Klinik für Diagnostik (R. Schwerdtfeger, K. M. Josten); Wilhelmshaven: St Willehad-Hospital (W. Augener); Wuppertal: Klinikum (L. Greiner, H. Hemeling); Würzburg: Universitätsklinik (K. Wilms, M. Wilhelm); Zwickau: Klinikum Heinrich Braun (G. Schott, U. Kreibich, W. Zschille).
Supported by the Deutsche Krebshilfe, Federal Republic of Germany, contract M84/92Hol and the Federal Ministry of Education and Research, Federal Republic of Germany, contract 01ZP88045.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Beate Gleißner, Department of Hematology, Oncology, and Transfusion Medicine, Hindenburgdamm 30, 12200 Berlin, Germany; e-mail: gleisner@zedat.fu-berlin.de.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal