Development of resistance to cytarabine (AraC) is a major problem in the treatment of patients with acute myeloid leukemia (AML). Inactivation of deoxycytidine kinase (dCK) plays an important role in AraC resistance in vitro. We have identified inactive, alternatively spliced dCK forms in leukemic blasts from patients with resistant AML. Because these dCK-spliced variants were only detectable in resistant AML, it was hypothesized that they might play a role in AraC resistance in vivo. In the current study, the biologic role of the alternatively spliced dCK forms in AraC resistance was further investigated by retroviral transductions in rat leukemic cells. Introduction of inactive, alternatively spliced dCK forms into AraC-resistant K7 cells, with no endogenous wild-type (wt) dCK activity, could not restore AraC sensitivity, whereas wt dCK fully restored the AraC-sensitive phenotype. Transfection of alternatively spliced dCK forms into AraC-sensitive KA cells, as well as in human leukemic U937 cells and in phytohemagglutinin-stimulated T cells, did not significantly change sensitivity toward AraC. In addition, cotransduction of wt dCK with alternatively spliced dCK in K7 cells did not result in altered sensitivity to AraC compared with K7 cells only transduced with wt dCK. These data indicate that the alternatively spliced dCK forms cannot act as a dominant-negative inhibitor on dCK wt activity when they are coexpressed in a single cell. However, a cell expressing alternatively spliced dCK forms that has lost wt dCK expression is resistant to the cytotoxic effects of AraC.
Introduction
Cytarabine (1-β-arabinofuranosylcytosine [AraC]), a deoxycytidine (dC) analogue, is the most effective cytotoxic agent in the treatment of acute myeloid leukemia (AML). In the cytoplasm of a cell, AraC is phosphorylated into a triphosphate form (Ara-CTP), which competes with dCTP for incorporation into DNA. Whenever it is incorporated into DNA, it blocks DNA synthesis by inhibiting the function of DNA and RNA polymerases.1,2AraC phosphorylation is catalyzed by 3 different kinases. The most essential step in the AraC activation is phosphorylation into the monophosphate form, which is catalyzed by deoxycytidine kinase (dCK; EC2.7.1.74).3 dCK functions as a 60-kd homodimer, consisting of 2 identical subunits of 30.5 kd each. The gene encoding human dCK consists of 7 exons4 under the control of ubiquitously expressed transcription factors such as Sp1 and E2F.5-7 dCK is expressed in the cytoplasm of most mammalian cells, with highest expression in thymus and T-lymphocyte lineages.8 9
Patients with AML are treated with combination chemotherapy consisting of AraC and an anthracycline antibiotic (eg, daunorubicin, idarubicin), occasionally supplemented by a third drug.10 Combination chemotherapy treatment induces complete remission (CR) in 30% to 80% of patients with previously untreated AML. However, only approximately 30% to 40% of patients who achieve CR have prolonged leukemia-free survival.11 Multiple drug resistance to AraC and anthracyclines is thought to explain the lack of long-term leukemia-free survival in patients with AML.
Resistance to AraC in vitro has primarily been correlated with mutational inactivation of dCK, resulting in a block in the phosphorylation of AraC to AraCTP.12-18 This leads to the inability of AraC to incorporate into DNA. Mutational inactivation of dCK in patients with refractory or relapsed AML is, however, rarely observed,19-21 indicating that a different resistance mechanism might be responsible for AraC resistance in vivo. Recently, we demonstrated the expression of alternatively spliced dCK fragments in coexpression with wild-type (wt) dCK in purified leukemic blasts and phytohemagglutinin (PHA)–stimulated T cells from patients with resistant AML.22 Four different alternatively spliced dCK variants were detected in 7 of 12 purified leukemic blast samples from patients with clinically resistant AML and in 6 of 12 PHA-stimulated T cells, generated from bone marrow (BM) samples from patients with resistant AML. The 4 alternatively spliced dCK variants did show deletions of exon 5, exons 3 to 4, exons 3 to 6, or exons 2 to 6. These spliced variants of dCK code for inactive dCK proteins in vitro, with lower molecular weights. Alternatively spliced dCK forms with deletion exons 2 to 3 and exons 2 to 5 were also detected in an AraC-resistant rat leukemic cell line.22 Aberrant dCK fragments with deletion exon 5 were previously described by others in 2 human AraC-resistant cell lines.12 15 Given that the alternatively spliced dCK forms were not detected in purified leukemic blasts from patients with clinically sensitive AML or in BM and PHA T cells from healthy donors, we hypothesize that these inactive, alternatively spliced dCK forms may contribute to the process of AraC resistance in patients with AML.
In this study we further investigate the biologic role of the alternatively spliced dCK forms in AraC resistance. Sole expression of inactive, alternatively spliced dCK forms probably make a cell resistant to the cytotoxic effects of AraC. This was analyzed by retroviral transduction of human alternatively spliced dCK forms or wt dCK into AraC-resistant rat leukemic cells (K7 cells with no endogenous dCK expression). Because the alternatively spliced dCK forms were always found in coexpression with wt dCK, alternatively spliced dCK forms may also be coexpressed with wt dCK in a single cell. This might result in sequestration of wt dCK monomers from the cell by the formation of heterodimers between alternatively spliced monomers and wt dCK monomers. In this heterodimer, the alternatively spliced dCK forms might function as a dominant-negative inhibitor of wt dCK activity, probably resulting in the reduced expression of active dCK enzyme reflected in decreased sensitivity to AraC. This hypothesis was tested by retroviral transduction of human alternatively spliced dCK forms into AraC-sensitive rat leukemic cells (KA cells with wt dCK expression) and by double transductions of human wt and human alternatively spliced dCK in resistant K7 cells. In addition, alternatively spliced dCK forms were transduced in a human leukemia cell line, U937, and in PHA-stimulated T cells generated from a patient with resistant AML. Obviously, the alternatively spliced dCK forms might not be directly involved in AraC resistance; they may be an epi-phenomenon and not play a primary role in AraC resistance development. In this report we describe the results of AraC sensitivity studies in cells transduced with alternatively spliced dCK forms elucidating the possible biologic role of alternatively spliced dCK forms in AraC resistance.
Materials and methods
Chemicals
Cytarabine (2-chloro-2′-deoxyadenosine, 1-β-D-arabinofuranosylcytosine), adenosine 5′-triphosphate magnesium salt, uridine 5′-triphosphate sodium salt, and bovine serum albumin (BSA) were purchased from Sigma (Sigma Chemical, St Louis, MO). Creatine kinase and creatine phosphate were obtained from Boehringer (Mannheim, Germany), and NaF was obtained from Merck (Darmstadt, Germany).
Cell lines and culture conditions
AraC-sensitive rat leukemic cell line RCL/0 was originally purchased from TNO (Rijswijk, The Netherlands).23AraC-sensitive cell line RO/1 (designated KA in this article) was derived from the RCL/0 cell line by limiting dilution. An AraC-resistant cell line was derived after limiting dilution of an ex vivo–generated AraC-resistant leukemic cell line, designated K7 in this article.24 In this cell line AraC resistance was caused by the deletion of dCK.
Rat leukemic cell lines were cultured in HEPES-buffered RPMI 1640 medium supplemented with 10% fetal calf serum, 4 mM L-glutamine, 50 μg/mL (mM) streptomycin, 50 U/mL penicillin, and 0.5 μg/mL (mM) amphotericin-B. The K7 cell line was cultured in the presence of additional 5% rat serum from brown Norway rats or Wistar rats. The resistant cell line was frequently tested for the resistance phenotype by culturing the cells in the presence of 10−5 M AraC.
Human myelomonocytic leukemic U937 (CRL-1593.2; American Type Culture Collection [ATCC], Rockville, MD) cells were cultured in HEPES-buffered RPMI 1640 medium supplemented with 10% fetal calf serum, 4 mM L-glutamine, 50 μg/mL (mM) streptomycin, 50 U/mL penicillin and 0.5 μg/mL (mM) amphotericin-B.
Generation of PHA-stimulated T cells of a patient with clinically resistant AML
After Ficoll-Hypaque (Sigma, St Louis) density-gradient centrifugation, a bone marrow sample from a patient with clinically resistant AML was thawed and cultured in the presence of 120 U/mL interleukin-2 (IL-2) (Roussel-Uclaf, Paris, France) and 0.8 μg/mL (mM) PHA (Murex Diagnostics, Dartford, United Kingdom) in HEPES-buffered RPMI 1640 medium supplemented with 10% human AB Rh-negative serum, 4 mM L-glutamine, 50 μg/mL (mM) streptomycin, 50 U/mL penicillin, and 0.5 mM amphotericin-B. After 3 days of PHA stimulation, PHA was washed away and stimulated T cells were maintained in medium with fresh IL-2 (120 U/mL).25 T cells were stimulated every 2 weeks with a mixture of irradiated allogeneic peripheral blood mononuclear cells, Epstein-Barr virus–transformed B cells, 0.8 μg/mL (mM) PHA, and 100 U/mL IL-2.
Construction of retroviral vectors and generation of retroviral supernatants
Briefly, the complete coding region of human dCK and different alternatively spliced dCK forms were amplified by nested reverse transcription–polymerase chain reaction (RT-PCR) amplification using human specific dCK primers (A7 × B5 and T5-BamHI × B6-BamHI, Table1). PCR products were sequenced to exclude mutations and were cloned into retroviral vectors. Moloney murine leukemia virus–based retrovirus vector LZRS and packaging cell φ–NX-A were kindly provided by G. Nolan (Stanford University, Palo Alto, CA).26 Two bicistronic retroviral vectors were constructed as described by Heemskerk at al.27 Wild-type dCK was cloned in the pLZRS vector with truncated nerve growth factor receptor (ΔNGF-R) as the marker gene, and cDNA encoding alternatively spliced dCK forms missing exon 5, exons 3 to 4, or exons 3 to 6 were cloned in pLZRS vectors with green fluorescence protein (GFP) as the marker gene. Retroviral vectors encoding for GFP alone were used as control vectors. Constructs were transfected into φ–NX-A cells using calcium phosphate (Life Technologies, Gaithersburg, MD) and were cultured in the presence of 2 μg/mL (mM) puromycin (Clontech Laboratories, Palo Alto, CA). Between 10 and 14 days after transfection, 6 × 106 cells were plated per 10-cm Petri dishes (Becton Dickinson, San Jose, CA), in 10 mL Iscoves modified Dulbecco medium (BioWhittaker Europe, Verviers, Belgium) supplemented with 10% fetal bovine serum without puromycin. After 24 hours, medium was refreshed, and the next day retroviral supernatants were harvested and frozen at −70°C.
PCR primers with corresponding annealing temperatures
| Primer . | 5′-3′ Primer sequence . | Annealing temperature, °C . |
|---|---|---|
| Human dCK | ||
| Forward | ||
| A7 | TCT TTG CCG GAC GAG CTC TG | 65 |
| T5-BamHI | GGA AGG ATC CAC CAT GGC CAC CCC GCC CAA | 70 |
| A6 | TAA GGA ATG GCC ACC CCG CC | 55 |
| Reverse | ||
| B5 | TGG AAC CAT TTG GCT GCC TG | 65 |
| B6-BamHI | CGC TGG ATC CAA GAT CAC AAA GTA CTC AA | 70 |
| B6 | CAA GAT CAC AAA GTA CTC AA | 55 |
| Rat dCK | ||
| Forward | ||
| A10 | ATG GCC ACC CCA CCT AAG AGG TT | 65 |
| T7 | GGA TCC TAA TAC GAC TCA CTA TAG GAA CAG ACC ACC ATG GCC ACC CCA CCT AAG AGG | 55 |
| Reverse | ||
| B5 | TTG CCT GTT GTC TCC TGT GC | 65 |
| B6 | TGC AAT CAC AAA GTA CTC AA | 55 |
| Primer . | 5′-3′ Primer sequence . | Annealing temperature, °C . |
|---|---|---|
| Human dCK | ||
| Forward | ||
| A7 | TCT TTG CCG GAC GAG CTC TG | 65 |
| T5-BamHI | GGA AGG ATC CAC CAT GGC CAC CCC GCC CAA | 70 |
| A6 | TAA GGA ATG GCC ACC CCG CC | 55 |
| Reverse | ||
| B5 | TGG AAC CAT TTG GCT GCC TG | 65 |
| B6-BamHI | CGC TGG ATC CAA GAT CAC AAA GTA CTC AA | 70 |
| B6 | CAA GAT CAC AAA GTA CTC AA | 55 |
| Rat dCK | ||
| Forward | ||
| A10 | ATG GCC ACC CCA CCT AAG AGG TT | 65 |
| T7 | GGA TCC TAA TAC GAC TCA CTA TAG GAA CAG ACC ACC ATG GCC ACC CCA CCT AAG AGG | 55 |
| Reverse | ||
| B5 | TTG CCT GTT GTC TCC TGT GC | 65 |
| B6 | TGC AAT CAC AAA GTA CTC AA | 55 |
Retroviral transduction of leukemic cells with dCK variants
Exponentially growing cells (KA, K7, U937, and PHA T cells) were transduced with retroviral supernatants based on the method described by Hanenberg et al28 with minor modifications, previously described,27 using recombinant human fibronectin fragments CH-296 (RetroNectin; Takara, Otzu, Japan) or 10 mg/mL DOTAP N-[1-(-2,3-Dioleoyloxy)propyl]-N,N,N-trimethylammoniummethylsulfat (Roche, Indianapolis, IN). Briefly, 3 × 106 KA cells were cultured on CH-296–coated Petri dishes together with 1 mL thawed virus supernatant for 6 hours, washed, and transferred to 24-wells culture plates. For DOTAP transfections, 1 mL supernatant was preincubated with 10 mg/mL DOTAP on ice for 10 minutes. Virus–DOTAP mixture was added to 3 × 106 K7 cells, transferred to 24-well plates, and incubated at 37°C.
Three to 5 days after transduction, transduction efficiencies were measured by the expression of the marker genes GFP orΔNGF-R using flow cytometry. ΔNGF-R expression was detected using murine antihuman NGF-R monoclonal antibody (mAb) 20.4 (ATCC). As second antibody, goat antimouse phycoerythrin-labeled polyclonal antibodies (Immunotech, Marseilles, France) were used. In all cases comparable expression levels of the marker genes were observed irrespective of the transduction efficiencies. Transduced cells were purified by fluorescence-activated cell sorter (FACS) analysis on a FACSVantage (Becton Dickinson, Mountain View, CA) on the basis of marker gene expression to obtain pure populations of transduced cells.
Cell viability assays and cell division time
Cell metabolic activity was measured with the cell proliferation reagent WST-1 (Boehringer) to determine IC50 values, which were determined at least 3 different times in triplicate experiments. Cells (5 × 104 cells per 96-well plate) were incubated in triplicate experiments in the presence of different concentrations of AraC at 37°C for 24, 48, and 72 hours. After 20-, 44-, or 68-hour incubation in the presence of AraC, cell viability was analyzed by the addition of 10 μL WST-1 solution. After 2- and 4-hour incubation with WST-1, colorimetric changes were quantified by measuring the absorbance in a spectrophotometer at 450 nm.
Doubling time of the cells was calculated from eosin counting after 24, 48, and 72 hours of exponential growth at 37°C in duplicate experiments. Cell division times were always determined in parallel with cytotoxicity assays.
dCK reverse transcription–polymerase chain reaction
RNA isolations and cDNA synthesis were performed as described previously.24 Briefly, total cellular RNA was isolated from 106 cells by using TRIzol (Gibco BRL, Life Technologies, Gaithersburg, MD) according to the manufacturer's protocol. Two micrograms total RNA was reverse transcribed into single-strand cDNA. The cDNA yield was determined by performing PCR on cDNA derived from the GAPDH housekeeping gene, generating 450 bp.
The amount of cDNA for dCK RT-PCR amplification was standardized to the GAPDH–PCR yield. Full-length rat dCK was amplified in a nested PCR using rat-specific PCR primers. The first PCR was performed with rat primers A10 × B5 and T7 × B6 (Table 1). For the detection of human-specific DNA sequences, human-specific PCR primers (A6 × B6; Table 1) were used that could amplify cDNA sequences cloned into the pLZRS vector. PCR reactions were performed in a reaction mixture containing 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl, pH 8.4, 0.2 mg BSA, 0.25 mM each dNTP, 50 pmol each primer, and 1 U Taq Polymerase (Perkin-Elmer-Cetus, Foster City, CA). PCR was started after denaturation for 5 minutes at 95°C, followed by 30 cycles consisting of 48 seconds at 95°C, 48 seconds at primer-specific annealing temperatures (Table 1), 48 seconds at 72°C, and a final elongation at 72°C for 5 minutes.
dCK activity assay
dCK activities were measured in duplicate experiments using a dCK protocol as originally described by Cheng et al29 with minor modification. Briefly, 107 cells were lysed in 70 μL lysis buffer containing 20 mM Tris-HCl, pH 7.5, 100 mM KCl, 20 mM NaCl, 4 mM MgCl2, 1 mM CaCl2, 2 mM dithiothreitol, and 10% glycerol. Protein concentrations were determined by Bio-Rad protein assay30 (Bio-Rad, Munich, Germany). dCK activity was estimated in 20 μL cellular extracts using 0.01 mM 3H-labeled 2-chlorodeoxyadenosine (2-CdA) as substrate. Duplicate experiments were performed in reaction mixture containing 20 mM Tris-HCl, pH 7.4, 5 mM MgUTP, 27 U/mL creatine phosphokinase, 7.5 mM creatine phosphate, 7.03 × 103Bq/mL 3H-labeled 2-CdA (specific activity, 1.48 × 1011 Bq/mmol), 10 mM unlabeled 2-CdA, 7 mM NaF, 0.2% BSA, and 0.2 mM tetrahydrouridine to block cytidine-deaminase activity. Reactions were initiated by the addition of ± 0.1 mg total protein per reaction (20 μL cell extract) and were incubated at 37°C for 20, 40, and 60 minutes. At each time point, 50-μL aliquots were spotted on DEAE-coated paper discs (Whatman DE-81; Whatman International, Maidstone, United Kingdom). Filters were dried and washed 4 times in 1 mM ammonium formate. Phosphorylated substrates bound to the filters were eluted from the filters by 0.6 M HCl–1.5 M NaCl, and 3H-labeled reaction products were determined by scintillation counting in Atomlight (Packard Bioscience, Groningen, The Netherlands) using an LKB Rackbeta scintillation counter. Enzyme kinetic properties were calculated by linear regression analysis and given in pmol/min × mg total protein.
dCK Western blot analysis
dCK protein expression was detected by Western blot analysis using a wt-specific dCK-pep monoclonal antibody (mAb), which was kindly provided by Prof I. Talianidis (Institute of Molecular Biology and Biotechnology, Heraklion, Greece).8 Western blot analysis was performed as described previously.31 Briefly, 5 × 106 cells were lysed in 100 μL lysis buffer (50 mM Tris-HCl, pH 7.6, 5 mM dithiothreitol, 20% vol/vol glycerol, 0.5% vol/vol Nonidet P40, and 25% vol/vol protease inhibitor cocktail (Boehringer) by freeze-thawing. Protein concentrations were determined by Bio-Rad protein assay.30 Electrophoresis of 30 μg protein was carried out in a 12.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis mini-gel at 100 V for 2 hours. Proteins were transferred to nitrocellulose membranes (0.45 μm; Bio-Rad) and were blocked overnight in 1% enhanced chemiluminescence-blocking reagent (BM Chemiluminescence Blotting Substrate; Boehringer). dCK protein was detected by staining with dCK-pep mAb (1:5000) for 2 hours in blotting buffer containing 10 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 0.05% Tween-20, followed by horseradish peroxidase–conjugated anti–rabbit immunoglobulin G (1:40 000; Promega, Madison, WI). Immunocomplexes were visualized by chemiluminescence reaction using BM Chemiluminescence Blotting Substrate (Boehringer) and were detected on Fuji Super RX film.
Statistical analysis
Statistical differences were established by the Wilcoxon rank sum test for unpaired data (Mann-Whitney U test).
Results
Generation of rat leukemic cell lines expressing human wild-type dCK or alternatively spliced dCK
Amphotropic retroviral producer lines from the packaging cell line φ–NX-A were established after calcium phosphate transfections with pLZRS-wt dCK-IRES-ΔNGFR, pLZRS-deletion exon 5 dCK-IRES-GFP, pLZRS-deletion exons 3-4 dCK-IRES-GFP, and pLZRS-deletion exons 3-6 dCK-IRES-GFP. Transduced cells were enriched by FACS sorting on the basis of ΔNGFR or GFP marker gene expression. More than 97% positive cell populations were obtained after 1 or 2 rounds of FACS sorting.
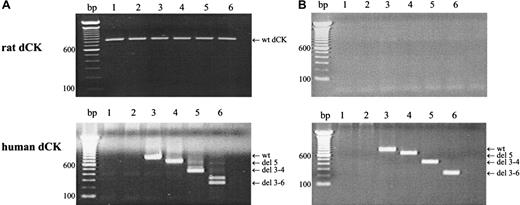
Target gene expression in the different transduced cell lines was analyzed by RT-PCR analysis, using human- or rat-specific dCK PCR primers. As can be seen in Figure 1, wt rat dCK was observed in the nontransduced cell line KA and in transduced KA cells (Figure 1A, upper figure). No rat dCK could be demonstrated in the AraC-resistant K7 cells or in retrovirally transduced K7 cells (Figure 1B, upper panel). PCR amplifications using human-specific dCK primers, confirmed wt human dCK expression in cells transduced with pLZRS-wt dCK-IRES-ΔNGFR. Human spliced variants of dCK were observed in cells that were transduced with alternatively spliced dCK variants (Figure 1A-B, lower panel).
Target gene expression in retrovirally transduced cells.
Target gene expression after retroviral transfection was analyzed by RT-PCR amplifications using rat-specific (upper panel) or human-specific (lower panel) dCK PCR-primers. (A) KA cells. (B) K7 cells. Lane 1, parental cells; lane 2, cells transduced with empty vector controls; lane 3, cells transduced with human wt dCK; lane 4, cells transduced with deletion exon 5 human dCK; lane 5, cells transduced with deletion exons 3 to 4 human dCK; lane 6, cells transduced with deletion exons 3 to 6 dCK. PCR fragments were separated on a 1.5% agarose gel and visualized by ethidium bromide staining.
Target gene expression in retrovirally transduced cells.
Target gene expression after retroviral transfection was analyzed by RT-PCR amplifications using rat-specific (upper panel) or human-specific (lower panel) dCK PCR-primers. (A) KA cells. (B) K7 cells. Lane 1, parental cells; lane 2, cells transduced with empty vector controls; lane 3, cells transduced with human wt dCK; lane 4, cells transduced with deletion exon 5 human dCK; lane 5, cells transduced with deletion exons 3 to 4 human dCK; lane 6, cells transduced with deletion exons 3 to 6 dCK. PCR fragments were separated on a 1.5% agarose gel and visualized by ethidium bromide staining.
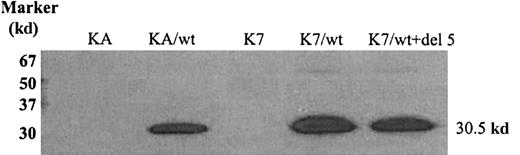
Because antibodies recognizing alternatively spliced dCK variants are unavailable, we were only able to demonstrate wt human dCK protein expression in AraC-sensitive and -resistant cells transduced with wt human dCK. As can been seen in Figure 2, wt dCK was properly translated into proteins of 30.5 kd in the KA and the K7 cell lines transduced with human wt dCK.
Wild-type human dCK protein detection by Western blot analysis.
Proteins (30 μg) were separated by 12.5% SDS-PAGE and transferred to nitrocellulose membranes. Human dCK proteins were detected by staining with human dCK-pep mAb.
Wild-type human dCK protein detection by Western blot analysis.
Proteins (30 μg) were separated by 12.5% SDS-PAGE and transferred to nitrocellulose membranes. Human dCK proteins were detected by staining with human dCK-pep mAb.
dCK activity in retrovirally transduced cells
dCK activity was analyzed in cellular extracts of retrovirally transduced rat leukemic cell lines resistant or sensitive to AraC. Results of dCK activity measurements are presented in Table2. Introduction of human wt dCK into AraC-sensitive KA cells increased dCK activity by a factor 50 compared with the nontransduced KA cells (201.5 ± 10.9 pmol/min × mg total protein versus 4.15 ± 1.37 pmol/min × mg total protein, respectively). A 2-fold increase in dCK activity was observed in KA cells transduced with alternatively spliced dCK variants (mean dCK activity in transduced cells, 9.36 ± 1.93 pmol/min × mg total protein versus 4.15 ± 1.37 pmol/min × mg total protein for the nontransduced KA cell line). A similar 2- to 3-fold increase in dCK activity was detected in cells transduced with control vector (GFP) (11.6 ± 0.99 pmol/min × mg total protein versus 4.15 ± 1.37 pmol/min × mg total protein, respectively).
dCK activities in retrovirally transduced rat leukemic cell lines
| Cells . | dCK activity . |
|---|---|
| KA | 4.15 ± 1.37 |
| KA/GFP | 11.6 ± 0.99 |
| KA/wt | 201.5 ± 10.9 |
| KA/del 5 | 9.81 ± 3.95 |
| KA/del 3-4 | 9.09 ± 3.27 |
| KA/del 3-6 | 6.94 ± 1.64 |
| K7 | — |
| K7/GFP | — |
| K7/wt | 941.3 ± 207.6 |
| K7/del 5 | — |
| K7/del 3-4 | — |
| K7/del 3-6 | — |
| Cells . | dCK activity . |
|---|---|
| KA | 4.15 ± 1.37 |
| KA/GFP | 11.6 ± 0.99 |
| KA/wt | 201.5 ± 10.9 |
| KA/del 5 | 9.81 ± 3.95 |
| KA/del 3-4 | 9.09 ± 3.27 |
| KA/del 3-6 | 6.94 ± 1.64 |
| K7 | — |
| K7/GFP | — |
| K7/wt | 941.3 ± 207.6 |
| K7/del 5 | — |
| K7/del 3-4 | — |
| K7/del 3-6 | — |
dCK activities were measured in cellular extracts using 0.01 mM3H-labeled 2-CdA as substrates in duplicate experiments. dCK activities are given in pmol/min × mg total protein (mean ± SD).
In AraC-resistant K7 cells with no detectable endogenous dCK activity, high dCK activity was restored after transduction with human wt dCK (dCK activity 941.3 ± 207.6 pmol/min × mg total protein). More important, no dCK activity could be measured in K7 cells transduced with different alternatively spliced variants of dCK.
AraC sensitivity of retrovirally transduced rat leukemic cells
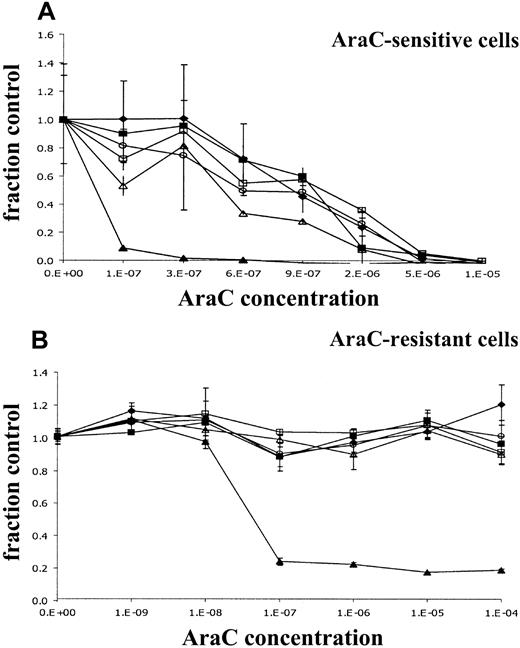
We analyzed the effects of the introduction of different dCK spliced variants on sensitivity for AraC by WST-1 assay. Metabolic activities were measured in cells exposed for 24, 48, and 72 hours to increasing concentrations of AraC. As can be seen in Figure3A, the incubation of KA cells in the presence of increasing concentrations of AraC resulted in decreased metabolic activity. Transfection of KA cells with human wt dCK highly increased AraC sensitivity after 72-hour AraC incubation. This increased sensitivity for AraC resulted in a decrease of the AraC IC50 concentration by a factor 18 after 72 hours of AraC incubation compared with the nontransduced cell line KA (IC50 concentration 0.69 ± 0.26 μM for the nontransduced cell line KA and 0.037 ± 0.007 μM for KA/wt dCK; Table 3). Introduction of 1 of 3 alternatively spliced dCK forms did change the sensitivity to AraC by a factor 2 (Table 3). However, a similar increase in IC50concentration was observed in KA cells transduced with empty vector (KA/GFP).
Dose-response curves of retrovirally transduced rat leukemic cell lines.
Metabolic activities of the cells in the presence of increasing concentrations of AraC for 72 hours was analyzed by the cell proliferation assay WST-1. Y-axis (fraction control) represents the metabolic activity of the cells in the presence of AraC divided by the metabolic activity of cells grown in the absence of AraC. (A) Retrovirally transduced KA cells. (B) Retrovirally transduced K7 cells. (♦) untransduced control cells; (▪) cells transduced with empty vectors; (▴) cells transduced with wt dCK; (○) cells with deletion exon 5 dCK; (▵) cells with deletion exons 3 to 4 dCK; (■) cells with deletion exons 3 to 6.
Dose-response curves of retrovirally transduced rat leukemic cell lines.
Metabolic activities of the cells in the presence of increasing concentrations of AraC for 72 hours was analyzed by the cell proliferation assay WST-1. Y-axis (fraction control) represents the metabolic activity of the cells in the presence of AraC divided by the metabolic activity of cells grown in the absence of AraC. (A) Retrovirally transduced KA cells. (B) Retrovirally transduced K7 cells. (♦) untransduced control cells; (▪) cells transduced with empty vectors; (▴) cells transduced with wt dCK; (○) cells with deletion exon 5 dCK; (▵) cells with deletion exons 3 to 4 dCK; (■) cells with deletion exons 3 to 6.
IC50 concentrations of retrovirally transduced rat leukemic KA cells
| Cells . | IC50 concentration, μM . |
|---|---|
| KA | 0.69 ± 0.26 |
| KA/GFP | 1.73 ± 0.99 |
| KA/wt | 0.037 ± 0.007 |
| KA/del 5 | 1.53 ± 1.38 |
| KA/del 3-4 | 1.67 ± 1.32 |
| KA/del 3-6 | 1.47 ± 1.05 |
| Cells . | IC50 concentration, μM . |
|---|---|
| KA | 0.69 ± 0.26 |
| KA/GFP | 1.73 ± 0.99 |
| KA/wt | 0.037 ± 0.007 |
| KA/del 5 | 1.53 ± 1.38 |
| KA/del 3-4 | 1.67 ± 1.32 |
| KA/del 3-6 | 1.47 ± 1.05 |
IC50 values (mean ± SD) were determined by 3 independent WST-1 assays, each performed in triplicate experiments, after 72-hour AraC incubation.
In the AraC-resistant K7 cell line, no reduced metabolic activity was measured at high concentrations of AraC (up to 100 μM AraC) (Figure3B). No change in the resistance phenotype of the cells could be detected in cells transduced with human alternatively spliced dCK variants or in the cells transduced with control vector. Introduction of human wt dCK restored sensitivity to AraC at the same levels observed in the KA cells transduced with human wt dCK (IC50concentration, K7 > 100 μM; IC50 concentration, K7 + wt dCK 0.040 ± 0.029 μM; IC50 concentration KA/wt dCK 0.037 ± 0.007 μM; Table 4; Figure 3B). Similar results were observed after determining3H-thymidine incorporation in untransduced cells or transduced KA and K7 cells exposed to increasing concentrations of AraC for 24, 48, and 72 hours (data not shown).
IC50 concentrations of retrovirally transduced rat leukemic K7 cells
| Cells . | IC50 concentration, μM . |
|---|---|
| K7 | > 100 |
| K7/GFP | > 100 |
| K7/wt | 0.040 ± 0.029 |
| K7/del 5 | > 100 |
| Kt/del 3-4 | > 100 |
| Kt/del 3-6 | > 100 |
| Cells . | IC50 concentration, μM . |
|---|---|
| K7 | > 100 |
| K7/GFP | > 100 |
| K7/wt | 0.040 ± 0.029 |
| K7/del 5 | > 100 |
| Kt/del 3-4 | > 100 |
| Kt/del 3-6 | > 100 |
IC50 values were determined by 4 independent WST-1 assays, each performed in triplicate experiments. IC50concentrations are presented as mean ± SD.
Because AraC is S-phase specific, we determined the cell division times in parallel with AraC sensitivity assays. No changes in cell-doubling times could be observed among cells that were or were not retrovirally transduced (16.8 ± 2.7 hours versus 16.9 ± 2.8 hours for KA and transduced KA cells, respectively, and 15.5 ± 1.3 hours versus 17.5 ± 1.8 hours for K7 and transduced K7 cells, respectively).
Cotransduction of wild-type dCK and deletion exon 5 alternatively spliced dCK
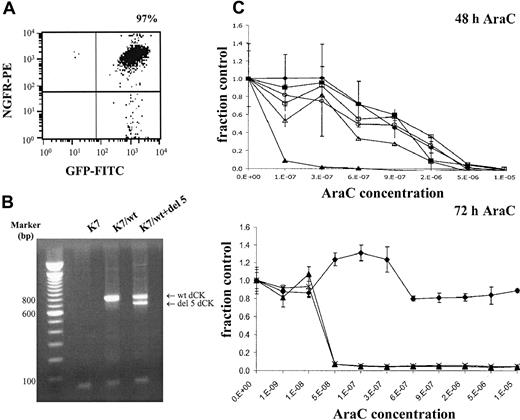
To investigate the possible dominant-negative effect of alternatively spliced dCK fragments on wt dCK activity, K7/wt cells were cotransduced with deletion exon 5 dCK spliced variant. Cotransduced cells were FACS sorted on the basis of ΔNGFR and GFP expression, which resulted in 97% pure populations (Figure4A). Target gene expression was analyzed by RT-PCR using human-specific dCK primers. Wild-type human dCK and deletion exon 5 constructs were equally expressed in these cells (Figure 4B).
Double-transduced K7 cells with wt dCK and deletion exon 5 dCK.
(A) Transduced cells were enriched by FACS sorting on the basis of truncated NGFR and GFP as marker genes. (B) Target gene expression was determined by RT-PCR analysis using human-specific dCK PCR primers. PCR fragments were separated on 1.5% agarose gels and visualized by ethidium bromide staining. (C) Dose-response curves were generated by WST-1 assays after 48 and 72 hours of AraC incubation. Metabolic activities of the cells in the presence of increasing concentrations of AraC (x-axis) were analyzed by the cell proliferation assay WST-1. Y-axis (fraction control) represents the metabolic activity of the cells in the presence of AraC divided by the metabolic activity of cells grown in the absence of AraC. (♦) untransduced K7 cells; (▴) K7/wt cells; and (×) K7/wt+del 5 double-transduced cells.
Double-transduced K7 cells with wt dCK and deletion exon 5 dCK.
(A) Transduced cells were enriched by FACS sorting on the basis of truncated NGFR and GFP as marker genes. (B) Target gene expression was determined by RT-PCR analysis using human-specific dCK PCR primers. PCR fragments were separated on 1.5% agarose gels and visualized by ethidium bromide staining. (C) Dose-response curves were generated by WST-1 assays after 48 and 72 hours of AraC incubation. Metabolic activities of the cells in the presence of increasing concentrations of AraC (x-axis) were analyzed by the cell proliferation assay WST-1. Y-axis (fraction control) represents the metabolic activity of the cells in the presence of AraC divided by the metabolic activity of cells grown in the absence of AraC. (♦) untransduced K7 cells; (▴) K7/wt cells; and (×) K7/wt+del 5 double-transduced cells.
dCK activity in double-transduced cells was determined on cellular extracts and compared with K7 cells only transduced with human wt dCK expression. No difference in dCK activity could be observed between these 2 cell lines, as can be seen in Table5 (dCK activity 941.3 ± 207.6 pmol/min × mg for K7/wt cells and 1007.5 ± 483.9 pmol/min × mg for K7/wt+del 5 cells).
dCK activity of double-transduced K7 cells
| Cells . | dCK activity . |
|---|---|
| K7 | — |
| K7/wt | 941.3 ± 207.6 |
| K7/wt + del 5 | 1007.5 ± 483.9 |
| Cells . | dCK activity . |
|---|---|
| K7 | — |
| K7/wt | 941.3 ± 207.6 |
| K7/wt + del 5 | 1007.5 ± 483.9 |
dCK activities were determined in cellular extracts in duplicate experiments using 0.01 mM 3H-labeled 2-CdA as a substrate. dCK activities are given in pmol/min × mg (mean ± SD).
Sensitivity of double-transduced cells (K7/wt+del 5 cells) for AraC (Figure 4C) was analyzed in WST-1 assays, as previously described. IC50 concentrations of the K7 cell line and transduced K7 cells (K7/wt and K7/wt+del 5 cells) are presented in Table 6. A modest, nonsignificant decrease in AraC sensitivity was detected in the double-transduced cells after 48 hours of AraC incubation (P = .40). This minor decreased sensitivity was abolished, however, after 72-hour incubation with increasing AraC concentrations. After 72-hour AraC incubation, no significant decrease in AraC sensitivity was observed in the K7/wt+del 5 double-transduced cells compared with K7/wt cells (P = .56). The reduction of metabolic activity in the presence of increasing concentrations of AraC was similar in K7/wt cells and the double-transduced cells K7/wt+del 5 (Figure 4C). In addition, no changes in cell division times were observed during the WST-1 assays (mean doubling time, 15.5 ± 1.3 hours for K7 cells, 16.8 ± 2.4 hours for K7/wt cells, and 16.8 ± 2.0 hours for double-transduced cells K7/wt+del 5).
IC50 concentrations of double-transduced K7 cells
| AraC incubation . | IC50 (μM) . | |
|---|---|---|
| 48 h . | 72 h . | |
| K7 | > 100 | > 100 |
| K7/wt | 0.032 ± 0.0116-150 | 0.040 ± 0.0296-151 |
| K7/wt + del 5 | 0.078 ± 0.078 | 0.029 ± 0.009 |
| AraC incubation . | IC50 (μM) . | |
|---|---|---|
| 48 h . | 72 h . | |
| K7 | > 100 | > 100 |
| K7/wt | 0.032 ± 0.0116-150 | 0.040 ± 0.0296-151 |
| K7/wt + del 5 | 0.078 ± 0.078 | 0.029 ± 0.009 |
IC50 concentrations (presented as mean ± SD) of double-transduced cells and untransduced cells were calculated from 3 independent WST-1 assays, each performed in duplicate.
P = .40.
P = .56.
Effects of alternatively spliced dCK forms on AraC sensitivity in human leukemic cells
To investigate the influence of the alternatively spliced human dCK forms on AraC sensitivity in human cells, human myelomonocytic leukemic cells (U937) were transduced with different alternatively spliced forms of dCK and human wt dCK. Wild-type dCK protein expression was detected by Western blot analysis in all transduced cells, with highly increased wt dCK expression in U937 cells transduced with human wt dCK (data not shown). Endogenous dCK activity was high in nontransduced U937 cells (158 ± 6 pmol/min × mg total protein; Table 7), as determined in cellular extracts. No major changes in dCK activity were observed when cells were transduced with empty vector or alternatively spliced dCK forms, whereas transduction of human wt dCK increased dCK activity more than 6 times (Table 7). Introduction of alternatively spliced dCK forms did not change the IC50 concentration for AraC compared with nontransduced cells. Unexpectedly, the overexpression of human wt dCK in U937 with endogenous human dCK expression did not increase the sensitivity for the cytotoxic effects of AraC in these cells compared with U937 cells (Table 7). No major changes in cell division times between nontransduced U937 cells and transduced U937 cells could be observed by eosin counting (32.7 ± 2.2 hours versus 27.7 ± 3.5 hours for U937 and transduced U937 cells, respectively).
dCK activities and IC50 concentrations in retrovirally transduced U937 cells
| Cells . | dCK activity . | IC50, μM . |
|---|---|---|
| U937 | 158 ± 0.006 | 2.97 ± 2.23 |
| U937/GFP | 135 ± 0.002 | 4.39 ± 2.70 |
| U937/wt | > 1000 | 2.42 ± 1.96 |
| U937/del 5 | 110 ± 0.002 | 4.05 ± 3.40 |
| U937/del 3-4 | 142 ± 0.008 | 2.94 ± 1.79 |
| U937/del 3-6 | 124 ± 0.000 | 2.69 ± 2.06 |
| Cells . | dCK activity . | IC50, μM . |
|---|---|---|
| U937 | 158 ± 0.006 | 2.97 ± 2.23 |
| U937/GFP | 135 ± 0.002 | 4.39 ± 2.70 |
| U937/wt | > 1000 | 2.42 ± 1.96 |
| U937/del 5 | 110 ± 0.002 | 4.05 ± 3.40 |
| U937/del 3-4 | 142 ± 0.008 | 2.94 ± 1.79 |
| U937/del 3-6 | 124 ± 0.000 | 2.69 ± 2.06 |
dCK activities (pmol/min × mg) were calculated from dCK phosphorylation assays using 0.01 mM 2-CdA as a substrate and were performed in duplicate experiments. IC50 concentrations for AraC were calculated from 3 independent WST-1 assays, each performed in triplicate experiments and presented as mean ± SD.
Discussion
Resistance to chemotherapy is a major problem in the treatment of patients with AML. The exact mechanisms of acquired resistance to the cytotoxic effects of AraC in patients with resistant AML are still unknown. Regarding AraC resistance in vitro, alterations in dCK activity, either by mutational inactivation or genomic rearrangements, are most frequently associated with resistance to AraC. Mutational inactivation of dCK is, however, not thought to confer resistance to AraC in patients with AML because mutations in the dCK gene are rarely found in patients with refractory or relapsed AML.19,20 In a recent study we demonstrated 4 different alternatively spliced dCK variants in coexpression with wt dCK in purified leukemic blasts from 7 of 12 patients with resistant AML and in 6 of 12 PHA-stimulated T cells generated from patients with resistant AML. These 4 alternatively spliced dCK forms code for dCK proteins with lower molecular weights and are shown to be inactive in vitro.22 Because these alternatively spliced dCK variants were not detectable in patients with sensitive AML, we hypothesized that these alternatively spliced dCK forms might have contributed to the process of AraC resistance in patients with AML. Given that the alternatively spliced dCK forms were always found in coexpression with wt dCK in purified blast cell populations, it is plausible that alternatively spliced dCK forms are coexpressed with wt dCK in a single cell. It is also conceivable, because of the heterogeneous characteristics of AML, that alternatively spliced dCK forms are exclusively expressed in a specific population of leukemic cells with no endogenous wt dCK expression. Expression studies of the alternatively spliced dCK forms are unavailable because no antibodies recognizing the alternatively spliced dCK forms have been generated thus far. Therefore, we cannot exclude that these alternatively spliced dCK forms are instable in vivo.
To investigate the effects of the expression of alternatively spliced dCK in a cell with no endogenous wt dCK expression on AraC sensitivity, alternatively spliced dCK forms were transduced into AraC-resistant K7 cells. Introduction of human wt dCK into K7 cells restored full AraC sensitivity, implicating that the retroviral vectors were properly translated. This is consistent with findings of our previous study in which restored sensitivity to AraC was described in an AraC (and Decitabine)–resistant rat leukemic cell line after transfection of rat wt dCK.32 Introduction of different alternatively spliced dCK variants into K7 cells could not restore the AraC sensitive phenotype, indicating that the alternatively spliced dCK proteins are inactive in vivo, which is consistent with the in vitro data previously reported.22 These observations imply that a cell that expresses alternatively spliced dCK forms but that has lost wt dCK expression is still resistant to the cytotoxic effects of AraC.
The possible dominant-negative effect of alternatively spliced dCK forms on endogenous wt dCK activity by the formation of heterodimers was analyzed by the transduction of alternatively spliced dCK forms into rat leukemic cells with endogenous wt dCK expression (KA cells). Overexpression of wt dCK increased AraC sensitivity by a factor 18. Two previous studies have already described an increase in AraC sensitivity in AraC-sensitive cells after retroviral transduction with wt dCK. Transduction of wt dCK increased AraC sensitivity by factors from 2 to 100 in 3 different carcinoma cell lines,33 whereas a decrease in the IC50 concentration by a factor 10 was observed in gliosarcoma cells.34 In contrast to wt dCK transductions, the introduction of alternatively spliced human dCK forms into KA cells did not alter the IC50 concentrations for AraC compared with cells that were transduced with control vector or with nontransduced cells. Although a relatively high degree of identity between rat and human dCK (89.1% identity at the nucleotide level and 91.9% identity at the amino acid level) is present,35 we previously observed different alternatively spliced dCK forms in an AraC-resistant rat leukemic cell line than in human leukemic cells.22 To exclude the possible inability of heterodimerization between human and rat dCK, we cotransduced human wt dCK with human alternatively spliced dCK into rat leukemic cells (K7) with no endogenous dCK expression. The minor, nonsignificant decrease in AraC sensitivity detected after 24 to 48 hours of AraC incubation in K7/wt+del 5 dCK cells, compared with K7/wt cells, suggests a delayed effect of AraC cytotoxicity on double-positive cells. This decreased sensitivity for AraC was abolished after 72-hour incubation with AraC (Figure 2), implying that the alternatively spliced dCK forms do not exhibit a long-term dominant-negative effect on the activity of wt dCK proteins when coexpressed in a single cell.
In addition, in human leukemic cells (U937) expressing high levels of endogenous wt dCK, no decreased sensitivity for AraC was observed when these cells were transduced with the alternatively spliced dCK forms. Unexpectedly, no increased sensitivity for AraC was observed in U937 cells transduced with human wt dCK, possibly because of the high level of endogenous wt dCK activity (higher than KA cells by a factor of 38). Maximal AraC phosphorylation that cannot be increased by the introduction of exogenous wt dCK might already occur at short time intervals in the nontransduced cells.
Previously, we detected alternatively spliced dCK forms in PHA-stimulated T cells from patients with resistant AML.22After the transduction of alternatively spliced dCK forms in PHA-stimulated T cells, no changes in sensitivity for AraC could be demonstrated (data not shown).
Since alternatively spliced dCK forms are observed in nonleukemic (PHA T) cells from resistant AML, they might be a result of a more general defect in the splicing machinery playing a role in the susceptibility of a patient with AML to respond to chemotherapy or even to contract leukemia. Alternative splicing of the folypolyglutamate synthetase (EFGS), which catalyzes the glutamination of folate antimetabolites in mammalian cells and tumors, is thought to play a role in refractoriness of AML cells for antifolates such as methotrexate (MTX).36,37 These data indicate that the alternative splicing of genes playing a key role the biosynthesis of precursors of DNA and RNA, may result in resistance to antimetabolites such as AraC and methotrexate. Previously, a more general defect in the splicing machinery was shown to be induced by the TLS-ERG myeloid leukemia–associated fusion gene frequently observed in AML.38 This fusion gene has been shown to alter the splicing of CD44 mRNA,39 whose splice variants have been implicated in tumor progression.40 In addition, this fusion protein was shown to initiate leukemogenesis in normal hematopoietic cells,41 suggesting that a defect in the splicing machinery can affect the expression of genes involved in leukemogenesis and perhaps in sensitivity to chemotherapy.
In conclusion, in this study we show that the alternatively spliced dCK forms cannot restore sensitivity to AraC when they are exclusively expressed in a cell with no endogenous wt dCK expression. In addition, we demonstrate that inactive, alternatively spliced dCK forms do not confer a long-term dominant-negative effect on wt dCK activity when they are coexpressed in a single cell. In this situation, alternatively splicing of dCK might be an epi-phenomenon in resistant AML caused by a defect in the splicing machinery and not directly involved in the development of AraC resistance in patients with AML.
We thank Prof Dr S. Eriksson (Department of Veterinary Medical Chemistry, Swedish University of Agricultural Sciences, Biomedical Centre, Uppsala, Sweden) and Prof Dr I. Talianidis (Institute of Molecular Biology and Biotechnology, Fo.R.T.H., Heraklion, Greece) for kindly supplying us with the dCK-pep monoclonal antibody.
Supported by the Dutch Cancer Society (grant RUL96-1347).
Submitted July 9, 2001; accepted October 10, 2001.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Marjan J. T. Veuger, Laboratory of Experimental Hematology, Dept of Hematology, Leiden University Medical Center, The Netherlands; e-mail: m.j.t.veuger@lumc.nl.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal