Abstract
Four patients from 3 Saudi Arabian families had delayed onset of immune deficiency due to homozygosity for a novel intronic mutation, g.31701T>A, in the last splice acceptor site of the adenosine deaminase (ADA) gene. Aberrant splicing mutated the last 4 ADA amino acids and added a 43-residue “tail” that rendered the protein unstable. Mutant complementary DNA (cDNA) expressed inEscherichia coli yielded 1% of the ADA activity obtained with wild-type cDNA. The oldest patient, 16 years old at diagnosis, had greater residual immune function and less elevated erythrocyte deoxyadenosine nucleotides than his 4-year-old affected sister. His T cells and Epstein-Barr virus (EBV) B cell line had 75% of normal ADA activity and ADA protein of normal size. DNA from these cells and his whole blood possessed 2 mutant ADA alleles. Both carried g.31701T>A, but one had acquired a deletion of the 11 adjacent base pair, g.31702-12, which suppressed aberrant splicing and excised an unusual purine-rich tract from the wild-type intron 11/exon 12 junction. During ADA replacement therapy, ADA activity in T cells and abundance of the “second-site” revertant allele decreased markedly. This finding raises an important issue relevant to stem cell gene therapy.
Introduction
Deficiency of adenosine (Ado) deaminase (ADA), a 41-kd monomeric zinc enzyme that catabolizes Ado and deoxyadenosine (dAdo), accounts for about 15% of cases of severe combined immunodeficiency (SCID).1,2 Typical patients are diagnosed by age 6 months and rarely survive beyond 1 to 2 years unless immune function is restored by stem cell transplantation or enzyme replacement therapy. About 20% of patients have some residual immune function and present later in childhood or beyond (“delayed” or “late/adult” onset), and some healthy children and adults with “partial” ADA deficiency have been identified by screening. Fibroblasts or lymphoblastoid cell lines (LCLs) from “partials” have 5% to 70% of normal ADA activity versus less than 1% to 2% in cell lines from immunodeficient patients. The level of total dAdo nucleotides (dAXP) in erythrocytes is elevated by 300- to 2000-fold in SCID, 30- to 300-fold in delayed/late onset, and 0- to 30-fold in “partial” deficiency.2 3
About 70 known mutations, the majority missense, span the 32-kb, 12 exon ADA gene on chromosome 20q.2,3 The association of certain mutations with milder phenotypes, and the correlation with dAXP level, suggested that alleles providing more than some critical level of functional ADA can ameliorate phenotype.4,5 When expressed in Escherichia coli, 28 ADA complementary DNAs (cDNAs) with missense mutations from immunodeficient patients yielded less than 0.005% to 0.6% of the ADA activity obtained with wild-type cDNA versus 1% to 28% for 5 mutations from healthy “partials.”6,7 Of 31 patients with SCID, 28 had genotypes composed of alleles that expressed 0.05% or less of wild-type activity, compared with only 2 of 21 patients with milder phenotypes.6 ADA splicing mutations have been associated with relatively mild or variable severity, because normal splicing is not always disrupted completely.5 8
Because of selection, any mechanism that increases ADA expression in lymphoid cells can potentially moderate phenotype. This is exemplified by the reversion of an inherited ADA missense mutation in B LCLs from a patient who had undergone a spontaneous clinical remission.9 A similar phenomenon involving the cytokine receptor common γ-chain gene in T cells was found in a patient with X-linked SCID.10 Revertant hematopoietic cells have also been found in patients with Fanconi anemia, Bloom syndrome, Wiskott-Aldrich syndrome, and in target organs in some other disorders.11-19 In addition to back mutation, allele function has been restored by mitotic recombination or gene conversion, which can eliminate the original mutation, and by “second-site” events that restored reading frame or led to an amino acid substitution better tolerated than the original. In Bloom syndrome, intragenic recombination or gene conversion are the usual mechanisms, consistent with reversion being much more common in heteroallelic than in homozygous patients.12 Whether this reflects an underlying DNA recombination defect or applies to other disorders is unclear.
We have discovered somatic reversion by a unique “second-site suppression” mechanism in a progenitor of T and B lymphocytes in 1 of 4 ADA-deficient patients homozygous for a novel splicing mutation. Our findings highlight the unusual purine-rich character of the terminal splice acceptor site of the ADA gene, and uncertainty about the structure and function of the C-terminus of the ADA protein. Other observations provide insight into an issue of importance to gene therapy, the influence of ADA replacement therapy on selection for rare ADA-expressing T lymphocytes.
Materials and methods
Lymphoid cells and LCLs
Interleukin 2 (IL-2)–dependent T lymphocytes were cultured from peripheral blood lymphocytes (PBLs) as described.20 Human T-lymphotropic virus 1 (HTLV-1)– transformed T LCLs were established as described,21 using irradiated TJF2 T-LCL (ADA-deficient) as the source of HTLV-1 (kindly provided by Dr Fabio Candotti, National Institutes of Health). Epstein-Barr virus (EBV) B-LCLs for Sib A and Sib B were established at the time of their diagnosis at King Faisal Hospital and later in the Pediatric Immunology Laboratory at Duke University Medical Center. T cells and T and B LCLs were maintained in AIM-V serum-free medium (Life Technologies, Grand Island, NY). ADA activity in extracts of lymphoid cells and LCLs was determined by radiochemical assay.20 ADA protein was detected by Western blot analysis of aliquots of cell extracts containing 50 μg total protein, performed with the 1C5 mouse monoclonal antibody (mAb) to human T-lymphoblast ADA.22
ADA activity and adenine nucleotide content of erythrocytes and dried blood spots
Activity of ADA in hemolysates was determined by radiochemical assay.20 Total erythrocyte AXP and dAXP were measured by high-performance liquid chromatography (HPLC) as the Ado and dAdo formed by treating neutralized acid extracts with alkaline phosphatase and venom phosphodiesterase.23 24 For testing patients from Saudi Arabia, we adapted these methods for using dried blood spots instead of erythrocytes (M.S.H. and P.B., unpublished method, January 1998-January 2000). Briefly, fresh EDTA or heparin-anticoagulated whole blood is applied to a “Guthrie filter card” (used widely for neonatal screening). For analysis, dried blood spots are extracted on ice with 25 mM Tris-HCl, pH 7.4, 15 mM KCl, 1 mM EDTA, and 1 mM dithiothreitol. Extracted ADA (and purine nucleoside phosphorylase) activity and total protein are assayed, as with hemolysates. Another aliquot of the extract is treated with perchloric acid, neutralized, and analyzed for AXP and dAXP, as for erythrocytes. The “percent dAXP,” that is, (dAXP/(AXP + dAXP) × 100, is used to assess dAXP elevation.
Analysis of ADA gene mutations
These studies were approved by the Duke University Institutional Review Board. ADA cDNA and genomic sequences are as reported,25,26 and preceded by “c.” and “g.,” respectively. ADA messenger RNA (mRNA; cDNA) numbering is relative to the start of transcription (ie, the ATG start codon begins at c.96). Standard methods were used to prepare, amplify, and clone genomic DNA and cDNA,27,28 using conditions recommended by the suppliers of reagents used in these procedures. Polymerase chain reaction (PCR) products cloned in pCR2.1 (Invitrogen, Carlsbad, CA) were sequenced using the ABI 377 PRISM Instrument. Direct sequencing of PCR products was performed by ddNTP chain termination with Sequenase T7 polymerase (Amersham Pharmacia Biotech, Piscataway, NJ). DdeI restriction enzyme was obtained from New England Biolabs (Beverly, MA). For Northern analysis, 50 μg total cellular RNA prepared using the Tri-Reagent Method (Molecular Research Center, Cincinnati, OH) was electrophoresed, blotted, hybridized, and probed as described.20
The ADA cDNA was prepared from cellular RNA using the reverse transcription (RT)–PCR kit (Amersham Pharmacia Biotech or Qiagen, Valencia, CA). The ADA coding region (c.96-1189) and “full-length” ADA cDNA (c.35-1498) were amplified as described.5 The cDNA segment c.904 to 1359 (exon 9-exon 12) was amplified with the primers (+)5′ACACGGAGCATGCAGTCAT and (−)5′ACATAATCAGAGAAGTG. Genomic DNA fragment g.31016-31920 (intervening sequence [IVS] 10-exon 12) was amplified using the primers (+)5′CACTACATTGCTAAGAGCTGCC and (−)5′ACATAATCAGAGAAGTG.
Detection of genomic DNA clones carrying the MutA and MutAΔ11 alleles
Genomic fragment g.31016-31920 (IVS 10-exon 12) amplified from DNA of lymphoid cells and LCLs was cloned in pCR2.1. Limited sequencing of random clones was performed using only ddTTP or ddCTP, which allowed scoring for the wild-type sequence and the 2 mutations of interest, g.31701T>A (MutA), and g.31701T>A; g.31702-31712del11 (MutAΔ11) (see “Results”).
Detection of a 13-nt insertion at the exon 11/exon 12 junction in uncloned and cloned ADA cDNA
Uncloned “full-length” ADA cDNA was subjected to nested PCR (primers: (+)5′TCCCAGAAGATGAAAAGAG, (−)5′ATTGAGATCATGGTCTTCTTGG) to obtain the fragment c.1099-1290 (exon 11-exon 12). The size of PCR products was analyzed on a 3% agarose/TAE gel. Full-length cDNA was also cloned into pCR2.1 and the segment c.1099-1402 (exon 11-exon 12) was then amplified from random boiled colonies (primers: (+)5′TCCCAGAAGATGAAAAGAG, (−)5′ACATAATCAGAGAAGTG). The PCR products were separated on 2.2% agarose gels. In these experiments, standards were run on each gel, consisting of PCR products obtained with authentic wild-type or mutant cDNA clones. Random PCR products were also sequenced.
Expression in E coli SØ3834
The coding regions of wild-type human ADA cDNA, and of the MutA cDNA (see “Results”) were ligated into the NcoI site of pZ.29 pZ/ADA plasmids were used to transform E coli SØ3834, which has a deletion of the bacterial ADAgene.29 The conditions used for constitutive expression and assay of ADA activity are as reported.6,22 Aliquots of lysates containing 30 μg total protein were analyzed by Western blotting with the 1C5 mAb to human ADA.22
Three cDNAs encoding other mutations at the C-terminus of human ADA were also expressed: G360X, with a stop signal at codon 360 to eliminate the last 4 amino acids; 360-363EPTS, changing the last 4 amino acids from GQNL to EPTS; and 360-363EPTS+20, changing residues 360-363 to EPTS and adding codons 364-383 of the MutA cDNA, encoding RAEPLKTPLLQAFTLWSHPN (single-letter amino acid codes). PCR mutagenesis was performed essentially as described.6 30 The forward primer for all 3 mutations was (+)5′CGCGCGAATTCGGGCACCATGGCCCAGACGCCCGCCTTCGAC. Reverse primers were: for G360X, (−)5′GCGCAAGCTTCGGGCCATGGTCTTCATGCAGAGGCTGAAGGTGG; for 360 to 363EPTS, (−)5′GCGCAAGCTTCGGGCCATGGTCTTCAGCTCGTTGGTTCTGCAGAGGC; and for 360 to 363EPTS+20, (−)5′GCGCAAGCTTCGGGCCATGGTCTTCAGTTGGGGTGACTCCACAG. The templates for the G360X and 360-363EPTS mutagenesis were the wild-type and MutA cDNAs; the MutA cDNA was the template for 360-363EPTS+20. The final PCR products were cloned into pBluescript II KS, fully sequenced, and then ligated into the NcoI site of pZ.
In vitro transcription-translation
[35S]Methionine-labeled ADA was generated in vitro from ADA cDNA constructs in pBluescript, using the TNT Coupled Rabbit Reticulocyte Lysate System (Promega, Madison, WI). Translation products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and fluorography, and were also electrophoresed on cellulose acetate and stained for ADA activity in situ, as described.20
Results
Case presentations
Family 1.
The 4-year-old proband, Sib A was the fifth child of second cousins from Saudi Arabia. Prior to age 3 years she had frequent bronchitis and one episode of pneumonia, but neither thrush nor chronic diarrhea. In her fourth year she had bacterial meningitis without sequelae and was hospitalized for extensive, but uncomplicated varicella. Her height and weight were in the 75th and 50th percentiles, respectively, for age. She had no tonsils or palpable lymph nodes; lungs were clear. Lymphopenia and other abnormalities suggesting combined immunodeficiency (Table 1) prompted testing for ADA deficiency (Table 2).
Immune function studies
| . | Sib A . | Sib B . | Patient Y . | Patient R . |
|---|---|---|---|---|
| Age at diagnosis, y | 4 | 16 | 5.5 | 1.3 |
| WBC, 109/L | 6.16 | 6.82 | 3.43 | 7.15 |
| Total lymphs | 1 060 | 1 570 | 274 | 787 |
| (2 000-8 000) | (1 000-4 800) | (2 000-8 000) | (4 000-10 500) | |
| CD3 | 974 | 1 522 | 181 | 449 |
| (1 600-2 700) | (782-2 834) | (1 600-2 700) | (2 200-4 100) | |
| CD4 | 115 | 235 | 110 | 181 |
| (1 000-1 700) | (322-1 750) | (1 000-1 700) | (1 400-2 800) | |
| CD8 | 827 | 1 130 | 82 | 244 |
| (600-1 000) | (338-1 086) | (600-1 000) | (800-1 800) | |
| CD19 | 21 | 31 | 8 | 71 |
| (400-800) | (67-555) | (400-800) | (700-1 600) | |
| CD16/56 | 31 | < 16 | 37 | 110 |
| (200-400) | (200-400) | (200-400) | (200-600) | |
| PHA* | 23 781/24% | 80 884/67% | ||
| Con A* | 18 369/27% | 68 741/75% | ||
| Allogeneic cells* | 15 954/36% | 41 209/81% | ||
| IgG, g/L | 13.7 | 11.2 | 12.3 | 12.3 |
| (4.6-12.4) | (6.4-13.5) | (4.6-12.4) | (3.5-12.1) | |
| IgA, g/L | 1.4 | 1.5 | 1.5 | 0.9 |
| (0.24-1.5) | (0.7-3.1) | (0.24-1.5) | (0.14-1.06) | |
| IgM, g/L | 1.05 | 1.13 | 0.62 | 1.95 |
| (0.43-1.96) | (0.56-3.52) | (0.43-1.96) | (0.43-1.73) | |
| IgE, IU/L | 3 820 | 347 | 8 580 | |
| (8-250) | (5-500) | (1.6-30) | ||
| Tetanus antibody | 2.07 | 10.4 | 2.8 | 10.8 |
| Pneumococcus antibody | 8.06 | 9.5 |
| . | Sib A . | Sib B . | Patient Y . | Patient R . |
|---|---|---|---|---|
| Age at diagnosis, y | 4 | 16 | 5.5 | 1.3 |
| WBC, 109/L | 6.16 | 6.82 | 3.43 | 7.15 |
| Total lymphs | 1 060 | 1 570 | 274 | 787 |
| (2 000-8 000) | (1 000-4 800) | (2 000-8 000) | (4 000-10 500) | |
| CD3 | 974 | 1 522 | 181 | 449 |
| (1 600-2 700) | (782-2 834) | (1 600-2 700) | (2 200-4 100) | |
| CD4 | 115 | 235 | 110 | 181 |
| (1 000-1 700) | (322-1 750) | (1 000-1 700) | (1 400-2 800) | |
| CD8 | 827 | 1 130 | 82 | 244 |
| (600-1 000) | (338-1 086) | (600-1 000) | (800-1 800) | |
| CD19 | 21 | 31 | 8 | 71 |
| (400-800) | (67-555) | (400-800) | (700-1 600) | |
| CD16/56 | 31 | < 16 | 37 | 110 |
| (200-400) | (200-400) | (200-400) | (200-600) | |
| PHA* | 23 781/24% | 80 884/67% | ||
| Con A* | 18 369/27% | 68 741/75% | ||
| Allogeneic cells* | 15 954/36% | 41 209/81% | ||
| IgG, g/L | 13.7 | 11.2 | 12.3 | 12.3 |
| (4.6-12.4) | (6.4-13.5) | (4.6-12.4) | (3.5-12.1) | |
| IgA, g/L | 1.4 | 1.5 | 1.5 | 0.9 |
| (0.24-1.5) | (0.7-3.1) | (0.24-1.5) | (0.14-1.06) | |
| IgM, g/L | 1.05 | 1.13 | 0.62 | 1.95 |
| (0.43-1.96) | (0.56-3.52) | (0.43-1.96) | (0.43-1.73) | |
| IgE, IU/L | 3 820 | 347 | 8 580 | |
| (8-250) | (5-500) | (1.6-30) | ||
| Tetanus antibody | 2.07 | 10.4 | 2.8 | 10.8 |
| Pneumococcus antibody | 8.06 | 9.5 |
Lymphocyte counts are expressed as 106 cells/L. Normal ranges for age are shown in parentheses.
WBC indicates white blood cell; PHA, phytohemagglutinin; and Con A, concanavalin A.
cpm/% response (patient cpm, mitogen-patient cpm, media)/(control cpm, mitogen-control cpm, media) × 100.
Diagnostic studies
| . | Dried blood spot . | Erythrocytes . | |||
|---|---|---|---|---|---|
| ADA activity nmol/h/mg . | Percent dAXP . | ADA activity nmol/h/mg . | dAXP nmol/mL . | Percent dAXP . | |
| Sib A | 0.15 | 7.1 | 2.2 | 140 | 9.6 |
| Sib B | 0.30 | 2.1 | 1.5 | 56 | 5.8 |
| Brother | 51.6 | < 0.1 | 58.4 | ||
| Brother | 15.5 | < 0.1 | |||
| Mother | 19.5 | < 0.1 | 26.9 | < 1 | < 0.1 |
| Father | 19.1 | < 0.1 | 27.8 | < 1 | < 0.1 |
| Patient Y | 0.1 | 6.0 | |||
| Patient R | 0.0 | 8.5 | |||
| SCID | 0.38 ± 0.5 | 41.9 ± 17 | < 0.5 | 766 ± 344 | 50.3 ± 18 |
| Delayed onset | 186 ± 55 | 15.5 ± 4 | |||
| Control | 32.5 ± 14 | < 0.1 | 63.0 ± 41 | < 2 | < 0.2 |
| . | Dried blood spot . | Erythrocytes . | |||
|---|---|---|---|---|---|
| ADA activity nmol/h/mg . | Percent dAXP . | ADA activity nmol/h/mg . | dAXP nmol/mL . | Percent dAXP . | |
| Sib A | 0.15 | 7.1 | 2.2 | 140 | 9.6 |
| Sib B | 0.30 | 2.1 | 1.5 | 56 | 5.8 |
| Brother | 51.6 | < 0.1 | 58.4 | ||
| Brother | 15.5 | < 0.1 | |||
| Mother | 19.5 | < 0.1 | 26.9 | < 1 | < 0.1 |
| Father | 19.1 | < 0.1 | 27.8 | < 1 | < 0.1 |
| Patient Y | 0.1 | 6.0 | |||
| Patient R | 0.0 | 8.5 | |||
| SCID | 0.38 ± 0.5 | 41.9 ± 17 | < 0.5 | 766 ± 344 | 50.3 ± 18 |
| Delayed onset | 186 ± 55 | 15.5 ± 4 | |||
| Control | 32.5 ± 14 | < 0.1 | 63.0 ± 41 | < 2 | < 0.2 |
Control values are mean ± SD. Control blood spots were obtained from 42 healthy individuals used as “simultaneous controls” for immunodeficient patients. Control values for erythrocytes are from > 200 non–ADA-deficient immunodeficient and healthy individuals tested in this laboratory at Duke University. Values for ADA activity and dAXP concentration in erythrocytes of ADA-deficient patients with SCID and delayed-onset phenotypes are as reported in Arredondo-Vega et al.6 It should be noted that ADA activity eluted from blood spots and present in hemolysates is normalized to total protein derived from different sources (red cells versus whole blood). Therefore, these values cannot be directly compared.
Sib B was found to be ADA deficient at age 16 years, when he was tested after the diagnosis was made in the proband. He was well until age 15 months, when he had bacterial meningitis and was comatose for a month. He recovered intellectually, but developed progressive weakness and severe kyphoscoliosis. During childhood he had recurrent otitis and pneumonia, which did not require hospitalization, and he developed restrictive lung disease. In the year prior to diagnosis he had uncomplicated varicella. His height and weight were less than fifth percentile for age. He lacked palpable lymph nodes, and had kyphoscoliosis, rales over lung bases, digital clubbing, and general muscle atrophy. Chest x-ray showed bronchiectasis.
Sib A and Sib B each had 1% of normal whole blood ADA activity, assayed after elution from dried blood spots, and slightly higher activity in hemolysates (Table 2). Blood and erythrocyte dAXP levels were elevated, but were less than those in patients with SCID. Both parents and one healthy brother had about half-normal ADA activity, and another healthy brother had normal activity. Two other siblings, who had died at 1 and 4 years of age, had histories of recurrent infection but had never been tested for ADA deficiency.
Both patients had combined immunodeficiency, but less profound than in SCID (Table 1). Total T-cell counts were low-normal, but CD4 cells were low and CD8 were normal, reversing the CD4/CD8 ratio; B and natural killer (NK) cells were decreased. Lymphocyte responses to mitogens and allogeneic cells were low but significant in the proband, and low-normal in Sib B. Levels of serum IgG, IgA, and IgM were normal, but IgE was elevated and specific antibody titers were low.
Because they lacked HLA-identical siblings and were considered poor candidates for a partially mismatched transplant, both patients were treated by enzyme replacement with polyethylene glycol (PEG)–ADA (Adagen, Enzon, Piscataway, NJ), initiated at Duke University Medical Center. Within 6 months, erythrocyte dAXP, lymphocyte counts, and mitogen responses normalized. After reimmunization both patients developed normal diphtheria and tetanus antibody titers; in vitro lymphocyte proliferative responses to candida and tetanus antigens, which were absent before PEG-ADA therapy, became significant in Sib A and completely normal in Sib B (Koleilat, Hershfield, and Buckley, unpublished observations, February 2000-October 2000). They have had no significant infections.
Patients Y (family 2) and R (family 3).
Soon after the diagnosis of patients in family 1, 2 other children with suggestive histories, each born to parents who were second-degree relatives and distantly related to family 1, were evaluated and found to be ADA deficient (Tables 1 and 2). Patient Y (family 2), aged 5.5 years, had been hospitalized several times for respiratory infections and thrombocytopenia. Growth parameters were less than third percentile and she had severe restrictive lung disease. Patient R (family 3) had repeated respiratory infections and diarrhea, but was first hospitalized at age 1 year, shortly before diagnosis of ADA deficiency, for myoclonic seizures attributed to viral encephalitis. A brother with a history of chest infections from 1 year of age, who had been diagnosed with common variable immunodeficiency, and a sister with a history of chest infections and diarrhea from age 4 months, had both died at age 7 years.
Patients Y and R were lymphopenic, but had normal or low normal serum immunoglobulins; patient R had elevated IgE (Table 1). Whole blood dAXP levels were elevated, but substantially lower than in SCID (Table 2). Both patients have received marrow transplants from HLA-identical siblings, performed at King Faisal Specialist Hospital, after conditioning with busulphan and cyclophosphamide. Despite good engraftment, patient Y continues to have respiratory symptoms related to chronic lung disease, and patient R continues to have seizures.
ADA activity in lymphocytes of Sib A and Sib B
Erythrocytes and whole blood of both patients in family 1 showed a similar degree of ADA deficiency, but dAXP levels were substantially lower in Sib B (Table 2). This suggested that greater residual ADA activity in a nonerythroid lineage of Sib B might be limiting the dAdo available for forming red blood cell dAXP. This proved to be the case (Table 3). ADA activity was less than 1% to 5% of normal in cultured T cells, T LCL, and B LCL from Sib A. By contrast, ADA activity was about 75% of normal in pretreatment T cells from Sib B, and also in a B LCL established after he had received PEG-ADA for 7 months. However, PBLs, T cells, and a T LCL derived from Sib B after 7 months of therapy had substantially less ADA activity, 19%, 8.5%, and 4% of normal, respectively.
ADA activity in lymphoid cells of Sib A and Sib B, family 1
| . | Sib A . | Sib B . | Normal3-150 . |
|---|---|---|---|
| Pretreatment | |||
| T cells | 99 | 1576 | |
| After 7 mo of treatment with PEG-ADA | |||
| PBL | 100 | 229 | 1197 ± 516 |
| T cells | 119 | 173 | 2047 ± 1360 |
| HTLV-T LCL | 31 | 98 | 2280 |
| EBV-B LCL | 63 | 5423 | 7435 ± 3285 |
| . | Sib A . | Sib B . | Normal3-150 . |
|---|---|---|---|
| Pretreatment | |||
| T cells | 99 | 1576 | |
| After 7 mo of treatment with PEG-ADA | |||
| PBL | 100 | 229 | 1197 ± 516 |
| T cells | 119 | 173 | 2047 ± 1360 |
| HTLV-T LCL | 31 | 98 | 2280 |
| EBV-B LCL | 63 | 5423 | 7435 ± 3285 |
Values shown (nmol/h/mg extract protein) have been corrected by subtracting activity resistant to the specific ADA inhibitor deoxycoformycin (90%-99% of ADA activity in patient cells was inhibited).
Genotype analysis
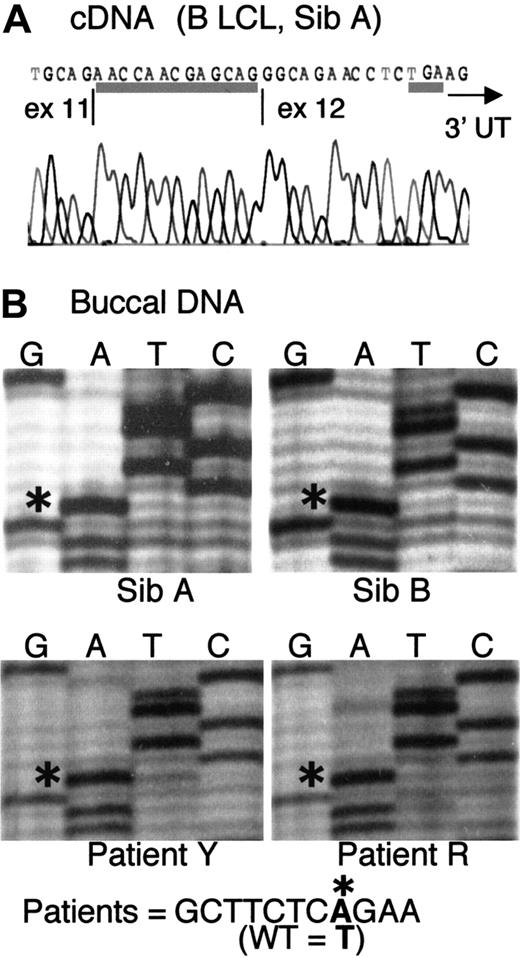
We initially found that 6 ADA cDNA clones prepared from a B LCL derived from Sib A had a 13-nt insertion at the junction of exons 11 and 12 (Figure 1A). The insert corresponds to g.31703-31715, the last 13 base pair (bp) of IVS 11. This finding led to the discovery of a novel mutation of the last splice acceptor site of the ADA gene, a T>A transversion at g.31701, located in IVS 11, 15 bp upstream of the start of exon 12 (Figure 1B). Presumably by changing a TG dinucleotide to AG, this mutation activates splicing at g.31703 instead of the normal g.31716.
Cryptic splicing due to a point mutation in IVS 11.
(A) Sequence of ADA cDNA clone derived from B LCL of Sib A, showing a 13-nt insert at the exon 11/12 junction (underlined). The insert corresponds to g.31703-31715, normally the last 13 nt of IVS 11. The TGA stop codon (underlined) and the start of the 3′ untranslated region (3′UTR, arrow) are indicated. (B) Partial sequences of fragment g.31016-31920 (IVS 10-exon 12) amplified from uncloned DNA from buccal brushings of patients, as indicated. The sequence g.31694-31704 is shown. Asterisks indicate an A at nt g.31701 in all 4 patients, instead of T found in wild-type (WT) human ADA.
Cryptic splicing due to a point mutation in IVS 11.
(A) Sequence of ADA cDNA clone derived from B LCL of Sib A, showing a 13-nt insert at the exon 11/12 junction (underlined). The insert corresponds to g.31703-31715, normally the last 13 nt of IVS 11. The TGA stop codon (underlined) and the start of the 3′ untranslated region (3′UTR, arrow) are indicated. (B) Partial sequences of fragment g.31016-31920 (IVS 10-exon 12) amplified from uncloned DNA from buccal brushings of patients, as indicated. The sequence g.31694-31704 is shown. Asterisks indicate an A at nt g.31701 in all 4 patients, instead of T found in wild-type (WT) human ADA.
Only the mutant A was found at g.31701 on direct sequencing of the fragment g.31016-31920 (spanning IVS 10-exon 12) amplified from DNA isolated from buccal brushings of Sib A and Sib B in family 1, as well as from buccal DNA of the distantly related patients Y and R (Figure1B). Consistent with homozygosity, buccal DNA of all 4 patients also showed complete digestion at a new DdeI restriction site in IVS 11 created by the g.31701T>A mutation (not shown). Patients Y and R were not studied further because they had received marrow transplants shortly after diagnosis.
Complex genotype of Sib B of family 1
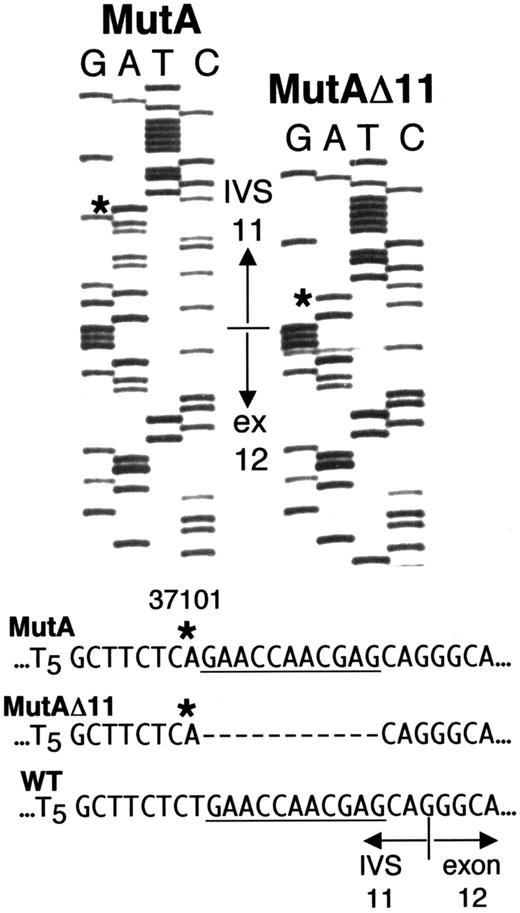
Direct sequencing (Figure 2A) andDdeI digestion (Figure 2B) of the IVS 10-exon 12 fragment amplified from whole blood DNA of Sib A also indicated homozygosity for g.31701T>A. Each parent was heterozygous (maternal sequence, Figure2A; DdeI digest for both parents, Figure 2B). Unlike results with Sib A, direct sequencing of the g.31016-31920 fragment from blood DNA of Sib B gave a “double ladder” in the vicinity of the IVS 11-exon 12 boundary (Figure 2A), and his DdeI digest indicated heterozygosity for g.31701T>A (Figure 2B, lane 3). To resolve the ambiguous results with Sib B, we amplified and cloned the g.31016-31920 fragment from DNA of his PBLs. Of 6 clones sequenced, 3 showed only the g.31701T>A mutation; the other 3 had “A” at g.31701, but each also had a deletion of 11 nt immediately downstream, that is, g.31702-31712 (Figure 3). For clarity, g.31701T>A is hereafter designated “MutA,” and the compound mutation g.31701T>A; g.31702-31712del11 is designated “MutAΔ11.”
Analysis of IVS 11 mutations in whole blood DNA.
(A) Sequence of segment g.31689-31704 (IVS 11) in PCR fragment g.31016-31920 (IVS 10-exon 12) from normal control, Sib A, Sib B, and their mother, as indicated. Arrows indicate g.31701. Sib A is homozygous, and her mother is heterozygous, for g.31701T>A. Blood DNA of Sib B gives a “double ladder.” (B) Ethidium bromide–stained agarose gel of a DdeI digest of fragment g.31016-31920 (IVS 10-exon 12) amplified from normal control DNA (lane 1), and from blood DNA of Sib A (lane 2), Sib B (lane 3), and their parents (lane 4, father; lane 5, mother). The 234- and 85-nt products are due to a novelDdeI site introduced by the g.31701T>A mutation.
Analysis of IVS 11 mutations in whole blood DNA.
(A) Sequence of segment g.31689-31704 (IVS 11) in PCR fragment g.31016-31920 (IVS 10-exon 12) from normal control, Sib A, Sib B, and their mother, as indicated. Arrows indicate g.31701. Sib A is homozygous, and her mother is heterozygous, for g.31701T>A. Blood DNA of Sib B gives a “double ladder.” (B) Ethidium bromide–stained agarose gel of a DdeI digest of fragment g.31016-31920 (IVS 10-exon 12) amplified from normal control DNA (lane 1), and from blood DNA of Sib A (lane 2), Sib B (lane 3), and their parents (lane 4, father; lane 5, mother). The 234- and 85-nt products are due to a novelDdeI site introduced by the g.31701T>A mutation.
PBL from Sib B possess 2 different ADA mutations.
Sequences from 2 clones of ADA fragment g.31016-31920 amplified from PBL DNA of Sib B. The sequence shown spans g.31684 in IVS 11 to g.31738 in exon 12. The normal IVS 11/exon 12 junction is indicated. Asterisks indicate the “A” at g.31701 (MutA) in both clones, instead of the wild type “T.” The clone labeled MutAΔ11 also has an 11-nt deletion of g.31702-31712 (dashes). For reference, the deleted segment is underlined in the wild-type and MutA sequences.
PBL from Sib B possess 2 different ADA mutations.
Sequences from 2 clones of ADA fragment g.31016-31920 amplified from PBL DNA of Sib B. The sequence shown spans g.31684 in IVS 11 to g.31738 in exon 12. The normal IVS 11/exon 12 junction is indicated. Asterisks indicate the “A” at g.31701 (MutA) in both clones, instead of the wild type “T.” The clone labeled MutAΔ11 also has an 11-nt deletion of g.31702-31712 (dashes). For reference, the deleted segment is underlined in the wild-type and MutA sequences.
The presence of both MutA and MutAΔ11 ADA alleles explains the “double-ladder” sequence and heterozygous DdeI pattern obtained with blood DNA of Sib B. MutAΔ11 can also account for the ADA activity found in his T cells and B LCL. Thus, the 11-nt deletion converts the AG created by MutA to AC, which should “deactivate” the cryptic splice site. It also restores a “normal,” though mutated, IVS 11-exon 12 splice acceptor site (discussed below).
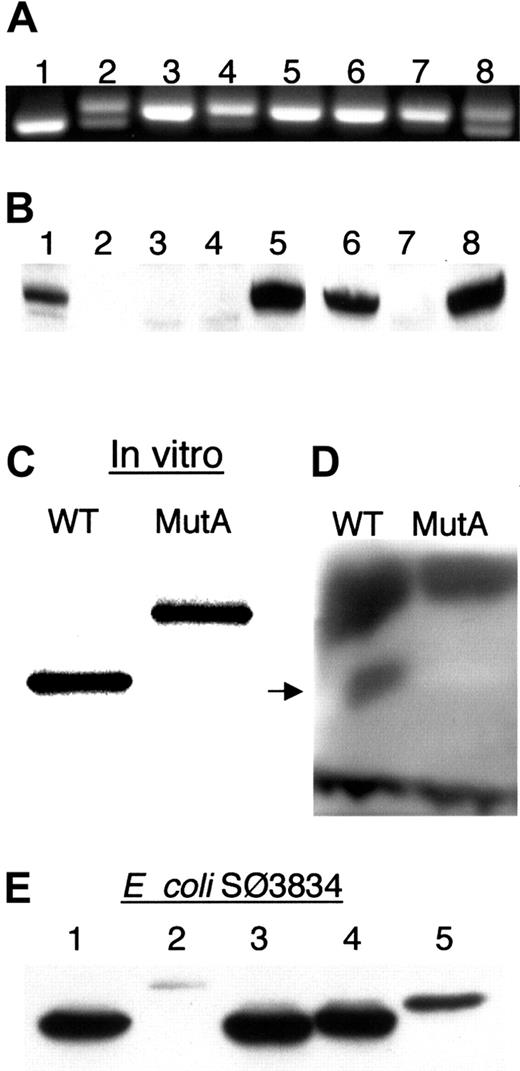
Northern analysis revealed no obvious differences from normal controls in the amount or size of ADA mRNA from T or B LCLs from Sib A and Sib B (data not shown). To better evaluate the region of interest, we amplified the exon 11/12 junction from uncloned cDNA prepared from their lymphoid cells (Figure 4A). Post–PEG-ADA treatment B LCLs and pre–PEG-ADA T cells from Sib B gave 2 products, a major band of the size obtained with cloned authentic MutA cDNA as template and a minor band of the size obtained with cloned wild-type ADA cDNA as template (Figure 4A, lanes 2 and 4). The T LCLs derived from Sib B after PEG-ADA treatment, which had low ADA activity, and T cells and T and B LCLs of Sib A, showed only the larger MutA product (Figure 4A, lanes 3 and 5-7).
Expression of ADA mRNA and ADA proteins.
(A) Ethidium bromide–stained agarose gel of fragment c.1099-1290 amplified from uncloned ADA cDNA. A 191-bp product is expected from both MutAΔ11 and wild-type ADA cDNA; MutA-derived cDNA gives a 204-bp product. Control lanes: 1, wild-type ADA cDNA; 8, equal mixture of wild-type and MutA ADA cDNAs. Sib B (lanes 2-4): 2, B LCL; 3, posttreatment T LCL; 4, pretreatment T cells. Sib A (lanes 5-7): 5, B LCL; 6, T LCL; 7, T cells. (B) Western blot of ADA in extracts of lymphoid cells. Lanes 1-5, T cells and T LCLs: 1, Sib B pretreatment T cells; 2, Sib A pretreatment T cells; 3, Sib B posttreatment T LCL; 4, Sib A posttreatment T LCL; 5 control T LCL. Lanes 6-8, B LCLs: 6, Sib B; 7, Sib A; 8, control. (C-E) Expression of recombinant MutA-derived ADA protein. (C) SDS-PAGE of 35S in vitro translation products resulting from the wild-type (WT) and MutA cDNA. (D) In situ assay for ADA activity of the translation products shown in panel C after nondenaturing electrophoresis. Arrow indicates position of human ADA. Dark bands at the top and bottom are, respectively, rabbit ADA and rabbit hemoglobin. (E) Western blot of ADA proteins expressed in E coli SØ3834. Lanes: 1, wild type; 2, MutA; 3, G360X; 4, 360-363EPTS; 5, 360-363EPTS+20 (see “Materials and methods” for description).
Expression of ADA mRNA and ADA proteins.
(A) Ethidium bromide–stained agarose gel of fragment c.1099-1290 amplified from uncloned ADA cDNA. A 191-bp product is expected from both MutAΔ11 and wild-type ADA cDNA; MutA-derived cDNA gives a 204-bp product. Control lanes: 1, wild-type ADA cDNA; 8, equal mixture of wild-type and MutA ADA cDNAs. Sib B (lanes 2-4): 2, B LCL; 3, posttreatment T LCL; 4, pretreatment T cells. Sib A (lanes 5-7): 5, B LCL; 6, T LCL; 7, T cells. (B) Western blot of ADA in extracts of lymphoid cells. Lanes 1-5, T cells and T LCLs: 1, Sib B pretreatment T cells; 2, Sib A pretreatment T cells; 3, Sib B posttreatment T LCL; 4, Sib A posttreatment T LCL; 5 control T LCL. Lanes 6-8, B LCLs: 6, Sib B; 7, Sib A; 8, control. (C-E) Expression of recombinant MutA-derived ADA protein. (C) SDS-PAGE of 35S in vitro translation products resulting from the wild-type (WT) and MutA cDNA. (D) In situ assay for ADA activity of the translation products shown in panel C after nondenaturing electrophoresis. Arrow indicates position of human ADA. Dark bands at the top and bottom are, respectively, rabbit ADA and rabbit hemoglobin. (E) Western blot of ADA proteins expressed in E coli SØ3834. Lanes: 1, wild type; 2, MutA; 3, G360X; 4, 360-363EPTS; 5, 360-363EPTS+20 (see “Materials and methods” for description).
In the experiment with uncloned cDNA (Figure 4A), the smaller, wild-type PCR product is underestimated because a heteroduplex forms that migrates with the larger MutA product (these were resolved on a 4% agarose gel, not shown). Therefore, we analyzed cloned genomic DNA and cDNA PCR products to better estimate the relative abundance of the MutA and MutAΔ11 alleles and their transcripts (Table4). MutAΔ11 accounted for about 35% of genomic DNA clones from ADA-expressing pretreatment Sib B T cells. About 40% of ADA cDNA clones from his ADA-expressing B LCL had a normal exon 11/12 junction (ie, were derived from MutAΔ11). By contrast, these values were 7% and 6% for ADA genomic and cDNA clones, respectively, derived from his posttreatment (PEG-ADA) T cells, which had about 8% of normal ADA activity (Tables 3 and 4). In a similar analysis of 25 cDNA clones derived from B LCL from Sib A, and of 36 from her T LCL, all were found to contain the 13-bp insert due to MutA.
Relative abundance of ADA alleles and ADA transcripts in lymphoid cells derived from Sib B
| Source . | Genomic DNA . | cDNA . | ||||
|---|---|---|---|---|---|---|
| Clones examined . | MutA (%) . | MutAΔ11 (%) . | Clones examined . | 13-nt insert (%) . | No insert (%) . | |
| T cells (before treatment) | 20 | 13 (65) | 7 (35) | |||
| T cells (after treatment) | 14 | 13 (93) | 1 (7) | |||
| EBV B LCL | 15 | 9 (60) | 6 (40) | |||
| T cells (after treatment) | 70 | 66 (94) | 4 (6) | |||
| Source . | Genomic DNA . | cDNA . | ||||
|---|---|---|---|---|---|---|
| Clones examined . | MutA (%) . | MutAΔ11 (%) . | Clones examined . | 13-nt insert (%) . | No insert (%) . | |
| T cells (before treatment) | 20 | 13 (65) | 7 (35) | |||
| T cells (after treatment) | 14 | 13 (93) | 1 (7) | |||
| EBV B LCL | 15 | 9 (60) | 6 (40) | |||
| T cells (after treatment) | 70 | 66 (94) | 4 (6) | |||
See “Materials and methods” for details.
Recombinant MutA cDNA expression
Exon 12 of human ADA includes 2 bp of codon 360, codons 361-363, the TGA stop codon, and the 3′ untranslated region (UTR). The MutA-induced 13-nt insertion is predicted to change residues 360-363 from GQNL to EPTS, and to extend the reading frame by 43 additional codons, to give a 406-residue protein of 45 540 Da (dalton) instead of the normal 40 724 Da (Figures 1A and 5). Only ADA protein of normal size was detected by Western analysis in extracts of the pretreatment T cells and posttreatment B LCL from Sib B, whereas no ADA of either size was detected in his posttreatment T cells or in T cells and B LCL from Sib A (Figure 4B).
Normal and mutant ADA C-terminal amino acids and final splice acceptor sites.
(A) C-termini of wild-type (WT) and MutA proteins (single-letter amino acid code). (B) IVS 11 splice acceptor sites. The consensus splice site58 is shown schematically to locate the conserved AG dinucleotide, the polypyrimidine tract Pyn, and the branchpoint A. In the wild-type (WT), MutA, and MutAΔ11 transcripts (below) nt removed by splicing are in lower case; nt retained in exon 12 are in capitals. The positions of nt −1 are indicated; arrows indicate g.31701 (mutated in MutA and MutAΔ11), and possible branchpoint at g.31686. The 13-nt insertion at the exon 11/12 junction due to MutA is in italics. Dashes indicate the 11-nt deletion in MutAΔ11.
Normal and mutant ADA C-terminal amino acids and final splice acceptor sites.
(A) C-termini of wild-type (WT) and MutA proteins (single-letter amino acid code). (B) IVS 11 splice acceptor sites. The consensus splice site58 is shown schematically to locate the conserved AG dinucleotide, the polypyrimidine tract Pyn, and the branchpoint A. In the wild-type (WT), MutA, and MutAΔ11 transcripts (below) nt removed by splicing are in lower case; nt retained in exon 12 are in capitals. The positions of nt −1 are indicated; arrows indicate g.31701 (mutated in MutA and MutAΔ11), and possible branchpoint at g.31686. The 13-nt insertion at the exon 11/12 junction due to MutA is in italics. Dashes indicate the 11-nt deletion in MutAΔ11.
When expressed in vitro in rabbit reticulocytes, MutA cDNA gave rise to a 35S-protein of the predicted size (Figure 4C). After nondenaturing electrophoresis of equal amounts of translation product on cellulose acetate, ADA activity was detected by in situ assay of the wild-type, but not the MutA protein (Figure 4D). Although this assay is relatively insensitive, the MutA in vitro translation product, though stable, is clearly much less active than the wild-type.
We also expressed the MutA cDNA, and some related constructs, inE coli SØ3834. This strain lacks the bacterialADA gene, permitting quantitation of as little as 0.005% of the ADA activity obtained with wild-type human ADA cDNA. When expressed under defined conditions, and normalized to total extract protein, MutA-transformed cells had 1% of the ADA activity obtained with wild-type cDNA (Table 5). Immunoreactive MutA protein (Figure 4E, lane 2) was estimated by Western blot to be about 3% of wild- type human ADA protein, based on comparison with serial dilutions of the wild-type extract, and by blotting with a polyclonal antiserum to human ADA as well as with the 1C5 mAb (data not shown).
Expression of ADA cDNAs in E coli SØ3834
| ADA cDNA . | ADA activity nmol/h/mg . | Percent of wild-type . |
|---|---|---|
| Wild-type | 276 458 ± 34 428 | 100.0 |
| MutA | 3 392 ± 2 157 | 1.2 ± 0.8 |
| G360X | 270 752 ± 79 502 | 97.9 ± 28.7 |
| 360-363EPTS | 274 683 ± 40 232 | 99.3 ± 14.5 |
| 360-363EPTS + 20 | 13 627 ± 2 418 | 4.9 ± 0.9 |
| ADA cDNA . | ADA activity nmol/h/mg . | Percent of wild-type . |
|---|---|---|
| Wild-type | 276 458 ± 34 428 | 100.0 |
| MutA | 3 392 ± 2 157 | 1.2 ± 0.8 |
| G360X | 270 752 ± 79 502 | 97.9 ± 28.7 |
| 360-363EPTS | 274 683 ± 40 232 | 99.3 ± 14.5 |
| 360-363EPTS + 20 | 13 627 ± 2 418 | 4.9 ± 0.9 |
ADA activity for the vector control is 3 nmol/h/mg.6
Wild-type levels of ADA activity (Table 5) and ADA protein (Figure 4E) were expressed with human ADA cDNA modified so as to delete the last 4 amino acids, or to change them from GQNL to EPTS, as in the MutA protein. A MutA cDNA truncated to encode only the first 24 of the 43 MutA-determined residues, yielded 5% of wild-type activity and 10% to 15% of the amount of ADA protein. These results show that the MutA-induced C-terminal extension, rather than mutation of residues 360-363, is responsible for instability and loss of catalytic activity.
In other studies not presented we found that recombinant MutA ADA had a Km for Ado of 70 μM, compared with 40 μM for wild-type ADA, and it showed neither greater thermolability than the wild-type enzyme, nor evidence of aggregation by size exclusion chromatography. The MutA protein formed a stable complex with the CD26/dipeptidyl peptidase IV glycoprotein, indicating that the CD26 binding site at the carboxy-end of the peripheral α2helix (amino acids 126-143)22 is apparently in a normal configuration.
Discussion
We report data on 4 ADA-deficient patients from 3 families within a large kindred, each with a delayed onset of combined immunodeficiency. These are the first patients from Saudi Arabia in whom ADA genotype has been examined, and possibly the first in whom ADA deficiency has been demonstrated. Buccal DNA of all 4 patients, and DNA from blood, T cells, and T and B LCLs of Sib A, the proband in family 1, showed homozygosity for a novel mutation, g.31701T>A (MutA), the most distal ADA mutation identified to date. By converting a TG to AG, MutA activates a cryptic splice site, inserting the last 13 nt of IVS 11 into ADA mRNA. This changes and extends the reading frame to mutate the last 4 amino acids of the ADA protein and add a 43-residue C-terminal tail (Figure 5A). No normally spliced ADA mRNA was detected in lymphoid cells of Sib A, and although a low level of ADA activity was measurable, neither the mutant protein nor ADA of normal size could be detected immunologically.
Expression of MutA cDNA in vitro and in E coli showed that instability and loss of catalytic activity are due to the C-terminal tail, not to mutation of amino acids 360-363, which were dispensable. This is not surprising because murine ADA, which is 83% identical in overall sequence, lacks the last 11 residues found in human ADA.31 Moreover, the murine ADA C-terminus (residues 337-351), though conserved, forms a peripheral helix that is not only remote from the active site, but makes few contacts with more proximal elements of the main α/β barrel structure.32The MutA-encoded C-terminal tail, which probably lacks a stable structure, may interfere with folding of the nascent ADA protein, leading to loss of activity and to rapid proteolysis. We have previously reported data on an ADA-deficient patient with a late-onset phenotype who was homozygous for a mutation in IVS 10, which resulted in mutation of the last 38 amino acids and addition of 100 extra residues.5 That protein was also inactive and unstable.
Expressing MutA cDNA under standardized conditions in E coliSØ3834 yielded 1% of the activity obtained with wild-type ADA cDNA. In previous studies, this was at the border between ADA activity expressed by mutant alleles from patients with a late/adult-onset phenotype and alleles from healthy subjects with “partial ADA deficiency.”6 7 In vivo, the steady-state level of ADA activity due to the MutA protein is insufficient for developing and sustaining normal immune function, but it prevents ADA substrates from rising to a level that causes the profound lymphopenia and lack of T- and B-cell function associated with SCID.
Mosaicism for a second-site suppressor of the effects of MutA on splicing
The natural history of the delayed-onset phenotype is more variable than for SCID, but progressive deterioration is expected. (4 undiagnosed children in 2 of the families studied, who also had histories compatible with delayed onset ADA deficiency, had died by age 7 years). Despite serious sequelae of early infections, Sib B in family 1 apparently stabilized at some time during childhood; he is one of only a few ADA-deficient patients to have survived, undiagnosed and without specific therapy, beyond the first decade.5,30,33At 16 years, his lymphocyte function was abnormal, but better preserved than in the other 3 patients when they were diagnosed at ages 1.3 to 5.5 years. dAXP in his erythrocytes were also less elevated, and among the lowest reported, other than in healthy subjects with “partial” ADA deficiency. These atypical features in Sib B are likely related to the greater ADA activity expressed by his lymphoid cells, owing to an unusual, previously unreported, form of somatic reversion: second-site suppression of a cryptic splice site. Reversion was evident in T cells and a B LCL, and must therefore have occurred in a common progenitor of both T and B lymphocytes. In previous reports of somatic reversion in ADA deficiency, only the T or B lineage has been involved.9 34
DNA from blood, T cells, and T and B LCLs of Sib B showed, to varying degrees, evidence of 2 different MutA alleles, one intact and one modified. The latter (MutAΔ11) had a contiguous deletion of 11 bp (g.31702-12), shown in bold in the sequence g.37100-23 CAGAACCAACGAGCAGGGCAGAAC. This deletion may have arisen by “slipped mispairing”35 during replication of one of the inherited MutA alleles, because the 5′ G of the deletion is part of a CAG trinucleotide (CTG in wild-type ADA) and the 3′ end is flanked by CAG; several other short direct repeats also occur within the larger segment shown. By converting the AG created by MutA to AC, the deletion inactivates the cryptic splice site. Remarkably, splicing at the normal position is restored, despite removal of 11 bp found in the wild-type IVS 11/exon 12 junction.
Figure 5B compares the IVS 11/exon12 junctions used in processing the wild-type, MutA, and MutAΔ11 transcripts. In each, the segment −1 to −4 matches the consensus mammalian splice acceptor sequence, and the A at g.31686, in the consensus sequence CTGAT, might serve as branchpoint. However, both mutants differ significantly from wild-type in the segment −5 to −14, which in the consensus is a pyrimidine-rich tract that plays an important role in branchpoint and splice site selection.36,37 In the wild-type, 7 of these 10 nt are purines, whereas in both MutA and MutAΔ11, 9 of 10 are pyrimidines. Using the system of Shapiro and Senapathy,38 in which the similarity of a particular splice acceptor site to the consensus is judged by a score of 0 to 100 over the interval +1 to −14, the wild-type splice site scores only 63.1, compared with 93.8 for MutA, and 94.9 for MutAΔ11.
We have previously described a rearrangement of the IVS 8/exon 9 junction in which 6 consecutive purines were interposed between nt −4 and the polypyrimidine tract. This mutation eliminated normal splicing and caused skipping of exon 9.8 Although the wild-type IVS 11/exon 12 junction has a similar purine-rich character, it is clearly functional. However, when juxtaposed with a “superior” MutA-induced splice site immediately upstream, the cryptic site is very strongly favored. It is worth noting that the splice acceptor site of the terminal intron, and removal of the terminal intron, have been shown to play a role in formation of the 3′ end of mRNA, which involves the cleavage and polyadenylation processing steps.39-42 In view of the differences in their IVS 11/exon 12 junctions it would be interesting to investigate the relative efficiency of ADA mRNA termination using substrates based on the wild type, MutA, and MutAΔ11 alleles.
Relationship of enzyme replacement therapy and relative abundance of MutAΔ11
The T cells derived from Sib B before therapy had much higher ADA activity and protein, and greater abundance of the MutAΔ11 allele and its transcript, than did T cells and a T LCL obtained after several months of treatment with PEG-ADA. By normalizing systemic levels of ADA substrates, PEG-ADA therapy permits ADA-deficient lymphoid cells to survive, proliferate, and function.2,43Given the improvement in his lymphocyte function, an expansion of homozygous MutA T cells can plausibly account for the apparent decline in heteroallelic MutA/MutAΔ11 T cells. Our findings in Sib B are similar to those in an SCID patient with unusually low red blood cell dAXP, in whom a T LCL established at diagnosis had half-normal ADA activity due to reversion of an inherited point mutation.34 ADA-expressing T LCLs could not be established from this patient after starting PEG-ADA therapy, leading to speculation that revertant T cells had been overgrown by nonrevertants. (A revertant B LCL was derived from Sib B during treatment. This may reflect stronger in vitro selection for, and expansion of, ADA-expressing cells [that arose in vivo] during the process of EBV transformation.)
These findings raise a related issue, namely, whether concomitant PEG-ADA therapy may contribute to the limited success of stem cell–targeted gene therapy for ADA deficiency.44-49 In addressing this question, 2 observations are relevant: (1) there is no evidence that PEG-ADA directly suppresses the growth or viability of either ADA-expressing or ADA-deficient T cells, and (2) to date, ex vivo transduction of human lymphohematopoietic stem cells with retroviral ADA vectors has been very inefficient.48 49 We suspect that, as in the case of somatic revertants, when metabolic toxicity is neutralized by PEG-ADA therapy, rescued ADA-deficient T cells become more abundant than transduced T cells simply because they arise from a much larger progenitor pool.
Performing stem cell gene therapy without concomitant use of PEG-ADA would minimize competition from untransduced lymphocytes, but this might or might not enhance clinical benefit. It is not known how long selection must operate before rare transduced stem cells can give rise to protective immune function, or how variable the response might be. Recent success with stem cell gene therapy for X-linked SCID is encouraging.50 However, ADA deficiency is a systemic metabolic disorder with a different pathogenesis. Injury to nonlymphoid organs has been reported in ADA-deficient humans and mice.51-55 PEG-ADA, a few milliliters of which has the ADA activity of 1012 normal T cells, acts in plasma to protect organs as well as lymphoid cells from toxic ADA substrates.43,54,56 Whether ADA expression limited to gene-transduced lymphoid cells can be as effective remains to be determined. A sharp rise in erythrocyte dAXP in patients undergoing gene therapy in whom PEG-ADA has been withheld (Kohn et al47 and unpublished observations, February 1999-October 1999), suggests that at the metabolic level, the therapies have to date not been equivalent.
We wish to thank Dr Rebecca Buckley of Duke University Medical Center for information regarding recovery of immune function during treatment with PEG-ADA.
Supported by National Institutes of Health grant RO1 DK20902 (M.S.H.) and a grant from Enzon, Inc. E.R. was supported by Fellowship 98/9329 from Fondo de Investigación Sanitaria, Instituto de Salud Carlos III, Ministerio de Sanidad y Consumo, Spain.
F.X.A-V. and I.S. contributed equally to this work.
M.S.H. is a consultant to Enzon whose product was discussed in the present work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Michael S. Hershfield, Box 3049, Duke University Medical Center, Durham, NC 27710; e-mail: msh@biochem.duke.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal