Abstract
Primary cutaneous lymphomas (CLs) constitute a spectrum of diseases characterized by a clonal accumulation of lymphocytes in the skin. Most CLs display a Th2 cytokine profile, including expression of interleukin-10 (IL-10). Because the up-regulation of HLA-G, a nonclassical class Ib molecule inducible by IL-10, might account for the immunescape of the malignant clone, HLA-G and IL-10 expression has been investigated in 45 cases of primary CL (10 of B-cell and 35 of T-cell origin) with quantitative polymerase chain reaction (PCR) and immunohistochemistry. HLA-G message was present in all cutaneous B-cell (CBCL) and T-cell (CTCL) lymphomas evaluated. Immunohistochemistry revealed HLA-G protein expression in 23 (51%) of 45 cases (7 of 10 CBCL, 16 of 35 CTCL). While in CBCL mostly indolent types displayed HLA-G positivity, in CTCL HLA-G expression was associated with high-grade histology and advanced stage of the disease. Except for neoplastic and infiltrating lymphocytes, other cells such as macrophages and dendritic cells showed HLA-G immunoreactivity. Furthermore, IL-10 protein expression was demonstrated in 16 (73%) of 22 HLA-G+ cases, which correlated with HLA-G protein presence (P < .001). HLA-G up-regulation together with IL-10 expression in CL might additionally contribute to the evasion of immunosurveillance and facilitate the transition from low- to high-grade lymphomas.
Introduction
Primary cutaneous lymphomas (CLs) comprise a spectrum of heterogeneous entities characterized by a clonal accumulation of lymphocytes initially restricted to the skin.1 Despite their rather benign course, cutaneous T-cell (CTCLs) and B-cell lymphomas (CBCLs) may spread to extracutaneous sites, confirming that they are systemic disease of the lymphoid system despite their clinical confinement to the skin.2 Decreased cell-mediated cytotoxity (in both natural killer [NK] cell and lymphokine-activated killer cell activity), decreased T-cell response to antigens, and several other immune phenomena imply the existence of general immune dysregulation in CTCL patients.3,4 Most of the abnormalities might be attributed to the pleiotropic immunosuppressive effects of interleukin-10 (IL-10), as demonstrated in different CL types.5-8
Despite the evidence of antitumor humoral and cell-mediated immune response in CL,9-11 the disease may evolve further, eventually killing the patient. Besides the production of immunosuppressive cytokines, alterations of HLA class I status responsible for the presentation of tumor peptides to T cells could be the reason for an ineffective antitumor response.12 So far only a few studies have been conducted investigating expression of HLA class I molecules, mainly in nodal lymphomas.13-15 The loss of major histocompatibility complex class I antigens was detected preferentially in large cell lymphomas and in total was less frequent (2.3%) in comparison to nonhematologic cancer (up to 50%).12 15 Therefore, this mechanism is less likely to take place in lymphomas.
The expression of one nonclassical HLA class I molecule, HLA-G, has been implicated as an additional way of tumor immunescape.16-19 HLA-G differs from other HLA class I molecules—hence the prefix “nonclassical”—by its quasimonomorphism and distribution restricted to immune privileged sites, such as fetomaternal interface and thymus.20,21 The HLA-G gene is transcribed in a variety of human adult tissues, peripheral blood B and T lymphocytes, and monocytes but not NK cells.21-25 Additionally, alternative splicing creates at least 7 different HLA-G isoforms, of which 4 are membrane-bound and 3 are coding for soluble proteins.21,26 The shift toward Th2 cytokine profile, dominated by IL-10, is thought to be responsible for HLA-G expression in placenta.27,28Initially, HLA-G was shown to protect from NK-mediated lysis of HLA-deficient targets through interactions with killer cell inhibitory receptors (KIR).20,29,30 The fact that other cells, such as lymphocytes, macrophages, and dendritic cells, harbor HLA-G–specific inhibitory ligands, such as p49, ILT-2, and ILT-4,31-33 explained HLA-G–mediated inhibition of a cytotoxic T-lymphocyte–specific response.34 HLA-G leader sequence-derived peptide stabilizes cell surface expression of another nonclassical class I molecule, HLA-E, which constitutes a preferential ligand for a widely expressed KIR, CD94/NKG2A.35Intratumoral-activated macrophages and dendritic cells have been described recently to express HLA-G protein,36 suggesting that HLA-G–mediated immune modulation might further contribute to the impairment and circumvention of an efficient immune response.
Because lymphocytes have the potential to transcribe and express HLA-G depending on the environmental conditions, we investigated HLA-G expression in primary CLs. We report here that HLA-G message and protein were expressed in both cutaneous CBCL and CTCL. In CBCL, mostly indolent types displayed HLA-G immunoreactivity, while in CTCL HLA-G expression was associated with large cell morphology and advanced stage of the disease. In addition, we have demonstrated IL-10 immunoreactivity in the proximity of HLA-G+ cells, suggesting that IL-10 was responsible for HLA-G up-regulation in CL.
Materials and methods
Tissue and blood samples; cell lines
Skin biopsy samples from 45 patients with primary CLs were taken primarily for diagnostic purposes after obtaining a prior informed consent of the patient. The specimens used in further investigations were a surplus material available after all routine diagnostic procedures. The healthy skin specimens were obtained from the Department of Plastic Surgery, University Hospital Zurich following breast reduction. The histologic diagnoses are summarized in Table 1 and were assessed in agreement with the European Organization for Research and Treatment of Cancer (EORTC) classification of primary CLs.37 The assignment to a certain stage was done according to the recommendations of EORTC Cutaneous Lymphoma Project Group.37 Peripheral blood mononuclear cells (PBMCs) were isolated from patients' blood and buffy coat preparation from healthy volunteers (obtained from a local blood bank) using a Ficoll-Hypaque density gradient centrifugation (Pharmacia Biotech, Uppsala, Sweden). Skin biopsy samples and PBMCs from 2 patients (CO) with secondary CBCL were included as controls. The HLA-G+ human choriocarcinoma cell line JEG-3 (ATCC, Manassas, VA) was cultured in Dulbecco modified Eagle medium (Biochrom, Berlin, Germany) supplemented with 10% heat-inactivated fetal calf serum with antibiotics-antimycotic (Gibco BRL, Life Technologies AG, Basel, Switzerland), 1 mM sodium pyruvate (Gibco BRL), and 2 mM L-glutamine (Biochrom). First trimester trophoblast tissue was obtained from elective termination of pregnancy after informed consent of the patient and used as an additional positive control.
Data of the CL patients evaluated for HLA-G and IL-10 protein expression
| Patient no. . | Gender . | Age, y . | Histologic classification (EORTC) . | Stage . | HLA-G protein expression . | IL-10 expression . |
|---|---|---|---|---|---|---|
| BL1 | M | 75 | Follicular B-cell lymphoma | ++ | + | |
| BL2 | M | 32 | Follicular B-cell lymphoma | ++ | − | |
| BL3 | M | 19 | Follicular B-cell lymphoma | + | − | |
| BL4 | M | 59 | Follicular B-cell lymphoma | + | ++ | |
| BL5 | M | 58 | MZBCL | ++ | ++ | |
| BL6 | F | 82 | Diffuse large B-cell lymphoma | + | + | |
| BL7 | F | 81 | Diffuse large B-cell lymphoma | − | ND | |
| BL8 | M | 49 | Diffuse large B-cell lymphoma | + | ND | |
| BL9 | F | 77 | Large B-cell lymphoma of the leg | − | − | |
| BL10 | M | 57 | T-cell-rich B-cell lymphoma | − | − | |
| TL1 | F | 36 | Lymphomatoid papulosis | − | − | |
| TL2 | M | 45 | Lymphomatoid papulosis | − | − | |
| TL3 | M | 65 | Lymphomatoid papulosis | − | − | |
| TL4 | M | 54 | Lymphomatoid papulosis | − | − | |
| TL5 | M | 52 | Lymphomatoid papulosis | − | − | |
| TL6 | M | 40 | MF | Ia | − | − |
| TL7 | M | 56 | MF | Ib | − | − |
| TL8 | F | 87 | MF | Ib | − | − |
| TL9 | F | 64 | MF | Ib | − | − |
| TL10 | M | 66 | MF | IIb | − | − |
| TL11 | M | 74 | MF | IIb | − | − |
| TL12 | F | 52 | MF | IIb | − | − |
| TL13 | M | 55 | MF | III | ++ | ++ |
| TL14 | F | 77 | MF | IVb | + | + |
| TL15 | F | 87 | Sézary syndrome | IVa | − | − |
| TL16 | F | 45 | CD8+ initial CTCL | Ia | + | + |
| TL17 | F | 50 | CD8+, TIA-1+, CD30− large cell CTCL | IIb | ++ | ++ |
| TL18 | F | 68 | CD30+ large cell CTCL | IIb | + | − |
| TL19 | F | 56 | CD30+ large cell CTCL | IIb | − | − |
| TL20 | F | 67 | CD30+large cell CTCL | IIb | + | − |
| TL21 | F | 29 | CD30+ large cell CTCL | IIb | − | − |
| TL22 | F | 45 | CD30+ large cell CTCL | IIb | − | − |
| TL23 | F | 33 | CD30−large cell CTCL | IIb | − | − |
| TL24 | M | 18 | CD30− large cell CTCL | IIb | ++ | − |
| TL25 | M | 65 | CD30− large cell CTCL‡ | IIb | ++ | + |
| TL26 | M | 76 | CD30− large cell CTCL‡ | IVb | ++ | ++ |
| TL27 | F | 36 | CD30− large cell CTCL‡ | IVb | − | − |
| TL28 | F | 50 | CD30− large cell CTCL‡ | IVb | ++ | + |
| TL29 | M | 69 | CD30+ pleomorphic CTCL | IIb | − | − |
| TL30 | M | 62 | CD30+pleomorphic CTCL | IVa | + | + |
| TL31 | M | 52 | CD30− pleomorphic small/medium-sized CTCL | IIb | ++ | + |
| TL32 | M | 80 | CD30− pleomorphic small/medium-sized CTCL | IIb | + | + |
| TL33 | M | 62 | CD30− pleomorphic small/medium-sized CTCL | IIb | + | + |
| TL34 | M | 50 | CD30− pleomorphic small/medium-sized CTCL | IVb | + | − |
| TL35 | M | 28 | GSS | IIb | ++ | ++ |
| Patient no. . | Gender . | Age, y . | Histologic classification (EORTC) . | Stage . | HLA-G protein expression . | IL-10 expression . |
|---|---|---|---|---|---|---|
| BL1 | M | 75 | Follicular B-cell lymphoma | ++ | + | |
| BL2 | M | 32 | Follicular B-cell lymphoma | ++ | − | |
| BL3 | M | 19 | Follicular B-cell lymphoma | + | − | |
| BL4 | M | 59 | Follicular B-cell lymphoma | + | ++ | |
| BL5 | M | 58 | MZBCL | ++ | ++ | |
| BL6 | F | 82 | Diffuse large B-cell lymphoma | + | + | |
| BL7 | F | 81 | Diffuse large B-cell lymphoma | − | ND | |
| BL8 | M | 49 | Diffuse large B-cell lymphoma | + | ND | |
| BL9 | F | 77 | Large B-cell lymphoma of the leg | − | − | |
| BL10 | M | 57 | T-cell-rich B-cell lymphoma | − | − | |
| TL1 | F | 36 | Lymphomatoid papulosis | − | − | |
| TL2 | M | 45 | Lymphomatoid papulosis | − | − | |
| TL3 | M | 65 | Lymphomatoid papulosis | − | − | |
| TL4 | M | 54 | Lymphomatoid papulosis | − | − | |
| TL5 | M | 52 | Lymphomatoid papulosis | − | − | |
| TL6 | M | 40 | MF | Ia | − | − |
| TL7 | M | 56 | MF | Ib | − | − |
| TL8 | F | 87 | MF | Ib | − | − |
| TL9 | F | 64 | MF | Ib | − | − |
| TL10 | M | 66 | MF | IIb | − | − |
| TL11 | M | 74 | MF | IIb | − | − |
| TL12 | F | 52 | MF | IIb | − | − |
| TL13 | M | 55 | MF | III | ++ | ++ |
| TL14 | F | 77 | MF | IVb | + | + |
| TL15 | F | 87 | Sézary syndrome | IVa | − | − |
| TL16 | F | 45 | CD8+ initial CTCL | Ia | + | + |
| TL17 | F | 50 | CD8+, TIA-1+, CD30− large cell CTCL | IIb | ++ | ++ |
| TL18 | F | 68 | CD30+ large cell CTCL | IIb | + | − |
| TL19 | F | 56 | CD30+ large cell CTCL | IIb | − | − |
| TL20 | F | 67 | CD30+large cell CTCL | IIb | + | − |
| TL21 | F | 29 | CD30+ large cell CTCL | IIb | − | − |
| TL22 | F | 45 | CD30+ large cell CTCL | IIb | − | − |
| TL23 | F | 33 | CD30−large cell CTCL | IIb | − | − |
| TL24 | M | 18 | CD30− large cell CTCL | IIb | ++ | − |
| TL25 | M | 65 | CD30− large cell CTCL‡ | IIb | ++ | + |
| TL26 | M | 76 | CD30− large cell CTCL‡ | IVb | ++ | ++ |
| TL27 | F | 36 | CD30− large cell CTCL‡ | IVb | − | − |
| TL28 | F | 50 | CD30− large cell CTCL‡ | IVb | ++ | + |
| TL29 | M | 69 | CD30+ pleomorphic CTCL | IIb | − | − |
| TL30 | M | 62 | CD30+pleomorphic CTCL | IVa | + | + |
| TL31 | M | 52 | CD30− pleomorphic small/medium-sized CTCL | IIb | ++ | + |
| TL32 | M | 80 | CD30− pleomorphic small/medium-sized CTCL | IIb | + | + |
| TL33 | M | 62 | CD30− pleomorphic small/medium-sized CTCL | IIb | + | + |
| TL34 | M | 50 | CD30− pleomorphic small/medium-sized CTCL | IVb | + | − |
| TL35 | M | 28 | GSS | IIb | ++ | ++ |
− indicates no expression detectable with 4H84 mAb, accordingly for IL-10; +, single-cell immunoreactivity with 4H84 mAb, accordingly for IL-10; ++, strong immunoreactivity with 4H84 mAb, accordingly for IL-10; and ND, not done.
Skin material was available for quantitative PCR.
PBMCs were available for quantitative PCR.
Transformed from MF.
RNA extraction; cDNA synthesis
Total RNA was extracted from 17 frozen tissue samples using a High Pure RNA Tissue Kit and from PBMCs of 10 patients using a High Pure RNA Isolation Kit (both from Roche Molecular Biochemicals [RMB], Mannheim, Germany) according to the manufacturer's recommendations. Approximately 1 μg of RNA was reverse transcribed using oligo-p(dT)15 priming and avian myeloblastosis virus reverse transcriptase (1st Strand cDNA Synthesis Kit for RT-PCR [avian myeloblastosis virus], RMB) at 42°C for 1 hour. Complementary DNAs (cDNAs) were then stored at −20°C until further use.
Real-time quantitative PCR
HLA-G–specific polymerase chain reaction (PCR) amplifications were carried out using following primer sets: U522 (exon 3) and L922 (exon 5) detecting mainly HLA-G1 isoform; G.257(exon 2) and G.1225 (3′-UT) screening for all alternatively spliced HLA-G messenger RNA (mRNA) forms, as previously described38 (Table2). Design of the primers and optimization for the use in the LightCycler quantification system was done with the OLIGO 5.0 Primer Analysis Software (Molecular Biology Insights, Cascade, CO). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers and plasmids pCRII.GAPDH were kindly provided by Andreas Trojan (Division of Oncology, Department of Internal Medicine, University Hospital Zurich, Switzerland). A Hot-Start PCR was performed using 2 μL ready-to-use mastermix (LightCycler-Faststart DNA Master SYBR Green I, RMB, containing termostable recombinant Taq polymerase, reaction buffer, dATP, dCTP, dGTP, dUTP), 0.5 μM of each oligonucleotide primer (desalted PCR grade, Microsynth, Balgach, Switzerland), variable free MgCl2 concentrations (Table 2), 2 μL cDNA, and a water to a final volume of 20 μL. After initial denaturation of 7 minutes, amplification occurred as a 3-step cycling procedure: denaturation, 95°C, 15 seconds, ramp rate 20°C per second; annealing (Table 2), 10 seconds, ramp rate 20°C per second; and elongation at 72°C (Table 1), ramp rate 2°C per second, for 40 cycles. The acquisition of fluorescence was done at 87°C to avoid contribution of nonspecific products to the overall signal. Finally, the temperature was raised gradually (0.2°C per second) starting from 70°C to 99°C for the melting curve analysis
Used oligonucleotides and PCR conditions
| Primers . | Sequence . | Amplicon size, base pairs . | Annealing temperature, elongation time . | Free MgCl2 . |
|---|---|---|---|---|
| U522 (exon 3) | 5′-CAA TGT GGC TGA ACA AAG GA-3′ | 420 | 60°C, 25 s | 4 mM |
| L922 (exon 5) | 5′-CCA GCA ACG ATA CCC ATG-3′ | |||
| G.257 (exon 2) | 5′-GGA AGA GGA GAC ACG GAA CA-3′ | G1-988; G2,4-712; G3-436; G5-1110; G6-834 | 60°C, 90 s | 2 mM |
| G.1225 (3′-UT) | 5′-TGA GAC AGA GAC GGA GAC AT-3′ | |||
| GAP1U19 | 5′-GAA GGT GAA GGT CGG AGT C-3′ | 224 | 55°C, 10 s | 4 mM |
| GAP206L19 | 5′-GAA GAT GGT GAT GGG ATT T-3′ |
| Primers . | Sequence . | Amplicon size, base pairs . | Annealing temperature, elongation time . | Free MgCl2 . |
|---|---|---|---|---|
| U522 (exon 3) | 5′-CAA TGT GGC TGA ACA AAG GA-3′ | 420 | 60°C, 25 s | 4 mM |
| L922 (exon 5) | 5′-CCA GCA ACG ATA CCC ATG-3′ | |||
| G.257 (exon 2) | 5′-GGA AGA GGA GAC ACG GAA CA-3′ | G1-988; G2,4-712; G3-436; G5-1110; G6-834 | 60°C, 90 s | 2 mM |
| G.1225 (3′-UT) | 5′-TGA GAC AGA GAC GGA GAC AT-3′ | |||
| GAP1U19 | 5′-GAA GGT GAA GGT CGG AGT C-3′ | 224 | 55°C, 10 s | 4 mM |
| GAP206L19 | 5′-GAA GAT GGT GAT GGG ATT T-3′ |
External standards for the GAPDH quantification consisted of 6 serial 1:10 dilutions (5 × 102 to 5 × 107molecules per reaction) of pCRII.GAPDH plasmids. Plasmids containing full-length HLA-G insert pLNCX.G139 were used as 6 external standards for HLA-G1 quantitative PCR, containing 102 to 107 copies per reaction (a kind gift from D.E. Geraghty, Fred Hutchinson Cancer Research Center, Seattle, WA). External standards were run concomitantly with patient samples under identical conditions. The PCR reactions were run as triplicates, and run data were analyzed with the quantification program V3.39 (RMB). The fluorescence signal was plotted against the cycle number for all samples and external standards. The Fit Points method option was used in the course of analyzing quantification data, allowing the definition of a noise band and subsequent background fluorescence subtraction and resulting in the display of only log-linear and plateau amplification phases. The point when the signal rose above the background level, the so-called crossing point, was then determined for each standard dilution. The standard curve was then generated for each run, plotting the crossing point against the log concentration of the standards. The mean crossing point value, SD, and coefficient of variation were than calculated for each standard and patient sample. The single HLA-G1 and GAPDH standard curves as a result of triplicate runs were then used as references to calculate the number of target molecules in each sample. Results were expressed initially as the absolute copy number per microliter. Normalization of the estimated HLA-G1 amount was achieved by calculating the ratios between HLA-G1 and GAPDH copy number in 1 μL cDNA to compensate the variations in quantity and quality of starting mRNA.40 The normalized values were then multiplied by the constant 104.
Ten microliters of each amplification product was analyzed for appropriate length by electrophoresis on 1.6% agarose gel stained with ethidium bromide. The estimated size of the amplified fragments matched the calculated size. In addition, product identity was confirmed by melting curve analysis, an application in the LightCycler analysis program. The specificity of the obtained PCR products was finally confirmed by sequencing.
Immunohistochemistry
The paraffin-embedded material was available for all 27 cases. The following antibodies were used: 4H84 (1:500 dilution), IgG1 mouse monoclonal antibody (mAb) raised against denatured HLA-G α1 domain (kindly provided by M. McMaster, University of California, San Francisco, CA); anti–major histocompatibility complex class I mAb TP25.99, reacting with HLA-A, -B, and -C but not HLA-G (kind gift from S. Ferrone, Roswell Park Cancer Institute, Buffalo, NY); anti–human IL-10 (E-10) mouse IgG2b mAb (Santa Cruz Biotechnology, Santa Cruz, CA); anti–human CD79a mouse IgG1 mAb and anti–human CD3 mouse polyclonal antibody (both from DAKO, Denmark); anti–human CD1a mouse IgG1 mAb (Immunotech, Marseille, France); and isotype-matched controls IgG1, IgG2a, and IgG2b (DAKO). Imunohistochemical staining was performed using the alkaline phosphatase–antialkaline phosphatase (APAAP) technique after microwave antigen recovery. To determine whether HLA-G expression coincides with IL-10 presence and to further determine the phenotype of HLA-G+ cells, paraffin-embedded tissue sections were double-stained with anti–IL-10 or anti-CD79a, -CD3, and anti-CD1a mAb and 4H84 (1:300 dilution) antibody employing the APAAP method. Briefly, after deparaffinization and antigen retrieval, nonspecific binding sites were blocked by incubating slides with 20% AB serum/phosphate–buffered saline for at least 15 minutes at room temperature. Tissue sections were incubated with different primary antibodies or isotype-matched control and, secondarily, always with 4H84 mAb for 60 minutes. Each specific antibody application was followed by 2 cycles of sequential incubations with rabbit anti–mouse IgG and APAAP complexes (DAKO). The immunoreaction was visualized with developing solutions containing blue-purple 5-bromo-4-chloro-3-indoxyl phosphate with nitroblue tetrazolium chloride (DAKO), which labeled primary antibody and red neufuchsin (DAKO) marking HLA-G. Finally, sections were counterstained with hematoxylin. All the incubations were performed at room temperature in a moist chamber.
Data analysis
Statistical analysis was performed using an SPSS statistical software (version 10.0; SPSS).
Results
Differential HLA-G gene expression in primary CLs
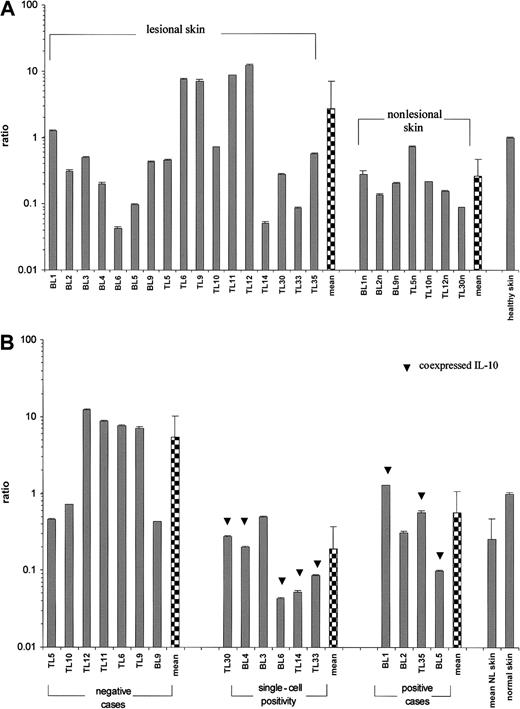
Amplification with the G.257/G.1225 primer pair screening for all HLA-G mRNA-spliced isoforms demonstrated variable HLA-G gene expression in all lymphomatous skin lesions available for reverse transcriptase (RT)–PCR as well as in nonlesional skin of 7 patients and healthy skin samples (Figure 1A). Furthermore, the full-length transcript HLA-G1 was present in all cases investigated (Figure 2A). Quantitative RT-PCR revealed up to 2 orders of magnitude difference in HLA-G1 transcriptional levels between lymphoma cases, being most pronounced in patients with mycosis fungoides (MF) (Figure 2A). The HLA-G gene expression in lymphoma samples did not statistically differ from the uninvolved skin group and from the healthy skin. Although the CTCL group transcribed HLA-G more abundantly in comparison to the CBCL group, the difference was not statistically significant and the correlation with the particular lymphoma subtype could not be established. When comparing HLA-G transcription grouped according to the pattern of protein expression evaluated by immunohistochemistry (Figure 2B), HLA-G1 transcription in cases negative for HLA-G protein expression was significantly higher from the ones expressing HLA-G (Kruskal- Wallis test, H = 9.167, P = .010). In addition, the absence of HLA-G immunoreactivity correlated with high HLA-G1 mRNA levels (one-tailed Spearman rank correlation test, rho = −.490,P = .023).
Results after RT-PCR amplification with G.257/G.1225 primer pair screening all HLA-G isoforms.
Results are shown in (A) lesional and nonlesional skin and in (B) PBMCs of CL patients. Due to the low sensitivity of ethidium bromide staining, for some samples the bands are not visible. Using a melting curve analysis (data not shown) and the primer pair amplifying HLA-G1, it was possible to confirm that all the cases were transcribing HLA-G. The cDNAs from JEG-3 cell line and first trimester placenta (Pla) were used as positive controls. Instead of cDNA template, water (–) was run as a negative control; hs indicates healthy skin; l, lesional skin; n, nonlesional skin; BC, buffy coat preparation; and MWM, molecular weight marker.
Results after RT-PCR amplification with G.257/G.1225 primer pair screening all HLA-G isoforms.
Results are shown in (A) lesional and nonlesional skin and in (B) PBMCs of CL patients. Due to the low sensitivity of ethidium bromide staining, for some samples the bands are not visible. Using a melting curve analysis (data not shown) and the primer pair amplifying HLA-G1, it was possible to confirm that all the cases were transcribing HLA-G. The cDNAs from JEG-3 cell line and first trimester placenta (Pla) were used as positive controls. Instead of cDNA template, water (–) was run as a negative control; hs indicates healthy skin; l, lesional skin; n, nonlesional skin; BC, buffy coat preparation; and MWM, molecular weight marker.
Relative HLA-G1 transcriptional levels in primary CLs.
Values are expressed as mean ± SD GAPDH-balanced ratios (as a result of triplicate runs) multiplied by 104. (A) Pattern of HLA-G1 expression in lesional (n = 17) versus nonlesional (n = 7) and a control healthy skin. The Student t test was performed; t = 2.54, P = .022. (B) Relative HLA-G1 transcriptional levels grouped according to the immunoreactivity with 4H84 mAb. Levels of HLA-G1 mRNA expression in cases showing no 4H84 immunoreactivity (n = 7) versus single-cell positivity (n = 5) versus HLA-G protein–positive lymphoma cases (n = 4). In some cases HLA-G immunoreactivity coincided with IL-10 expression (▾). NL indicates nonlesional skin.
Relative HLA-G1 transcriptional levels in primary CLs.
Values are expressed as mean ± SD GAPDH-balanced ratios (as a result of triplicate runs) multiplied by 104. (A) Pattern of HLA-G1 expression in lesional (n = 17) versus nonlesional (n = 7) and a control healthy skin. The Student t test was performed; t = 2.54, P = .022. (B) Relative HLA-G1 transcriptional levels grouped according to the immunoreactivity with 4H84 mAb. Levels of HLA-G1 mRNA expression in cases showing no 4H84 immunoreactivity (n = 7) versus single-cell positivity (n = 5) versus HLA-G protein–positive lymphoma cases (n = 4). In some cases HLA-G immunoreactivity coincided with IL-10 expression (▾). NL indicates nonlesional skin.
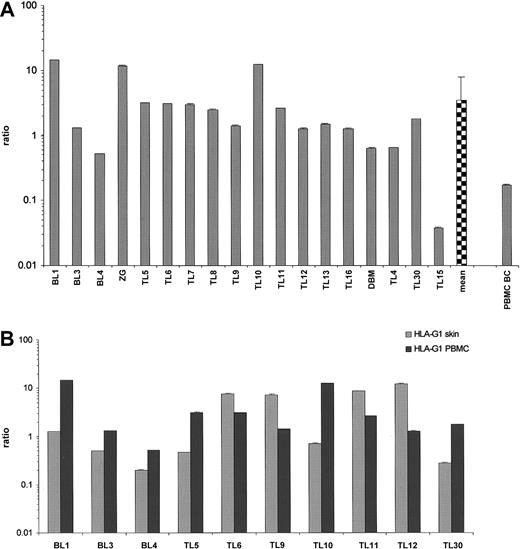
Further on, we sought to compare HLA-G transcriptional levels from the lesional skin with the one in patients' PBMCs, presuming that the malignant clone would be preferentially homing to the skin and exhibit difference in HLA-G gene expression due to the influence of microenvironmental factors. Again, HLA-G isoform screening revealed the positivity of all cases (Figure 1B). The full-length transcript was present in all cases within the patient group, varying significantly (Student t test, t = 2.693,P = .021), and in PBMCs obtained from a buffy coat preparation (Figure 3A). The levels of HLA-G1 mRNA did not statistically differ between the patient group and buffy coat sample. Of note, patients' own PBMCs demonstrated an average of 5 times higher HLA-G1 transcription than the lesional skin (Figure 3B). Interestingly, 4 MF patients (TL6, TL9, TL11, TL12) exhibited an average of 4 times higher levels of HLA-G1 in the skin (Figure3B). None of the 10 patients evaluated had a detectable B-cell or T-cell clone in the blood.
Relative HLA-G1 mRNA levels in PBMCs obtained from primary CL patients.
(A) Values are expressed as mean ± SD GAPDH-balanced ratios (as a result of triplicate runs) multiplied by 104. BC indicates buffy coat. (B) Relative HLA-G1 transcriptional levels in lesional skin and PBMCs of primary CL patients. Note that 4 patients with CTCL (TL6, TL9, TL11, TL12) exhibited an average 4 times higher HLA-G transcription in lesional skin than in PBMCs.
Relative HLA-G1 mRNA levels in PBMCs obtained from primary CL patients.
(A) Values are expressed as mean ± SD GAPDH-balanced ratios (as a result of triplicate runs) multiplied by 104. BC indicates buffy coat. (B) Relative HLA-G1 transcriptional levels in lesional skin and PBMCs of primary CL patients. Note that 4 patients with CTCL (TL6, TL9, TL11, TL12) exhibited an average 4 times higher HLA-G transcription in lesional skin than in PBMCs.
HLA-G protein is expressed preferentially in indolent CBCL or in high-grade CTCL
Immunohistochemical staining with 4H84 mAb showed HLA-G protein up-regulation in 23 (51%) of 45 cases (7 of 10 CBCL, 16 of 35 CTCL) (Table 1). According to the number of HLA-G–expressing cells, observed HLA-G positivity was described as a strong (numerous cells dispersed within the malignant infiltrate) or as a single-cell positivity (scant, scattered cells throughout the infiltrate). Correspondingly, strong HLA-G expression could be detected in half of the positive cases in both lymphoma groups (6 of 10 CBCL, 8 of 16 CTCL). None of the samples displayed alterations in total HLA class I expression, evaluated with TP25.99 mAb.
No particular pattern of HLA-G expression could be elucidated. However, in CL types prone to create the structures similar to their physiologic counterparts, such as follicular center cell lymphoma (FCCL) or marginal zone B-cell lymphoma (MZBCL), the presence of HLA-G was restricted to the mantle zone of pseudofollicular structures (Figures 4B-C). The tendency of HLA-G+ cells toward a particular dermal compartment was not observed, although there was occasional close contact with epidermis (Figure 4A).
HLA-G immunoreactivity in primary CLs.
(A) FCCL (patient BL2, original magnification [OM] × 100). (B) FCCL (patient BL1, OM × 160). (C) MZBCL (patient BL5, OM × 100). (D) Large cell CD30−, CD8+, TIA-1+ CTCL (patient TL17, OM × 200). (E) GSS (patient TL135., OM × 100). (F) A giant multinucleated HLA-G+ cell in GSS (OM × 200). (G) Double immunostaining for HLA-G (red) and CD79a (blue-purple) in FCCL (patient BL4, OM × 630). (H) Double immunolabeling of HLA-G (red) and CD3 (blue-purple) in FCCL (patient BL4, OM × 630). (I) HLA-G (red) and CD3 (blue-purple) immunoreactivity in CTCL (patient TL135., OM × 630). (J) Expression of HLA-G (red) and IL-10 (blue-purple) in CTCL, MF (patient TL13, OM × 200). Colocalization of HLA-G/IL-10 creates a mixed violet-purple pigment, as shown also in panel K (patient TL17, OM × 400). (L) Double immunostaining of HLA-G (red) and CD1a (blue-purple) in CTCL (patient TL4, OM × 400).
HLA-G immunoreactivity in primary CLs.
(A) FCCL (patient BL2, original magnification [OM] × 100). (B) FCCL (patient BL1, OM × 160). (C) MZBCL (patient BL5, OM × 100). (D) Large cell CD30−, CD8+, TIA-1+ CTCL (patient TL17, OM × 200). (E) GSS (patient TL135., OM × 100). (F) A giant multinucleated HLA-G+ cell in GSS (OM × 200). (G) Double immunostaining for HLA-G (red) and CD79a (blue-purple) in FCCL (patient BL4, OM × 630). (H) Double immunolabeling of HLA-G (red) and CD3 (blue-purple) in FCCL (patient BL4, OM × 630). (I) HLA-G (red) and CD3 (blue-purple) immunoreactivity in CTCL (patient TL135., OM × 630). (J) Expression of HLA-G (red) and IL-10 (blue-purple) in CTCL, MF (patient TL13, OM × 200). Colocalization of HLA-G/IL-10 creates a mixed violet-purple pigment, as shown also in panel K (patient TL17, OM × 400). (L) Double immunostaining of HLA-G (red) and CD1a (blue-purple) in CTCL (patient TL4, OM × 400).
Concerning the type of CL predominantly expressing HLA-G, in CBCL indolent types (FCCL and MZBCL) demonstrated prominent HLA-G protein expression (Figure 4C). Within the CTCL group, MF cases of a stage ranging from Ia to IIb were clearly negative for HLA-G. Conversely, in MF patients with advanced stage (III-IVb) as well as in patients with CD30- large or pleomorphic CTCL (Figure 4D,J), HLA-G immunoreactivity was a consistent phenomenon (Table 1). On the contrary, in CD30+ lesions HLA-G expression was either not detectable (lymphomatoid papulosis) or present as a rare, single-cell positivity (CD30+ large cell and pleomorphic lymphoma).
HLA-G protein expression was observed in case of granulomatous slack skin (GSS), a rare type of CTCL characterized by a dense granulomatous CD4+ T-cell infiltrate and by the presence of multinucleated giant cells involved in the destruction of elastic tissue (Figure 4E). Additionally, 2 cases of CTCL showing cytotoxic, CD8+ phenotype displayed a discrete but noticeable HLA-G protein presence.
We performed a series of double staining, labeling HLA-G together with common B-cell (CD79a) and T-cell (CD3) markers to characterize further HLA-G–expressing cells. In FCCL and MZBCL, small to medium-sized B lymphocytes expressed HLA-G, which was confirmed also by double staining with CD79a (Figure 4G). Moreover, infiltrating T cells (CD3+) were also shown to express HLA-G (Figure 4H). In diffuse large B-cell lymphoma, an additional HLA-G+ cell type was large cells with vesicular nuclei and prominent nucleoli. Macrophages and endothelial cells occasionally displayed HLA-G immunoreactivity independent of the CL histology type. Moreover, plasma cells were shown quite repeatedly to be HLA-G+ irrespective of the CL type. Whether it was an artifact or specific staining remains elusive.
In CTCL, overall scattered small to medium sized T-lymphocytes were the cells most frequently expressing HLA-G (Figure 4D,I,J). In large and pleomorphic CTCL, large and pleomorphic T cells, respectively, contributed to the HLA-G+ cell population (Figure 4D). A case of GSS demonstrated fine cytoplasmic HLA-G staining of multinucleated giant cells and accompanying spindle-shaped histiocytes in addition to scattered small lymphocytes (Figure 4E-F).
Simultaneous labeling of CD1a (Langerhans cells) and HLA-G demonstrated that HLA-G+ cells are often in close contact with CD1a-marked cells irrespective of the CL type. Apart from the morphology changes of Langerhans cells when they migrate to the dermis (more rounded, less dendritic in shape, decreased CD1a expression), we have noticed an occasional HLA-G coexpression (Figure 4L).
IL-10 expression coincides with HLA-G protein up-regulation
The intensity of IL-10 immunoreactivity was assessed according to the same criteria (see above) as for HLA-G. IL-10–producing cells were detected in 16 (73%) of 22 HLA-G+ CL cases (one-tailed Spearman rank correlation test, rho = . 761, P<.001) (Table 1). IL-10–secreting cells were either colabeled with HLA-G or located in the vicinity of HLA-G–expressing cells (Figure 4J-K). An inverse correlation between HLA-G1 transcription intensity and the expression of IL-10 could be demonstrated in the way that the highest HLA-G1 transcription was associated with absence of IL-10 in tumorous skin (one-tailed Spearman rank correlation test, rho = −.586,P = .007) (Figure 2B). Additionally, a statistical difference in HLA-G1 transcriptional levels could be shown between the groups exhibiting strong, single-cell, or no IL-10 expression (Kruskal-Wallis test, H = 6.954, P = .031).
Discussion
Among the HLA class I antigens, 2 families are distinguishable, both sharing homology at the sequence level consistent with a broadly similar secondary structure.41 These 2 groups consist of ubiquitously expressed, highly polymorphic class Ia (classical) HLA-A, -B and -C molecules and selectively expressed, rather monomorphic class Ib (nonclassical) molecules, principally HLA-E, -F, -G, all of them having features making them quite distinct from the members of class Ia group.21,30,42 HLA-G has gained interest because of its involvement in tumor immunescape and progression.16,18,19 43 We have investigated HLA-G expression in different CL subtypes, presuming that its expression on one side could account for the lesions' immunologic “stealthiness” and, on the other side, be an additional feature involved in the transition from low- to high-grade lymphoma.
We have demonstrated that HLA-G message was present in lesional and uninvolved skin of CL patients as well as in normal skin. A case-to-case diversity of the isoforms detected is probably due to a malignant transformation itself and a different cellular environment.23,24 We have shown the full-length HLA-G transcript in all of the samples, confirming that this is the most commonly transcribed HLA-G form in PBMCs and keratinocytes.24,44 Only in situations in which the translation of the HLA-G1 protein is impaired (ie, individuals homozygous for the null allele HLA-G*0105N), the other shorter isoforms might serve as a “biological back-up” undertaking necessary HLA-G functions, although the HLA-G transcription and splicing are not affected.45 Therefore, we have focused our quantitative PCR primarily on this isoform.
Comparison of the HLA-G1 transcription intensity of lesional skin samples and nonlesional ones with healthy skin showed no statistical difference, possibly due to the abundant HLA-G message in keratinocytes in healthy skin.44 Because the lymphocytes are the predominant cell population within the lymphomatous lesions, the end PCR result is likely to reflect the transcription of the infiltrate rather than the one of solely active epidermal basal cell layer. On the other hand, observed significant case-to-case differences in HLA-G1 levels are probably reflecting the interindividual differences of patients, unique biology of the lesions, or various clinical stages of the disease. With respect to that, the CTCL group exhibited the highest HLA-G1 transcriptional levels, particularly low-stage MF, but not the HLA-G protein expression. The inverse correlation between the HLA-G transcription and HLA-G (or IL-10) protein expression might be in part explained by the effects that IL-10 exerts on HLA-G transcription and protein expression. Namely, after IL-10 stimulation monocytes up-regulated cell surface HLA-G expression, but the mRNA levels remained unchanged.46 On the other hand, IL-10–induced HLA-G expression in purified trophoblast cells was followed by a significant increase in HLA-G mRNA, suggesting a cell type–dependable use of different regulatory mechanisms, ie, transcriptional or posttranscriptional regulation of HLA-G expression.46-48Another explanation for high HLA-G1 mRNA levels in HLA-G−early CTCL cases might be a different, interferon-γ–polarized cytokine profile in these lesions,49,50 knowing the capability of interferon-α to produce increased in HLA-G mRNA without protein up-regulation.25,51 52 It is conceivable that such a particular micromilieu could account for the difference between the skin and PBMC HLA-G1 transcriptional levels noted in 4 CTCL cases.
Irrespective of the CL type, the strongest HLA-G immunoreactivity accounted for the 20% of the cellular infiltrate. Two populations of cells expressing HLA-G were identified: lymphoid cells of B or T phenotype and cells of myeloid origin, such as macrophages and dendritic cells. Surprisingly, no change in total HLA class I pattern could be detected. This might be due to the central role of lymphocytes in immune response in a way that any change of HLA class I status would be a disadvantage, rendering them immediately susceptible to destruction. Moreover, coexpression of HLA-G on targets with intact HLA class I molecules would favor evasion of immunosurveillance, inhibiting NK- and cytotoxic T-lymphocyte–mediated antitumor response.53 54
Shift from a Th1 to Th2 cytokine profile and subsequent expression of IL-10 has been associated with the disease progression and impairment of tumor immunosurveillance in different malignancies as well as in advanced MF.1,7,55Additionally, IL-10 has been shown to be an autocrine growth factor in B-cell lymphomas.8 56 In our study HLA-G immunoreactivity was often associated with IL-10 expression in CTCL and CBCL, implying that the IL-10 might be one of the factors participating in the up-regulation of HLA-G. Moreover, the induction of HLA-G might serve as an additional way that IL-10 modulates and abrogates the CL-associated immune response.
In CBCL, HLA-G–expressing cells seemed to avoid “germinal centers” and to reside in the mantle zone. The mantle zone is a region of intensive immune interactions, such as antigen presentation and immunomodulation, where HLA-G+ cells might be also involved, because HLA-G is capable of presenting peptides57,58 and modulating the release of cytokines in PBMCs upon direct contact.59,60 Indeed, in our CL cases dendritic cells were often in close contact with HLA-G–bearing cells; however, the nature of this interaction remains unclear. Occasionally in CBCL, and according to the distribution of infiltrate in CTCL, HLA-G–expressing cells were located often subepidermally. Epidermal keratinocytes are potent producers of various cytokines,61 62 which could have a profound influence on the malignant infiltrate as well as on the HLA-G up-modulation.
HLA-G expression in CTCL was clearly evident in MF patients with advanced stage or in ones that underwent large cell transformation and also in CD30− large cell CTCL. All of these entities are associated with aggressive clinical behavior and poor survival.37,63,64 CD30− CTCL with the predominance of small or medium-sized pleomorphic T cells has been described as a separate entity, due to its better prognosis.37 Accordingly, HLA-G expression in these cases was not so pronounced, suggesting its true up-regulation only in highly malignant conditions such as CD30− large cell lymphomas or after a large-cell transformation in MF. Another group of primary CL, primary cutaneous CD8+ cytotoxic CTCL, was recently shown to express an HLA-G ligand, KIR molecule ILT-2.65Concurrent expression of HLA-G (as shown in 2 CD8+CTCL cases in our study) and interaction with clonal and host KIRs66 could account for growth/survival advantage of the dominant clone.65 Together with a shift toward a Th2 cytokine profile, HLA-G induction might be responsible for the circumvention of host immune response and subsequent disease aggressiveness and progression. HLA-G could be contributing as well to the tumor-associated angiogenesis, as hypothesized in placenta,67,68 due to its consistent expression by endothelial cells in our study. Within the group of CD30+primary cutaneous lymphoproliferative disorders, HLA-G expression could not be detected in self-healing lymphomatoid papulosis, while in CD30+ large cell CTCL it was limited to a sparse, single-cell positivity. This might be understandable, knowing that primary large CTCL expressing CD30 antigen has a significantly better prognosis than CD30− large cell CTCL.37 69
Apart from lymphoid cells, intralesional macrophages and dendritic cells displayed HLA-G immunoreactivity in many CL cases, initially shown in lung cancer.36 As a curiosity, large multinucleate giant cells in GSS, which are of monocyte-macrophage lineage,70 demonstrated strong HLA-G expression, a feature apparently inherent to monocyte lineage.71 72
Given the fact that B and T lymphocytes as well as macrophages and dendritic cells bear KIRs (ILT-2, ILT-4, p49),66,73some of which have been shown to bind soluble HLA-G1 tetrameric complexes,74 an open question remains as to whether HLA-G immunoreactivity in these cells is a phenomenon per se or an artifact due to the binding of soluble HLA-G molecules (ie, HLA-G5, -G6, -G7) to these receptors, particularly because the antibodies specific to the soluble protein are not yet available for use in immunohistochemistry. This might account for a strong HLA-G positivity in indolent, low-grade CBCL and of plasma cells in general, knowing that ILT-2 is present virtually on all B cells and its expression changes during the course of B-cell differentiation, ie, going up as the cells mature.31,35 75 The dedifferentiation is probably the reason why the large cell CBCL displayed a weak HLA-G positivity.
In conclusion, we have demonstrated for the first time expression of HLA-G and its association with IL-10 production in different types of primary CLs. The distinct pattern of HLA-G expression in CL suggests that this nonclassical molecule might be either interfering with the mounting of an efficient tumor-associated immune response in CL or facilitating the transition from low- to high-grade lymphoma. Whereas the role of class Ia antigens in immune recognition is well understood both in structural and functional terms, in the field of nonclassical HLA molecules and their putative ligands many questions remain to be answered.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Reinhard Dummer, UniversitätsSpital Zürich, Dermatologische Klinik, Gloriastrasse 31, CH-8091 Zürich, Switzerland; e-mail:dummer@derm.unizh.ch.


![Fig. 4. HLA-G immunoreactivity in primary CLs. / (A) FCCL (patient BL2, original magnification [OM] × 100). (B) FCCL (patient BL1, OM × 160). (C) MZBCL (patient BL5, OM × 100). (D) Large cell CD30−, CD8+, TIA-1+ CTCL (patient TL17, OM × 200). (E) GSS (patient TL135., OM × 100). (F) A giant multinucleated HLA-G+ cell in GSS (OM × 200). (G) Double immunostaining for HLA-G (red) and CD79a (blue-purple) in FCCL (patient BL4, OM × 630). (H) Double immunolabeling of HLA-G (red) and CD3 (blue-purple) in FCCL (patient BL4, OM × 630). (I) HLA-G (red) and CD3 (blue-purple) immunoreactivity in CTCL (patient TL135., OM × 630). (J) Expression of HLA-G (red) and IL-10 (blue-purple) in CTCL, MF (patient TL13, OM × 200). Colocalization of HLA-G/IL-10 creates a mixed violet-purple pigment, as shown also in panel K (patient TL17, OM × 400). (L) Double immunostaining of HLA-G (red) and CD1a (blue-purple) in CTCL (patient TL4, OM × 400).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/99/2/10.1182_blood.v99.2.609/6/m_h80222020004.jpeg?Expires=1769161287&Signature=In6uOgueFCTiY0SU8PUhzdiaYPg8CwVRAqzT-~McEJMr7X2MNYKMxNdyvYGsm2RQKmZNIPXDeiNcPWFebI1oPa8LJHg6301476Pp2IijvhQtWq1Yx7qBA0QNWe95rOlIDrtrTpKVr3d98SMEntlFp0bXfEqjr-qgziCD0096Mnne6adFHsBfgBDXf22Gg~aKlG3Zh2A1kvMlbeOIB407Xinv2Rk6KRayMgDGq01DmJDx0eFHlXIVqFYvlE-qlgELwaPofXEBLyjjJEddrljJzqGjG9jl1rfH8hDspWoGit9YDFke2BCMPng7VzUT1JDp03CYKiFa6FkOKW4k~eQp-g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal