Abstract
The unique clinicopathologic features of Hodgkin lymphoma (HL) are due to the multiple cytokines produced by its neoplastic cells, the Hodgkin and Reed-Sternberg (HRS) cells. Cytokine signaling is mediated through the signal transducer and activator of transcription (STAT) family of transcription factors. Immunoblotting and immunohistochemistry were used to examine cell lines and tissue sections derived from patients with HL and non-Hodgkin lymphoma (NHL) for expression of activated STAT proteins. Constitutive phosphorylation of STAT6 and STAT3 was common in HL. STAT6 was constitutively phosphorylated in 5 of 5 HL cell lines and in HRS cells from 25 of 32 (78%) classical HL cases. STAT3 was constitutively phosphorylated in 4 of 5 HL cell lines and in HRS cells from 27 of 31 (87%) classical HL cases. Only 4 of 24 NHL cases demonstrated constitutive STAT6 activation, whereas STAT3 activation was observed in 6 of 13 (46%) cases of B-cell NHL and 8 of 11 (73%) cases of T-cell NHL. Constitutive STAT5 phosphorylation was not a common feature of HL or NHL. STAT6 mediates signaling by interleukin 13 (IL-13), a cytokine frequently expressed by HRS cells. Antibody-mediated neutralization of IL-13 resulted in significant decreases in both cellular proliferation and levels of phosphorylated STAT6 of HL cell lines. In conclusion, constitutive STAT6 phosphorylation is a common and distinctive feature of HRS cells in classical HL, whereas STAT3 activation was regularly present in both HL and NHL. These results suggest that IL-13 signaling is largely responsible for the constitutive STAT6 activation observed in HRS cells and further implicate IL-13 as an important growth factor in classical HL.
Introduction
Hodgkin lymphoma (HL) has distinct pathologic and clinical features that distinguish it from non-Hodgkin lymphoma (NHL). In HL, the neoplastic Hodgkin and Reed-Sternberg (HRS) cells make up only a small proportion of the clinically detectable mass, the bulk of the tumor being composed of a reactive infiltrate of lymphocytes, eosinophils, plasma cells, histiocytes, and fibroblasts.1Patients with HL also often present with fever, night sweats, and weight loss, consistent with an abnormal pattern of cytokine expression. Studies of HL-derived cell lines and tissues involved by HL have shown that HRS cells produce multiple cytokines, including interleukin-5 (IL-5), IL-6, IL-7, IL-9, IL-10, granulocyte-macrophage colony-stimulating factor (GM-CSF), lymphotoxin α, and transforming growth factor β (TGF-β).2 In addition, we have recently demonstrated that IL-13 is expressed by HRS cells in a large majority of patients with HL.3,4 Moreover, HRS cells and cells within the reactive infiltrate express cytokine receptors such as the IL-2 receptor (IL-2R),5 IL-6R,6 and IL-13R,4 suggesting the potential for a response to these cytokines. HRS-produced cytokines may be acting as autocrine growth factors or may influence cells in the reactive infiltrate in a paracrine manner. However, the small amount of direct evidence implicating cytokines as growth factors in HL has been obtained from studies of cell lines in vitro, a situation that most likely does not reflect all aspects of the biology of HRS cells in vivo. Evidence for cytokine activity in actual HL-involved tissue has yet to be reported.
The interaction of cytokines with their specific cell surface receptors triggers the activation of intracellular signaling cascades that ultimately have effects on multiple cellular functions. The signal transducer and activator of transcription (STAT) family of proteins plays a central role in cytokine signaling.7 STAT proteins are latent transcription factors located in the cytoplasm that become activated by phosphorylation on a single tyrosine residue in response to cytokine receptor stimulation. This tyrosine phosphorylation leads to STAT dimerization and translocation to the nucleus, followed by activation of transcription resulting in changes to gene expression.7 The 7 members of the STAT family can be broadly divided into 2 groups based on their activation patterns in vivo. STAT1, STAT3, STAT5a, and STAT5b are activated by a wide variety of extracellular and intracellular stimuli, whereas STAT2, STAT4, and STAT6 show a more restricted pattern of activation and are triggered by only 1 or 2 extracellular factors.8 For example, STAT3 is activated by IL-2, IL-6, IL-7, IL-9, IL-10, and IL-15,8 as well as by intracellular tyrosine kinases such as src and abl.9,10 Similarly, STAT5 is activated in response to IL-2, IL-3, IL-5, IL-7, IL-9, and GM-CSF.8 In contrast, STAT6 is primarily activated by IL-4 and IL-13.11Importantly, constitutive activation of STATs, particularly STAT3 and STAT5, has been associated with oncogenic transformation.12
We previously identified IL-13 and IL-13R expression as common features of HRS cells in HL.3,4 IL-13 and IL-4 have similar biologic functions,13 and both cytokines activate STAT611,14; however, IL-4 is not typically expressed by HRS cells.15 We have therefore hypothesized that IL-13 expression by HRS cells and their autocrine response to this cytokine may be linked to HL pathogenesis. To pursue this line of investigation, we examined the activation of several STAT proteins in a series of lymphomas from patients with HL and NHL. Our results further implicate IL-13 signaling via STAT6 as a major factor in the pathogenesis of HL.
Materials and methods
Cell lines, tissues, and reagents
The Epstein-Barr virus-negative (EBV−) HRS cell lines L-428, KM-H2, HDLM-2, and L-540, the lymphoblastoid cell line LCL-HO, and the anaplastic large cell lymphoma (ALCL) cell line KARPAS-299 were purchased from the German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany). The EBV− HRS cell line L-1236 was provided by Dr V. Diehl (University of Cologne, Cologne, Germany). The lymphoblastoid cell line LCL-GK was a gift from Dr M. Dosch (Hospital for Sick Children, Toronto, ON, Canada), and the EBV− diffuse large B-cell lymphoma (DLBCL)–derived cell lines Ly2 and Ly10 were provided by Dr H. Messner (Ontario Cancer Institute/Princess Margaret Hospital, Toronto, ON, Canada). Daudi and TF-1 cells were purchased from American Type Culture Collection (ATCC; Rockville, MD). All cell lines were grown in RPMI 1640 supplemented with penicillin, streptomycin, and 10% fetal calf serum (FCS).
Formalin-fixed paraffin-embedded biopsy specimens representing the following diseases were investigated: 32 cases of classical HL, including 21 cases of nodular sclerosis HL (NSHL), 8 cases of mixed cellularity HL (MCHL), and 3 cases of lymphocyte-depletion HL (LDHL); 4 cases of nodular lymphocyte predominance HL (NLPHL); 5 cases of T-cell–rich B-cell lymphoma (TCRBCL); 8 cases of DLBCL; 6 cases of ALCL with a T/null cell phenotype; and 5 cases of peripheral T-cell lymphoma, unspecified (PTCL). All cases were diagnosed according to the Revised European-American Lymphoma Classification.16 All patients were negative for the human immunodeficiency virus. By immunohistochemistry, the Reed-Sternberg cells in classical HL cases were CD15+ and CD30+, and the lymphocytic and histiocytic cells in NLPHL cases were CD20+, CD15−, and CD30−.
Phosphospecific polyclonal antibodies that detect the corresponding STAT protein only when it is phosphorylated at the indicated tyrosine residue were used as follows: Phospho(Tyr705)-STAT3, Phospho(Tyr694)-STAT5, and Phospho(Tyr641)-STAT6 (Cell Signaling Technology, Beverly, MA). For immunoblotting, antibodies were used at a 1:1000 dilution. For immunohistochemistry, anti–P-STAT5 and anti–P-STAT6 were used at 1:100, and anti–P-STAT3 was used at 1:200. Antibodies detecting total STAT5 and STAT6 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and the antibody detecting total STAT3 was from Cell Signaling Technology. The antibody detecting EBV latent membrane protein-1 (LMP-1) was purchased from Dako (Carpinteria, CA).
Immunoblotting
Cells were lysed in ice-cold lysis buffer consisting of 1% Triton X-100, 20 mM Tris-HCl pH 8.0, 150 mM NaCl, 20 mM EDTA, 1 μg/mL leupeptin, 1 μg/mL aprotinin, 2 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 mM NaF, 0.4 mM NaVO4, and 20 mM β-glycerol phosphate. Whole-cell lysates were fractionated on 8% sodium dodecyl sulfate (SDS)–polyacrylamide gels (Novex, San Diego, CA). Proteins were transferred to polyvinylidene difluoride (PVDF) membranes (Roche Diagnostics, Indianapolis, IN) and immunoblotted with specific antibodies. Equal protein loading was verified by reprobing the blots with antiactin antibody (Santa Cruz Biotechnology). Total-cell extracts from interferon α (IFN-α)–treated and untreated serum-starved HeLa cells, which served as controls for P-STAT3 immunoblotting, were purchased from Cell Signaling Technology.
Immunohistochemistry
Phosphorylated STAT proteins were detected in formalin-fixed paraffin-embedded tissue sections using standard immunohistochemical techniques. Briefly, tissue slides were immersed in 10 mM sodium citrate buffer, pH 6.0, and heated to maximal pressure in a microwave pressure cooker (Dako). Slides were allowed to cook for an additional 2 minutes to complete heat-induced antigen retrieval. Slides were then incubated with the primary antibody overnight at room temperature. Antibody binding was detected using a biotinylated antirabbit IgG, Vectastain avidin-biotinylated enzyme complex kit, and 3,3′-diaminobenzidine (Vector Labs, Burlingame, CA). Cases were considered positive if 10% or greater of the malignant cell population demonstrated nuclear staining.
In some experiments, the specificity of immunohistochemical staining was confirmed by preabsorbing the phosphospecific antibody preparation for 1 hour at room temperature with a 10-fold excess (by weight) of the tyrosine-phosphorylated peptides against which the antibodies were raised (kindly provided by Cell Signaling Technology). The absorbed antibody preparation was then used for immunohistochemical staining as above.
In situ hybridization
Interleukin 4 messenger RNA (mRNA) in formalin-fixed paraffin-embedded tissue and IL-13 mRNA in cytospin preparations of cell lines was detected by in situ hybridization as previously described.4 Cytospins were prepared according to the manufacturer's directions (Shandon, Pittsburgh, PA), air-dried, and fixed in 50% methanol/50% acetone at room temperature for 6 minutes. The IL-4 probe was a 278-base pair (bp)EcoRI/EcoRV fragment of pcD-hIL-4 (ATCC) subcloned into pBluescript SK (Stratagene, La Jolla, CA). The probe for IL-13 has been previously described.3
Cytokine production
Supernatants of cell cultures (5 × 105/mL) were recovered 48 hours after medium exchange and assayed by enzyme-linked immunosorbent assay (ELISA) for IL-13 (Biosource International, Camarillo, CA) and IL-4 (R & D Systems, Minneapolis, MN). The sensitivities of IL-13 and IL-4 detection were 12 pg/mL and 10 pg/mL, respectively.
Analyses of cell proliferation and apoptosis
Cells were cultured in 96-well flat-bottomed plates at 3 × 104 cells/well for 24, 48, or 72 hours in the presence of varying concentrations of anti–IL-13 or isotype control antibodies (BD Pharmingen, San Diego, CA). Cells were also treated with varying concentrations of IL-13 (R & D Systems). [3H]-thymidine (1μCi [0.037 MBq]/well; Amersham Pharmacia Biotech, Piscataway, NJ) was added to each well for the last 16 hours of incubation. The cells were harvested on filters and the incorporation of [3H]-thymidine into cellular DNA was measured as previously described.17 Apoptosis was determined by FACS analysis using the TACS annexin V–fluorescein isothiocyanate (FITC) apoptosis detection kit (R & D Systems) following the manufacturer's instructions.
Results
HL-derived cell lines demonstrate variable STAT activation patterns
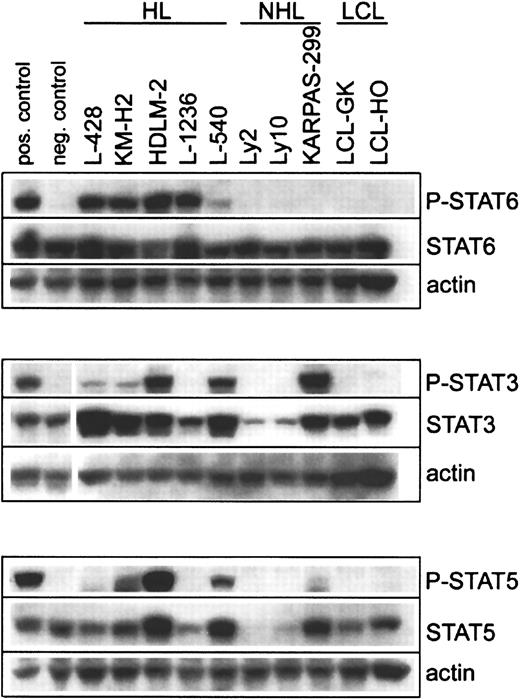
To investigate the activation status of STAT proteins in HL-derived cell lines, L-428, KM-H2, HDLM-2, L-1236, and L-540 cells were analyzed for phosphorylated STAT proteins (P-STAT) by immunoblotting. All 5 HL cell lines stained positively for P-STAT6, whereas the 2 DLBCL, 1 ALCL, and 2 B-lymphoblastoid cell lines examined were all negative (Figure 1). Four of 5 HL cell lines showed comparable levels of P-STAT6, but significantly lower levels were present in L-540 cells. Phosphorylation of STAT3 and STAT5 in HL cell lines was more variable (Figure 1). P-STAT3 was detected in 4 of 5 HL cell lines, with high levels in HDLM-2 and L-540 but significantly lower levels in L-428 and KM-H2. P-STAT5 was detected in 3 of 5 HL cell lines, with the highest levels seen in HDLM-2, and lower levels in L-540 and KM-H2. P-STAT3 and P-STAT5 were also detected in KARPAS-299, a cell line derived from an ALCL with t(2;5). The DLBCL and lymphoblastoid cell lines did not show any evidence of STAT3 or STAT5 tyrosine phosphorylation.
Phosphorylation of STAT proteins in HL and NHL cell lines.
Cell lysates were prepared from HL-derived cell lines (L-428, KM-H2, HDLM-2, L-1236, L-540), B-cell NHL-derived cell lines (Ly2, Ly10), a T-cell NHL-derived cell line (KARPAS-299), and B-cell lymphoblastoid cell lines (LCL-GK, LCL-HO). Equal amounts of whole-cell lysates were subjected to SDS–polyacrylamide gel electrophoresis followed by Western blot analysis. Phosphorylated STAT proteins were detected using phosphospecific antibodies recognizing Tyr641-phosphorylated STAT6, Tyr705-phosphorylated STAT3, and Tyr694-phsophorylated STAT5. These experiments were repeated twice with similar results. For P-STAT6, Daudi cells stimulated for 15 minutes with 10 ng/mL IL-4 served as the positive control, and unstimulated Daudi cells served as the negative control. For P-STAT3, serum-starved HeLa cells stimulated with IFN-α served as the positive control, and unstimulated serum-starved HeLa cells served as the negative control. For P-STAT5, TF-1 cells stimulated for 15 minutes with 25 ng/mL GM-CSF served as the positive control, and unstimulated TF-1 cells served as the negative control.
Phosphorylation of STAT proteins in HL and NHL cell lines.
Cell lysates were prepared from HL-derived cell lines (L-428, KM-H2, HDLM-2, L-1236, L-540), B-cell NHL-derived cell lines (Ly2, Ly10), a T-cell NHL-derived cell line (KARPAS-299), and B-cell lymphoblastoid cell lines (LCL-GK, LCL-HO). Equal amounts of whole-cell lysates were subjected to SDS–polyacrylamide gel electrophoresis followed by Western blot analysis. Phosphorylated STAT proteins were detected using phosphospecific antibodies recognizing Tyr641-phosphorylated STAT6, Tyr705-phosphorylated STAT3, and Tyr694-phsophorylated STAT5. These experiments were repeated twice with similar results. For P-STAT6, Daudi cells stimulated for 15 minutes with 10 ng/mL IL-4 served as the positive control, and unstimulated Daudi cells served as the negative control. For P-STAT3, serum-starved HeLa cells stimulated with IFN-α served as the positive control, and unstimulated serum-starved HeLa cells served as the negative control. For P-STAT5, TF-1 cells stimulated for 15 minutes with 25 ng/mL GM-CSF served as the positive control, and unstimulated TF-1 cells served as the negative control.
HRS cells in HL-involved tissue also demonstrate variable STAT activation patterns
The utility of the anti–P-STAT antibodies in fixed cellular material was first tested in cytospin preparations of HDLM-2 (positive for P-STAT3, P-STAT5, and P-STAT6 by immunoblotting) and LCL-GK (negative for P-STAT3, P-STAT5, and P-STAT6 by immunoblotting). HDLM-2 showed nuclear staining for all 3 P-STAT antibodies, whereas LCL-GK cells were negative (data not shown). To investigate the activation status of STAT proteins in lymphomas, immunohistochemical analysis of phosphorylated STAT proteins was performed on primary biopsy material from patients with HL and NHL and on normal lymphoid tissue. In a section of a benign reactive human tonsil, P-STAT6 (Figure2A) and P-STAT5 (Figure 2C) were present in only a few nuclei of scattered lymphocytes outside of the germinal center. Basal STAT3 activation in the normal tonsil was significantly higher, with numerous P-STAT3+ cells within and outside the germinal center (Figure 2B).
Expression of phosphorylated STAT proteins in normal lymphoid tissue.
Expression of P-STAT6 (A), P-STAT3 (B), and P-STAT5 (C) was examined by immunohistochemistry in formalin-fixed sections of a reactive human tonsil. P-STAT6 and P-STAT5 demonstrated low basal levels of expression, with rare nuclear staining of lymphocytes (arrows) outside of the germinal center (GC). Basal levels of P-STAT3 were significantly higher, with numerous positive cells within and outside of the GC. Original magnifications, × 500.
Expression of phosphorylated STAT proteins in normal lymphoid tissue.
Expression of P-STAT6 (A), P-STAT3 (B), and P-STAT5 (C) was examined by immunohistochemistry in formalin-fixed sections of a reactive human tonsil. P-STAT6 and P-STAT5 demonstrated low basal levels of expression, with rare nuclear staining of lymphocytes (arrows) outside of the germinal center (GC). Basal levels of P-STAT3 were significantly higher, with numerous positive cells within and outside of the GC. Original magnifications, × 500.
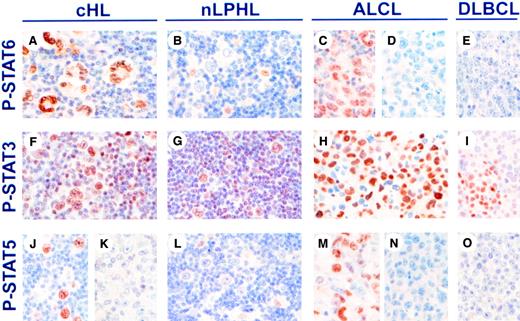
STAT activation was then studied in biopsies of malignant lymphomas. P-STAT6 was detected in nuclei of HRS cells in 25 of 32 (78%) of classical HL cases (Table 1 and Figure3A). Six cases showed fainter cytoplasmic staining in addition to nuclear staining. In contrast to classical HL, the lymphocytic and histiocytic cells in all 4 cases of NLPHL were P-STAT6− (Figure 3B). Similarly, the malignant cell population of only 2 of 6 ALCL cases (Figure 3C,D) and the malignant cells of 2 of 5 cases of TCRBCL expressed P-STAT6. Interestingly, the 2 P-STAT6+ ALCL cases were previously reported to express IL-13,4 whereas 3 P-STAT6− ALCL cases that were previously examined for IL-13 expression were found to be IL-13−.4All cases of DLBCL (Figure 3E) and PTCL were P-STAT6−.
Phosphorylated STAT protein expression in malignant lymphomas
| Diagnosis . | P-STAT6+ malignant cells (%) . | P-STAT3+ malignant cells (%) . | P-STAT5+ malignant cells (%) . |
|---|---|---|---|
| Classical HL | 25/32 (78) | 27/31 (87) | 8/31 (26) |
| NSHL | 20/21 (95) | 18/20 (90) | 4/20 (20) |
| MCHL | 4/8 (50) | 7/8 (88) | 4/8 (50) |
| LDHL | 1/3 (33) | 2/3 (67) | 0/3 |
| NLPHL | 0/4 | 2/4 (50) | 0/4 |
| DLBCL | 0/8 | 3/8 (38) | 0/8 |
| TCRBCL | 2/5 (40) | 3/5 (60) | 0/5 |
| ALCL | 2/6 (33) | 5/6 (83) | 3/6 (50) |
| PTCL, unspecified | 0/5 | 3/5 (60) | 0/5 |
| Diagnosis . | P-STAT6+ malignant cells (%) . | P-STAT3+ malignant cells (%) . | P-STAT5+ malignant cells (%) . |
|---|---|---|---|
| Classical HL | 25/32 (78) | 27/31 (87) | 8/31 (26) |
| NSHL | 20/21 (95) | 18/20 (90) | 4/20 (20) |
| MCHL | 4/8 (50) | 7/8 (88) | 4/8 (50) |
| LDHL | 1/3 (33) | 2/3 (67) | 0/3 |
| NLPHL | 0/4 | 2/4 (50) | 0/4 |
| DLBCL | 0/8 | 3/8 (38) | 0/8 |
| TCRBCL | 2/5 (40) | 3/5 (60) | 0/5 |
| ALCL | 2/6 (33) | 5/6 (83) | 3/6 (50) |
| PTCL, unspecified | 0/5 | 3/5 (60) | 0/5 |
P-STAT+ indicates cells expressing phosphorylated (activated) STAT protein.
Expression of phosphorylated STAT proteins in malignant lymphomas.
Expression of P-STAT6 (A-E), P-STAT3 (F-I), and P-STAT5 (J-O) was examined by immunohistochemistry in formalin-fixed sections of classical HL (cHL), nLPHL, ALCL, and DLBCL. Staining for nuclear P-STAT6 was positive in HRS cells of cHL (A). In nLPHL, lymphocytic and histiocytic cells were P-STAT6−, whereas scattered macrophage nuclei were P-STAT6+ (B). P-STAT6 was also positive in 33% cases of ALCL (C-D), and negative in DLBCL (E). Nuclear P-STAT3 was present in HRS cells and cells in the reactive infiltrate in cHL (F), lymphocytic and histiocytic cells and cells in the reactive infiltrate from nLPHL (G), ALCL (H), and DLBCL (I). Nuclear P-STAT5 was present in HRS cells in 26% of cHL (J-K), negative in lymphocytic and histiocytic cells in nLPHL (L), positive in 50% of ALCL (M-N), and negative in DLBCL (O). Original magnifications, × 720.
Expression of phosphorylated STAT proteins in malignant lymphomas.
Expression of P-STAT6 (A-E), P-STAT3 (F-I), and P-STAT5 (J-O) was examined by immunohistochemistry in formalin-fixed sections of classical HL (cHL), nLPHL, ALCL, and DLBCL. Staining for nuclear P-STAT6 was positive in HRS cells of cHL (A). In nLPHL, lymphocytic and histiocytic cells were P-STAT6−, whereas scattered macrophage nuclei were P-STAT6+ (B). P-STAT6 was also positive in 33% cases of ALCL (C-D), and negative in DLBCL (E). Nuclear P-STAT3 was present in HRS cells and cells in the reactive infiltrate in cHL (F), lymphocytic and histiocytic cells and cells in the reactive infiltrate from nLPHL (G), ALCL (H), and DLBCL (I). Nuclear P-STAT5 was present in HRS cells in 26% of cHL (J-K), negative in lymphocytic and histiocytic cells in nLPHL (L), positive in 50% of ALCL (M-N), and negative in DLBCL (O). Original magnifications, × 720.
Among the classical HL cases, P-STAT6 was differentially expressed according to subtype, with NSHL cases demonstrating a significantly higher rate of P-STAT6 expression compared to MCHL or LDHL (Table2). Of the NSHL cases, 20 of 21 (95%) contained P-STAT6+ HRS cells, whereas only 4 of 8 (50%) of MCHL cases and 1 of 3 (33%) of LDHL cases contained P-STAT6+ HRS cells (P = .003). The majority of cells expressing P-STAT6 demonstrated the morphology of HRS cells, whereas few cells in the reactive infiltrate were P-STAT6+. The same cases of classical HL were previously analyzed for IL-13 expression by in situ hybridization,4 and STAT6 activation was significantly associated with IL-13 expression. P-STAT6 was expressed in 24 of 27 (89%) of IL-13+ cases, but in only 1 of 5 (20%) of IL-13− cases (P = .004).
Phosphorylated STAT protein expression in patients with HL
| Case no. . | Diagnosis . | Age, y/sex . | Stage . | P-STAT6+ HRS cells . | P-STAT3+ HRS cells . | P-STAT5+ HRS cells . | EBV LMP-1+ HRS cells . |
|---|---|---|---|---|---|---|---|
| 1 | NSHL | 16/M | IV B | +++ | + | ++ | − |
| 2 | NSHL | 24/F | II A | ++++ | − | − | nd |
| 3 | NSHL | 15/F | II A | + | +++ | − | − |
| 4 | NSHL | 43/M | IV B | +++ | ++++ | − | − |
| 5 | NSHL | 33/M | III B | +++ | ++++ | − | − |
| 6 | NSHL | 30/M | II A | ++++ | ++ | − | − |
| 7 | NSHL | 32/F | II B | +++ | ++++ | − | − |
| 8 | NSHL | 27/F | II A | ++++ | +++ | − | − |
| 9 | NSHL | 22/M | III B | +++ | + | − | +++ |
| 10 | NSHL | 34/M | III A | +++ | +++ | + | − |
| 11 | NSHL | 19/M | III A | ++++ | +++ | − | − |
| 12 | NSHL | 36/M | III B | ++++ | + | − | − |
| 13 | NSHL | 31/F | IV A | ++++ | +++ | − | − |
| 14 | NSHL | na | na | ++++ | nd | nd | nd |
| 15 | NSHL | na | na | + | + | − | nd |
| 16 | NSHL, syn | 64/M | III B | − | − | − | − |
| 17 | NSHL, syn | 20/F | II A | ++++ | +++ | − | − |
| 18 | NSHL, syn | 35/M | I B | ++++ | ++++ | ++ | ++++ |
| 19 | NSHL, syn | 45/M | III A | +++ | +++ | ++++ | − |
| 20 | NSHL, syn | 41/F | II A | ++++ | +++ | − | − |
| 21 | NSHL, syn | 25/M | III B | ++++ | +++ | − | − |
| 22 | MCHL | na | na | ++ | +++ | ++ | ++ |
| 23 | MCHL | na | na | − | ++++ | − | ++++ |
| 24 | MCHL | 28/M | II A | ++++ | ++ | ++ | ++++ |
| 25 | MCHL | 28/M | II B | − | ++ | − | ++++ |
| 26 | MCHL | 42/M | I A | ++ | ++++ | ++++ | +++ |
| 27 | MCHL | 42/F | III A | − | ++ | − | − |
| 28 | MCHL | na/F | II A | + | ++++ | ++ | ++++ |
| 29 | MCHL | 88/M | I A | − | − | − | ++++ |
| 30 | LDHL | 54/F | IV B | − | +++ | − | ++++ |
| 31 | LDHL | 65/M | na | − | − | − | − |
| 32 | LDHL | 72/F | IV B | ++++ | +++ | − | ++++ |
| Case no. . | Diagnosis . | Age, y/sex . | Stage . | P-STAT6+ HRS cells . | P-STAT3+ HRS cells . | P-STAT5+ HRS cells . | EBV LMP-1+ HRS cells . |
|---|---|---|---|---|---|---|---|
| 1 | NSHL | 16/M | IV B | +++ | + | ++ | − |
| 2 | NSHL | 24/F | II A | ++++ | − | − | nd |
| 3 | NSHL | 15/F | II A | + | +++ | − | − |
| 4 | NSHL | 43/M | IV B | +++ | ++++ | − | − |
| 5 | NSHL | 33/M | III B | +++ | ++++ | − | − |
| 6 | NSHL | 30/M | II A | ++++ | ++ | − | − |
| 7 | NSHL | 32/F | II B | +++ | ++++ | − | − |
| 8 | NSHL | 27/F | II A | ++++ | +++ | − | − |
| 9 | NSHL | 22/M | III B | +++ | + | − | +++ |
| 10 | NSHL | 34/M | III A | +++ | +++ | + | − |
| 11 | NSHL | 19/M | III A | ++++ | +++ | − | − |
| 12 | NSHL | 36/M | III B | ++++ | + | − | − |
| 13 | NSHL | 31/F | IV A | ++++ | +++ | − | − |
| 14 | NSHL | na | na | ++++ | nd | nd | nd |
| 15 | NSHL | na | na | + | + | − | nd |
| 16 | NSHL, syn | 64/M | III B | − | − | − | − |
| 17 | NSHL, syn | 20/F | II A | ++++ | +++ | − | − |
| 18 | NSHL, syn | 35/M | I B | ++++ | ++++ | ++ | ++++ |
| 19 | NSHL, syn | 45/M | III A | +++ | +++ | ++++ | − |
| 20 | NSHL, syn | 41/F | II A | ++++ | +++ | − | − |
| 21 | NSHL, syn | 25/M | III B | ++++ | +++ | − | − |
| 22 | MCHL | na | na | ++ | +++ | ++ | ++ |
| 23 | MCHL | na | na | − | ++++ | − | ++++ |
| 24 | MCHL | 28/M | II A | ++++ | ++ | ++ | ++++ |
| 25 | MCHL | 28/M | II B | − | ++ | − | ++++ |
| 26 | MCHL | 42/M | I A | ++ | ++++ | ++++ | +++ |
| 27 | MCHL | 42/F | III A | − | ++ | − | − |
| 28 | MCHL | na/F | II A | + | ++++ | ++ | ++++ |
| 29 | MCHL | 88/M | I A | − | − | − | ++++ |
| 30 | LDHL | 54/F | IV B | − | +++ | − | ++++ |
| 31 | LDHL | 65/M | na | − | − | − | − |
| 32 | LDHL | 72/F | IV B | ++++ | +++ | − | ++++ |
++++ indicates 75% to 100% HRS cells positive; +++, 50% to 74% HRS cells positive; ++, 25% to 49% HRS cells positive; +, 10% to 24% HRS cells positive; −, less than 10% HRS cells positive. nd indicates not done; na, not available; syn, syncytial variant of NSHL.
The activation status of other STATs was determined in HL tissue samples. Nuclear P-STAT3 was detected in HRS cells in 27 of 31 (87%) of classical HL cases (Figure 3F), with no difference in P-STAT3 expression among subtypes of classical HL. Five cases showed fainter cytoplasmic staining in addition to nuclear staining. P-STAT3 was detected in a significant proportion of cells in the reactive infiltrate, including lymphocytes, macrophages, fibroblasts, endothelial cells, and neutrophils, as well as in HRS cells. STAT3 activation was not specific for classical HL, because it was also present in lymphocytic and histiocytic cells in 2 of 4 cases of NLPHL (Figure 3G), and in the malignant cell populations in 5 of 6 cases of ALCL (Figure 3H), 3 of 8 cases of DLBCL (Figure 3I), 3 of 5 cases of TCRBCL, and 3 of 5 cases of PTCL. P-STAT5 was detected in HRS cells in only 8 of 31 (26%) of classical HL cases (Figure 3J,K), and was absent in the lymphocytic and histiocytic cell population of NLPHL (Figure3L). Among NHLs, P-STAT5 was present only in the malignant cells of 3 of 6 cases of ALCL (Figure 3M-O).
Hodgkin lymphoma is associated with EBV in 30% to 50% of cases, and EBV LMP-1 is known to activate STAT proteins, particularly, STAT1 and STAT3.18 We therefore studied the relationship between LMP-1 expression and STAT protein phosphorylation in classical HL. LMP-1 was expressed by HRS cells in 11 of 29 (38%) cases of classical HL (Table 2). No correlation was found between LMP-1 expression and phosphorylation of STAT3, STAT5, or STAT6.
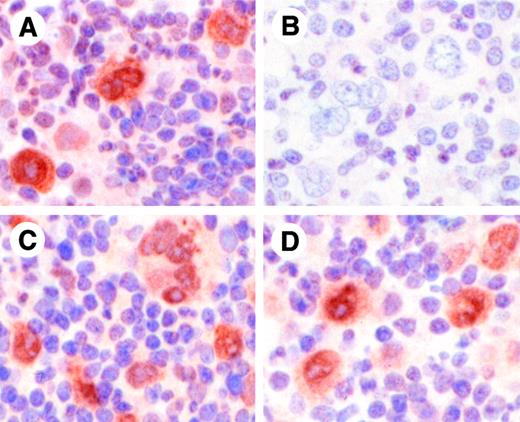
To confirm the specificity of the phosphospecific antibodies used for immunohistochemical analysis, the antibodies were preincubated with the phosphorylated peptides against which the antibodies were raised. HRS cells from a case of NSHL stained positively for P-STAT6 when antibody alone was used (Figure 4A); however, preincubation of the antibody with P-STAT6 peptide abrogated this staining (Figure 4B). Specific P-STAT6 staining was maintained when the anti–P-STAT6 antibody was preincubated with P-STAT3 peptide (Figure4C) or P-STAT5 peptide (Figure 4D). Similarly, preincubation of anti–P-STAT3 antibody with P-STAT3 peptide and preincubation of anti–P-STAT5 antibody with P-STAT5 peptide blocked specific staining with each antibody (not shown).
Specificity of antiphospho-STAT6 antibody for phospho-STAT6.
Antiphospho-STAT6 antibody was preincubated for 1 hour without peptide (A), with phospho-STAT6 peptide against which the antibody was raised (B), phospho-STAT3 peptide (C), or phospho-STAT5 peptide (D), prior to immunohistochemical staining of a case of classical HL. HRS cells demonstrate positive staining with antibody alone (A), which is completely abrogated by preincubation with the phospho-STAT6 peptide (B). Preincubation of antiphospho-STAT6 antibody with phospho-STAT3 (C) or phospho-STAT5 (D) peptides did not affect staining. Original magnifications, × 1340.
Specificity of antiphospho-STAT6 antibody for phospho-STAT6.
Antiphospho-STAT6 antibody was preincubated for 1 hour without peptide (A), with phospho-STAT6 peptide against which the antibody was raised (B), phospho-STAT3 peptide (C), or phospho-STAT5 peptide (D), prior to immunohistochemical staining of a case of classical HL. HRS cells demonstrate positive staining with antibody alone (A), which is completely abrogated by preincubation with the phospho-STAT6 peptide (B). Preincubation of antiphospho-STAT6 antibody with phospho-STAT3 (C) or phospho-STAT5 (D) peptides did not affect staining. Original magnifications, × 1340.
All 3 cases in which the HRS cells were negative for P-STAT3, P-STAT5, and P-STAT6 showed positive staining of cells within the reactive infiltrate for P-STAT3, excluding the possibility of false-negative results due to degradation of phosphorylated proteins before fixation or during tissue processing.
L-1236 is an IL-13–responsive HL-derived cell line
We previously studied the proliferative response of HL-derived cell lines to antibody-mediated neutralization of IL-13. The proliferation of HDLM-2 cells was found to be IL-13 dependent, whereas that of L-428 and KM-H2 cells could not be inhibited by the addition of anti–IL-13 antibody.3 We have extended these analyses to the HL-derived cell lines L-1236 and L-540. IL-13 expression was evaluated by ELISA and by in situ hybridization. IL-13 expression in L-1236 cells was detected by in situ hybridization with an IL-13–specific antisense probe (Figure5A); no signal was observed when a control sense probe was used (Figure 5B). Cytospin preparations of L428 (IL-13+ by Northern analysis3) and LCL-GK (IL-13− by Northern analysis3) served as positive and negative controls, respectively. ELISA (sensitivity limit 12 pg/mL) was not sensitive enough to detect IL-13 expression in L-1236 cells. IL-13 expression by L-540 cells could not be demonstrated by either method.
Expression of IL-13 and response to IL-13 of the HL-derived cell line L-1236.
(A-B) Cytospin preparations of L-1236 cells were examined by in situ hybridization with antisense (A) and sense (B) probes for IL-13 (original magnifications, × 1400). Black-colored silver grains denote mRNA expression. IL-13+ cells were detected with the antisense probe, whereas no signal was obtained with the control sense probe. (C-D) HL-derived cell lines L-1236 (●) and L-540 (○) and control LCL-GK (▾) cells were treated with either 20 μg/mL anti–IL-13 antibody (C) or 20 μg/mL of an isotype control (D). Proliferation was measured at 24, 48, and 72 hours by [3H]-thymidine incorporation. The y-axis shows the proliferation of the treated cells as a percentage of the corresponding untreated control, calculated as the mean of triplicate samples. (E) L-1236 cells were treated with various doses of anti–IL-13 antibody and [3H]-thymidine incorporation was measured at 72 hours. The value of 56 446 cpm (untreated control at 72 hours) was taken as 100% proliferation. (F) L-1236 cells were treated with various doses of exogenous IL-13 and [3H]-thymidine incorporation was measured at 48 hours. The value of 34 688 cpm (untreated control at 48 hours) was taken as 100% proliferation. These experiments were repeated twice with similar results.
Expression of IL-13 and response to IL-13 of the HL-derived cell line L-1236.
(A-B) Cytospin preparations of L-1236 cells were examined by in situ hybridization with antisense (A) and sense (B) probes for IL-13 (original magnifications, × 1400). Black-colored silver grains denote mRNA expression. IL-13+ cells were detected with the antisense probe, whereas no signal was obtained with the control sense probe. (C-D) HL-derived cell lines L-1236 (●) and L-540 (○) and control LCL-GK (▾) cells were treated with either 20 μg/mL anti–IL-13 antibody (C) or 20 μg/mL of an isotype control (D). Proliferation was measured at 24, 48, and 72 hours by [3H]-thymidine incorporation. The y-axis shows the proliferation of the treated cells as a percentage of the corresponding untreated control, calculated as the mean of triplicate samples. (E) L-1236 cells were treated with various doses of anti–IL-13 antibody and [3H]-thymidine incorporation was measured at 72 hours. The value of 56 446 cpm (untreated control at 72 hours) was taken as 100% proliferation. (F) L-1236 cells were treated with various doses of exogenous IL-13 and [3H]-thymidine incorporation was measured at 48 hours. The value of 34 688 cpm (untreated control at 48 hours) was taken as 100% proliferation. These experiments were repeated twice with similar results.
To investigate the effects of IL-13 on L-1236 cells, proliferation in the presence of neutralizing anti–IL-13 antibody was measured by determining [3H]-thymidine incorporation. After 72 hours, the proliferation of L-1236 cells was suppressed to 41% of that of untreated L-1236 cells (Figure 5C), whereas an isotype control antibody had no effect on proliferation (Figure 5D). In contrast, anti–IL-13 antibody had no effect on the proliferation of either L-540 cells or the lymphoblastoid cell line LCL-GK. Treatment of L-1236 cells with increasing concentrations of anti–IL-13 antibody revealed that the inhibition of proliferation was dependent on the dose (Figure 5E). The decreased proliferation induced by IL-13 neutralization was due to an increase in the apoptotic rate, as determined by annexin V staining. Treatment of L-1236 cells with 20 μg/mL anti–IL-13 for 48 hours resulted in a reduction in viable, nonapoptotic (annexin-V−) cells to 66% of the value for untreated L-1236 cells (data not shown). Treatment of L-1236 cells with exogenous IL-13 alone resulted in a dose-dependent increase in proliferation such that thymidine incorporation reached 174% of that of untreated L-1236 cells at 500 ng/mL IL-13 (Figure 5F). Exogenous IL-13 in concentrations up to 500 ng/mL had no effect on the proliferation of L-540 cells (data not shown).
STAT6 phosphorylation in HL-derived cell lines is largely IL-13 dependent
STAT6 is activated primarily in response to IL-4 and IL-13. However, previous reports have shown that IL-4 is rarely expressed in HL.15 In the present study, IL-4 was undetectable in ELISAs of culture supernatants from the HL-derived cell lines L-428, KM-H2, HDLM-2, L-1236, and L-540. Biopsy samples from tissues involved by HL were examined for IL-4 mRNA expression by in situ hybridization. HRS cells in all 16 cases examined were negative for IL-4 (data not shown). Two cases showed IL-4 signal in scattered lymphocytes within the reactive infiltrate. A case of peripheral T-cell lymphoma that showed specific signal with an IL-4 antisense probe and no signal with an IL-4 sense probe was used as a control.
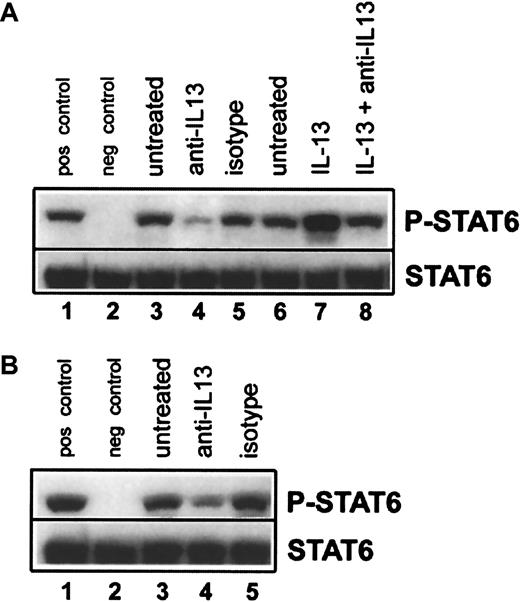
We next explored whether IL-13 was responsible for the constitutive STAT6 activation seen in HL cell lines. The IL-13–responsive HL cell lines L-1236 (Figure 6A) and HDLM-2 (Figure 6B) were incubated for 18 hours in medium alone, medium containing anti–IL-13 antibody, or medium containing the isotype control. Expression of P-STAT6 was then analyzed by immunoblotting. Treatment with neutralizing antibody to IL-13 resulted in the inhibition of basal STAT6 phosphorylation in both cell lines (Figure6A,B; lanes 1-5). To confirm the specificity of this effect, L-1236 cells were incubated in medium alone for 18 hours, followed by a 15-minute incubation with either medium alone, medium plus 5 ng/mL IL-13, or medium plus 5 ng/mL IL-13 that had been preincubated with 20 μg/mL anti–IL-13 antibody for 60 minutes. Exogenous IL-13 alone resulted in an increase in STAT6 phosphorylation above basal levels, whereas there was no increase when the exogenous IL-13 was neutralized with anti–IL-13 antibody (Figure 6A, lanes 6-8). Taken together, our data demonstrate a strong association of IL-13 expression and activation of its downstream signaling mediator STAT6 with the proliferation of HL-derived cells.
Inhibition of constitutive STAT6 phosphorylation in HL-derived cell lines by treatment with anti–IL-13.
(A) L-1236 cells were cultured for 18 hours in medium alone (lane 3), medium containing 20 μg/mL anti–IL-13 (lane 4), or medium containing 20 μg/mL isotype-matched control (lane 5). After 18 hours in medium alone, L-1236 cells were either left unstimulated for 15 minutes (lane 6), or treated for 15 minutes with 5 ng/mL IL-13 (lane 7) or 5 ng/mL IL-13 that had been preincubated for 1 hour with 20 μg/mL anti–IL-13 (lane 8). Equal amounts of total-cell lysates were then subjected to SDS–polyacrylamide gel electrophoresis, followed by Western blot analysis with antiphospho(Tyr641)–STAT6 (upper row). The membrane was stripped and reblotted with an antibody against total STAT6 to confirm equal loading (lower row). (B) HDLM-2 cells were cultured for 18 hours in medium alone (lane 3), medium containing 20 μg/mL anti–IL-13 (lane 4), or medium containing 20 μg/mL isotype-matched control (lane 5), and phospho-STAT6 levels were assessed as above. For both panels A and B, Daudi cells stimulated for 15 minutes with 10 ng/mL IL-4 served as the positive control (pos control, lane 1), and unstimulated Daudi cells served as the negative control (neg control, lane 2). These experiments were repeated twice with similar results.
Inhibition of constitutive STAT6 phosphorylation in HL-derived cell lines by treatment with anti–IL-13.
(A) L-1236 cells were cultured for 18 hours in medium alone (lane 3), medium containing 20 μg/mL anti–IL-13 (lane 4), or medium containing 20 μg/mL isotype-matched control (lane 5). After 18 hours in medium alone, L-1236 cells were either left unstimulated for 15 minutes (lane 6), or treated for 15 minutes with 5 ng/mL IL-13 (lane 7) or 5 ng/mL IL-13 that had been preincubated for 1 hour with 20 μg/mL anti–IL-13 (lane 8). Equal amounts of total-cell lysates were then subjected to SDS–polyacrylamide gel electrophoresis, followed by Western blot analysis with antiphospho(Tyr641)–STAT6 (upper row). The membrane was stripped and reblotted with an antibody against total STAT6 to confirm equal loading (lower row). (B) HDLM-2 cells were cultured for 18 hours in medium alone (lane 3), medium containing 20 μg/mL anti–IL-13 (lane 4), or medium containing 20 μg/mL isotype-matched control (lane 5), and phospho-STAT6 levels were assessed as above. For both panels A and B, Daudi cells stimulated for 15 minutes with 10 ng/mL IL-4 served as the positive control (pos control, lane 1), and unstimulated Daudi cells served as the negative control (neg control, lane 2). These experiments were repeated twice with similar results.
Discussion
The multiple cytokines produced by HRS cells in HL are thought to promote HRS cell growth and survival and to recruit the characteristic reactive infiltrate. To investigate cytokine signaling in HRS cells in vivo, we have examined the activation of STAT proteins, transcription factors that mediate cytokine signaling. STAT activation requires phosphorylation at specific tyrosine residues and translocation from the cytoplasm to the nucleus.7 Activation of STAT3, STAT5, and STAT6 was assessed in HL and NHL cell lines by immunoblotting, using antibodies that specifically recognize the tyrosine-phosphorylated form of a given STAT protein. The same antibodies were used in immunohistochemical analyses of various lymphoma and normal tissues, revealing both the phosphorylation and nuclear localization of an activated STAT protein. In addition to nuclear staining, a minority of cases demonstrated weaker staining in the cytoplasm, the site of STAT protein phosphorylation.
STAT6 was activated in all 5 HL-derived cell lines examined and in a variable proportion of HRS cells in 78% of classical HL cases. Strikingly, HRS cells were P-STAT6+ in 95% of cases of the nodular sclerosis subtype of HL. STAT6 is activated primarily in response to the Th2 cytokines IL-4 and IL-13.14 Mice deficient for STAT6 have defects in both IL-4– and IL-13–mediated functions, including IL-4–mediated B- and T-cell proliferation, Th2 cytokine expression, immunoglobulin class switching, and up-regulation of CD23 and major histocompatibility complex class II molecules.19-21 These data demonstrate the critical role of this transcription factor in the signaling pathway of these cytokines. We have previously shown that IL-13 and its receptor are commonly expressed by HRS cells,3,4 whereas HRS cells and cells within the reactive infiltrate express IL-4 at a significantly lower level compared to IL-13 (present study, Herbst et al,15 and Dukers et al22). We therefore hypothesized that STAT6 activation in HL is primarily due to expression of IL-13. Both the in vitro and in vivo data in the present study support this hypothesis. Antibody-mediated neutralization of IL-13 in 2 IL-13–responsive HL cell lines resulted in a significant decrease in P-STAT6. Furthermore, there was a correlation between antibody-mediated neutralization of IL-13, decreased STAT6 phosphorylation, and reduced HL cell proliferation. However, complete abrogation of STAT6 activity was not observed in the antibody neutralization experiments, and low levels of P-STAT6 were present in L-540 cells that do not express detectable levels of IL-13 or IL-4. These findings suggest that signals other than IL-4 and IL-13 may contribute to STAT6 activation in HL. In vivo, STAT6 activation correlated positively with IL-13 expression in HL-derived tissues. Our results indicate that IL-13 signaling is active in a majority of HL cases, and that this cytokine plays an important role in HRS cell growth.
It is possible that IL-13 is also acting in a paracrine manner contributing to the inflammatory infiltrate in HL.4 This may be through the IL-13–stimulated release of chemokines by HRS cells, or through direct effects of IL-13 on the reactive cells that express IL-13R, particularly B cells and macrophages. However, we did not detect significantly increased P-STAT6 levels within the reactive infiltrate compared to normal tonsillar lymphoid tissue. In contrast to the persistent STAT6 activation in HRS cells, STAT6 activation is a transient event under normal physiologic conditions. Immunohistochemistry, which provides only a snapshot of the activation pattern, may not be sensitive enough to detect subtle differences in STAT6 activation in normal cells. Therefore, our data do not permit us to determine the extent of the role of IL-13 in the pathogenesis of the reactive infiltrate.
In contrast to its frequent activation in HRS cells in classical HL cases, STAT6 was rarely activated in normal lymphoid tissue and in cases of NHL. Normal tonsillar tissue showed only a few P-STAT6+ cells, and a large majority of NHL tissue sections were P-STAT6−. Interestingly, 2 cases each of ALCL and TCRBCL contained P-STAT6+ cells, and 3 of these 4 cases were also IL-13+. IL-4, the other principal activator of STAT6, has previously been identified in TCRBCL,23 suggesting that this cytokine may also contribute to STAT6 activation in this type of lymphoma.
STAT3 is activated by several extracellular and intracellular stimuli, including IL-10, the IL-6 family of cytokines, IL-2, and other cytokines sharing the IL-2Rγ chain (including IL-7, IL-9, and IL-15), and oncogenic tyrosine kinases such as src andabl.8 Among these stimuli, IL-6,6IL-7,24 IL-9,25 and IL-1015 have been previously documented as expressed by various samples of HRS cells. In the present study, STAT3 activation was identified in most cases of classical HL, not only in the HRS cells but also in a large proportion of the reactive infiltrate. However, STAT3 activation was not specific to classical HL and was also observed in a variable proportion of cases of NLPHL, T-cell NHL, and B-cell NHL. Analysis of a section of a reactive tonsil revealed higher basal activation of STAT3 in normal lymphoid tissue compared to STAT5 and STAT6. From its in vivo activation patterns in both lymphomas and benign lymphoid tissue, it seems unlikely that STAT3 plays a defining role in the pathogenesis of classical HL.
Like STAT3, STAT5 is activated by several stimuli, including engagement of cytokine receptors sharing the IL-2Rγ chain (IL-2, IL-7, IL-9, IL-15).26 IL-3, IL-5, and GM-CSF also activate STAT5.27 Among these stimuli, IL-5,3,28GM-CSF,3 IL-7,24 and IL-925 have been identified as expressed by various HL cell lines and HL-involved tissues. In the present study, although STAT5 activation was observed in 3 of 5 HL cell lines, it was identified in only 26% of HL lymphomas examined. STAT5 activation was similarly uncommon in cases of NHL, being present in only 3 of 6 ALCL cases.
Mounting evidence indicates that STAT proteins, particularly STAT3 and STAT5, are involved in oncogenesis.12 Constitutive STAT3 activation has been identified in a wide variety of malignancies,12 and expression of a constitutively activated STAT3 molecule in immortalized fibroblasts leads to cellular transformation.29 STAT5 has been shown to be essential for transformation by bcr-abl in chronic myelogenous leukemia.30,31 Activation of STAT3 and STAT5 is thought to contribute to oncogenesis by preventing apoptosis through bcl-xl up-regulation,32,33 and by stimulating proliferation via cyclin D1 up-regulation.29 The activation of STAT6 in malignancies is less well-studied. Constitutive activation of STAT6 has been demonstrated in human T-cell leukemia virus- associated T-cell leukemia/lymphoma,34 as well as in leukemia associated with the p190bcr-abl.35A role for STAT6 in IL-4–mediated T-cell proliferation has also been reported,19,20,36,37 mediated through up-regulation of the IL-4Rα chain37 and direct effects on the cyclin-dependent kinase inhibitor p27Kip1.38 We believe that our study sheds light on a possible link between constitutive STAT6 activation and IL-13–induced HRS cell proliferation in cases of classical HL.
In conclusion, we have examined activation of STAT proteins in HL to study cytokine activity in this disease. Neither STAT3 nor STAT5 activation was specifically associated with HL, whereas STAT6 activation was a common and distinctive feature of HRS cells. We also showed that STAT6 activation in these cells is due in part to IL-13 signaling. Our previous work implicated IL-13 as an autocrine growth factor for HRS cells.3,4 Given the role of STAT6 in the proliferation of components of the normal immune system,19,20 it is likely that IL-13 is mediating its growth-promoting effects on HRS cells via STAT6. Constitutive IL-13 activity and STAT6 phosphorylation may also underlie other aspects of HL such as the recruitment of the reactive infiltrate. For example, IL-13 signaling through STAT6 is involved in the release of chemokines such as macrophage-derived chemokine,39,40 a molecule involved in Th2 cell recruitment that is frequently expressed by HRS cells in classical HL.41 Our results strongly imply that neutralization of IL-13 signaling by targeting either IL-13R or STAT6 activation may positively affect the course of disease in a majority of patients with HL. As previously discussed,4 inhibition of IL-13 signaling for therapeutic purposes may not be seriously detrimental to the immune system of patients, because it should not compromise a Th2 response mediated through IL-4, allowing the patient some measure of immune protection.
The authors thank J. Ho for excellent technical support, M. Bray for critical comments, and M. Saunders for scientific editing.
Supported in part by grants from the Canadian Institute of Health Research and the National Cancer Institute of Canada.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Tak W. Mak, Ontario Cancer Institute, Amgen Institute, 620 University Ave, Suite 706, Toronto, ON, Canada M5G 2C1; e-mail: tmak@oci.utoronto.ca.


![Fig. 5. Expression of IL-13 and response to IL-13 of the HL-derived cell line L-1236. / (A-B) Cytospin preparations of L-1236 cells were examined by in situ hybridization with antisense (A) and sense (B) probes for IL-13 (original magnifications, × 1400). Black-colored silver grains denote mRNA expression. IL-13+ cells were detected with the antisense probe, whereas no signal was obtained with the control sense probe. (C-D) HL-derived cell lines L-1236 (●) and L-540 (○) and control LCL-GK (▾) cells were treated with either 20 μg/mL anti–IL-13 antibody (C) or 20 μg/mL of an isotype control (D). Proliferation was measured at 24, 48, and 72 hours by [3H]-thymidine incorporation. The y-axis shows the proliferation of the treated cells as a percentage of the corresponding untreated control, calculated as the mean of triplicate samples. (E) L-1236 cells were treated with various doses of anti–IL-13 antibody and [3H]-thymidine incorporation was measured at 72 hours. The value of 56 446 cpm (untreated control at 72 hours) was taken as 100% proliferation. (F) L-1236 cells were treated with various doses of exogenous IL-13 and [3H]-thymidine incorporation was measured at 48 hours. The value of 34 688 cpm (untreated control at 48 hours) was taken as 100% proliferation. These experiments were repeated twice with similar results.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/99/2/10.1182_blood.v99.2.618/6/m_h80222004005.jpeg?Expires=1764959970&Signature=pWfL7uXiYLGc9eEcwmM9vWmEMfVKkzf0F3Bxazh-j0kmwJEZmLWNpaKtzFKOsZUZpQFHe~XNOtkFXgLEG-LdnkKKai~gtyC3akI5fGmbxYHhfA-WniHPJDN4qFT~8Hb9UltN6F4ETSK--3LsMBIqxPA~kHaRxKN3HziTGgp-FhZ8TLagoKTTtGZSpIEqu6HTouw3unac5GvMI3OOD3BqGNUOtCcrrdoUZYweHgqtu-g5JPqfxr0eKhxEst1DAj8E6dWe8QuBNSUS20DHVrpdZq6qHMUAFYy-3vyOmGkgRpgh09sUbmSiqYGhqwIn7s1n7O-llsY6sVCn4Vish71HiA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal