Reconstitution of human cytomegalovirus (HCMV)–specific cytotoxic T lymphocytes (CTLs), predominantly directed against pp65, provides protective immunity for the development of HCMV disease after allogeneic stem cell transplantation (SCT). To define pp65-derived CTL epitopes that would allow sensitive detection of HCMV-specific immune reconstitution, a computer-based epitope prediction was performed. Peptide-specific CTL responses were assessed by interferon-γ release. With this approach, pp65-derived epitopes presented by the HLA alleles A*0101, A*0201, A*1101, and B*0702 were identified. The frequency of CTLs in healthy HCMV-seropositive individuals ranged from about 0.1% to 3.3% of all CD8+ T cells. In patients at risk of HCMV infection after allogeneic SCT, HCMV-peptide–specific CTLs were found in 14 of 19 patients at a median of 90 days after SCT (range, 35-234 days) and HCMV-antigen–specific CD4+ T lymphocytes in 11 of 18 patients at a median of 90 days after SCT (range, 35->180 days). Peak counts of peptide-specific CD8+ T cells ranged from 0.14 to 60.6 cells/μL; those of protein-specific CD4+ T cells ranged from 0.64 to 18.97 cells/μL. Reconstitution of HCMV-peptide–specific CD8+ T cells and protein-specific CD4+ T cells was associated with clearance of HCMV infection (r2 = 0.89, P < .0001 and r2 = 0.61, P = .0045, respectively). HCMV infection recurred after documentation of HCMV-specific T-cell reconstitution (n = 4) when immunosuppression was intensified. Patients in whom late-onset HCMV disease developed lacked HCMV-protein–specific T cells at 3 months after SCT. In conclusion, prospective monitoring of HCMV-specific CD4+ and CD8+ T-cell reconstitution can be performed rapidly by using flow cytometry after specific stimulation with HCMV peptides and proteins and might help to further improve clinical management of HCMV infection after allogeneic SCT.
Introduction
Human cytomegalovirus (HCMV) infection continues to be one of the most important infections in patients undergoing allogeneic stem cell transplantation (SCT) and is associated with high morbidity and mortality despite the introduction of new antiviral treatment strategies.1-3 Whereas early-onset HCMV disease (that occurring during the first 100 days after transplantation) can be reduced significantly after introduction of pre-emptive or prophylactic antiviral therapy for HCMV infection, HCMV disease occurring after day 100 is now one of the leading causes of death after allogeneic SCT.3 4
Studies have demonstrated reconstitution of HCMV-specific immune responses after allogeneic SCT to be protective against the development of HCMV disease.5-8 In these studies, HCMV-specific T-cell responses were assessed by standard cytotoxicity assays using HCMV-infected autologous fibroblasts as target cells after repetitive in vitro stimulations or by lymphoproliferation assays measuring tritium-thymidine uptake after specific stimulation. These assays are labor intensive and time intensive and not applicable routinely. Novel flow cytometry–based techniques, as well as enzyme-linked immunospot (ELISPOT) assays, allow rapid and sensitive quantification of peptide-specific and protein-specific T cells.9-12However, both techniques depend on defined epitopes for the detection of HCMV-specific cytotoxic T lymphocytes (CTLs). Until now, only a limited number of HCMV-peptide epitopes were defined by use of cloning techniques13-15 and, more recently, by epitope prediction based on major histocompatibility complex (MHC) class I motifs.16 17
In this study, HCMV epitopes were defined by the epitope-prediction method provided by SYFPEITHI software18-20 and the ELISPOT technique. Reconstitution of HCMV-peptide–specific and HCMV-protein–specific T-cell responses was associated with resolution of HCMV infection. None of the patients with documented HCMV-specific T-cell responses had onset of HCMV disease, indicating reconstitution of protective immunity.
Patients, materials, and methods
Patients
Twenty-three patients were analyzed for reconstitution of HCMV-specific T-cell responses after allogeneic SCT. Reconstitution of HCMV-specific T cells was assessed prospectively in 19 patients on days 20 to 40, 50 to 70, and 80 to 100 after SCT. In patients lacking HCMV-specific T cells on day 80 to 100 after SCT, T-cell reconstitution was assessed beyond this time point at monthly intervals. In 8 patients, HCMV-specific T cells were analyzed more than 150 days after SCT.
Patient characteristics are shown in Table1. Conditioning therapy consisted of fractionated total-body irradiation (TBI) (3 × 2 × 2 Gy on 3 successive days), cyclophosphamide (2 × 60 mg/kg body weight [bw]), and etoposide (40 mg/kg bw). Alternatively, busulfan (16 mg/kg bw given orally) and cyclophosphamide with cytosine arabinoside (2 × 2 g/m2 body-surface area on 2 successive days) were administered. Five patients who had relapse after autografting for multiple myeloma received a reduced conditioning treatment consisting of 2 Gy TBI, 2 × 20 mg/kg bw cyclophosphamide, and 5 × 30 mg/m2 fludarabine. One patient received a second graft after immunosuppression therapy using total nodal irradiation and thiotepa (2 × 5 mg/kg bw) after rejection of a first T-cell–depleted allograft. Graft-versus-host disease (GVHD) prophylaxis consisted of cyclosporin A administered according to serum levels and antithymocyte globulin given for 3 days at a dose of 20 mg/kg bw 4 to 2 days before SCT in patients receiving a transplant from a matched sibling donor or for 4 days (day 4 to day 1 before SCT) in patients receiving a transplant from a matched unrelated donor. Five patients received a CD34+-selected graft obtained with use of the CliniMACS system (Miltenyi, Bergisch-Gladbach, Germany). All patients received polymerase chain reaction (PCR)–based antiviral therapy as reported previously.2 All patients gave informed consent to donate blood for analysis of immune reconstitution after allogeneic SCT.
Characteristics of patients (n = 23)
| Variable . | Value . |
|---|---|
| Median (range) age, y | 44 (26-66) |
| Sex: M/F | 11/12 |
| Underlying disease | |
| CML | 5 |
| AML | 7 |
| ALL | 3 |
| Multiple myeloma | 5 |
| CLL | 2 |
| MDS | 1 |
| Patient/donor HCMV serologic status | |
| +/+ | 15 |
| −/+ | 3 |
| +/− | 5 |
| Source of stem cells | |
| Peripheral blood | 15 |
| Bone marrow | 7 |
| Bone marrow and peripheral blood | 1 |
| Conditioning regimen | |
| TBI/CY ± VP16 | 12 |
| BU/CY ± ARA-C | 5 |
| TBI/FLU/CY | 5 |
| TT/TNI | 1 |
| Type of donor | |
| HLA-identical sibling | 10 |
| Matched unrelated | 8 |
| Mismatched family member | 5 |
| GVHD prophylaxis | |
| CSA/ATG | 13 |
| CSA | 5 |
| CD34+selection*±CSA/ATG | 5 |
| Acute GVHD | |
| Grade 0/I | 13 |
| Grade II | 7 |
| Grade III/IV | 3 |
| Variable . | Value . |
|---|---|
| Median (range) age, y | 44 (26-66) |
| Sex: M/F | 11/12 |
| Underlying disease | |
| CML | 5 |
| AML | 7 |
| ALL | 3 |
| Multiple myeloma | 5 |
| CLL | 2 |
| MDS | 1 |
| Patient/donor HCMV serologic status | |
| +/+ | 15 |
| −/+ | 3 |
| +/− | 5 |
| Source of stem cells | |
| Peripheral blood | 15 |
| Bone marrow | 7 |
| Bone marrow and peripheral blood | 1 |
| Conditioning regimen | |
| TBI/CY ± VP16 | 12 |
| BU/CY ± ARA-C | 5 |
| TBI/FLU/CY | 5 |
| TT/TNI | 1 |
| Type of donor | |
| HLA-identical sibling | 10 |
| Matched unrelated | 8 |
| Mismatched family member | 5 |
| GVHD prophylaxis | |
| CSA/ATG | 13 |
| CSA | 5 |
| CD34+selection*±CSA/ATG | 5 |
| Acute GVHD | |
| Grade 0/I | 13 |
| Grade II | 7 |
| Grade III/IV | 3 |
Values are numbers of patients unless otherwise indicated. CML indicates chronic myeloid leukemia; AML, acute myeloid leukemia; ALL, acute lymphoblastic leukemia; CLL, chronic lymphocytic leukemia; MDS, myelodysplastic syndrome; HCMV, human cytomegalovirus; TBI, total body irradiation; CY, cyclophosphamide; VP16, etoposide; BU, busulfan; ARA-C, cytosine arabinoside; FLU, fludarabine; TT, thiotepa; TNI, total nodal irradiation; GVHD, graft-versus-host disease; CSA, cyclosporin A; and ATG, antithymocyte globulin.
CD34+ selection was performed by using the CliniMACS system.
Peptides
The HCMV tegument protein pp65 has been identified as a major target antigen for CD8+ MHC class I–restricted, HCMV-specific CTLs.13,21 Therefore, potential HCMV pp65 peptides with a high probability of binding to HLA-A*0101, HLA-A*0201, HLA-A*0301, HLA-A*1101, HLA-B*0702, HLA-B*0801, and HLA-B*3501 were predicted by computer analysis as described previously.18Briefly, the pp65 protein was screened against a matrix pattern that evaluates every amino acid within nonamer or decamer peptides fitting to the above-mentioned HLA class I motifs. Anchor residues are valued 10 and other residues 0 to 10, reflecting amino acid preferences for certain positions within the peptide. The theoretical maximum score for a candidate peptide is 36; scores for abundant natural ligands are typically between 32 and 34. Such motif predictions are available by using the database SYFPEITHI (http://www.syfpeithi.de).18
The pp65-derived peptides were synthesized by using standard fluorenylmethyl chloroformate chemistry on an automated peptide synthesizer (432A; Applied Biosystems, Weiterstadt, Germany). Synthesis products were analyzed by reversed-phase high-performance liquid chromatography (HPLC; Varian Star; Zinsser, München, Germany) and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (G2025A, Hewlett-Packard, Palo Alto, CA). Peptides with a purity of less than 80% were purified by preparative HPLC.
For immune-reconstitution studies after allogeneic SCT, additional peptides derived from the pp65 protein (A*2402, AA 367-379, PTFTSQYRIQGKL; B*3502, AA 188-195, FPTKDVAL; and B*44XX, AA 512-521, EFFWDANDIY) and the pp150 protein (A*0301, AA 945-955, TTVYPPSSTAK) were synthesized.15
Cell lines
The HLA-A2–expressing cell line T2 (174xCEM.T2 hybridoma, deficient in transporter associated with antigen processing [TAP] 1 and TAP2) was grown in RP10 medium (RPMI 1640 [Biochrome, Berlin, Germany] supplemented with 10% heat-inactivated fetal-calf serum [FCS] and 2% penicillin/streptomycin) and used to provide stimulator cells in the ELISPOT assay.
Generation of monocyte-derived dendritic cells (DCs)
Generation of DCs from peripheral blood mononuclear cells (PBMNCs) was performed as described previously.22-26Briefly, PBMNCs were isolated by using Ficoll-Paque (Biochrome) density-gradient centrifugation of heparin-treated blood obtained from buffy-coat preparations of blood from healthy HCMV-seropositive blood donors registered at the local blood bank. All donors expressed the HLA class I alleles A*0101, A*0201, A*0301, A*1101, B*0702, B*0801, or B*3501.
PBMNCs were plated (1 × 107 cells/3 mL per well) into 6-well plates (Becton Dickinson, Heidelberg, Germany) in RP10 medium. After 2 hours of incubation at 37°C, nonadherent cells were removed and the adherent blood monocytes (purity > 80%) were cultured in RP10 medium supplemented with 100 ng/mL human recombinant granulocyte-macrophage colony-stimulating factor (Leucomax; Novartis, Basel, Switzerland), 1000 IU/mL interleukin (IL) 4 (R&D Systems, Wiesbaden, Germany), and 10 ng/mL tumor necrosis factor α (R&D Systems).
Fresh medium and cytokines were added to the DC cultures every other day, and cell differentiation was monitored by using light microscopy. Analysis of surface markers as assessed by flow cytometry after 7 days of culture showed high levels of MHC class I and II expression, as well as CD80, CD83, CD86, CD40, and CD54 (data not shown), corresponding to characteristics of mature DCs.
Short-term culture for expansion of peptide-specific CTLs in HCMV-seropositive individuals
Autologous monocyte-derived mature DCs (5 × 105/mL) were pulsed with a mixture of 3 of the predicted synthetic peptides (10 μg/mL each), irradiated with 30 Gy, and incubated with 3 × 106 autologous PBMNCs in minimal essential α medium (Biochrome) supplemented with 10% heat-inactivated FCS and antibiotics. After 7 days of culture, cells were restimulated weekly with irradiated autologous peptide-pulsed PBMNCs, and human recombinant IL-2 (Lymphocult-T-LF; Biotest, Dreieich, Germany) was added at a concentration of 10 U/mL every other day. After 2 restimulations, cells were analyzed for peptide specificity by intracellular interferon-γ (IFN-γ) staining by using flow cytometry after specific stimulation with individual peptides.
ELISPOT assay
To test the predicted epitopes in a biologic assay, CD8+ T lymphocytes were isolated from HCMV-seropositive healthy donors and analyzed for reactivity to HCMV peptides derived from the pp65 matrix protein in an IFN-γ–ELISPOT assay as described previously.17 T2 cells were used as antigen-presenting cells (APCs) for patients expressing HLA-A*0201, and autologous monocyte-derived mature DCs were used for all other HLA restrictions. Spot-forming cells were counted by using a KS-ELISPOT microscope (Zeiss, Jena, Germany).11
Cell-surface and intracellular cytokine staining
Intracellular cytokine staining for IFN-γ production was performed as described previously.9 White blood cells were obtained from healthy blood donors and patients after allogeneic SCT selected on the basis of HCMV serostatus and HLA type. PBMNCs (5 × 105) were stimulated overnight with peptide solutions in the presence of brefeldin A (Sigma, Deisenhofen, Germany) at a final concentration of 10 μg/mL each. To assess the precursor frequencies of HCMV-specific CD4+ lymphocytes, stimulation was alternatively performed by using HCMV antigen (1.25 μg/mL). The next morning, cells were stained with monoclonal antibodies (mAbs) directed against surface markers and fixed in Dulbecco phosphate-buffered saline (DPBS) containing 2% formaldehyde (Sigma) for 15 minutes at room temperature. After centrifugation at 400g for 5 minutes, cells were resuspended in DPBS containing 0.5% saponin (Roth, Karlsruhe, Germany), 1% bovine serum albumin (Roth) and anti–IFN-γ antibody (0.2 μg/well; Becton Dickinson). After intracellular IFN-γ staining, samples were analyzed on a fluorescence-activated cell-sorter scanner flow cytometer (FACSCalibur; Becton Dickinson) by using CellQuest software (Becton Dickinson).
Isotype control reagents were used to delineate positive and negative populations. Unstimulated samples were analyzed to verify the effect of the stimulation. Cell staining was performed by using fluorescein isothiocyanate–conjugated (FITC), peridinin chlorophyll protein–conjugated, or phycoerythrin (PE)-conjugated mouse mAbs against the following surface markers: CD3, CD4, CD8, and human IFN-γ (all from Becton Dickinson). Absolute numbers of peptide-specific and protein-specific T cells were calculated on the basis of the absolute numbers of CD3+CD4+ and CD3+CD8+ lymphocytes/μL blood and the relative numbers of these lymphocyte fractions that stained positive for IFN-γ after specific stimulation.
Cell staining with MHC-peptide tetrameric complexes
MHC-peptide tetrameric complexes (kindly provided by R. Handgretinger, St Jude Children's Research Hospital, Memphis, TN) were used to assess the frequency of peptide-specific CD8+ T cells in leukapheresis products from 6 HCMV-seropositive family-member donors. PBMNCs (2-5 × 105) were stained in a 96-well plate with 20 μg/mL PE-labeled HLA-A2 or HLA-A1 tetramers (in 50 μL phosphate-buffered saline [PBS] with 10% FCS) and incubated for 15 minutes at 37°C. Cells were washed twice with 200 μL PBS and 10% FCS at 37°C. After tetramer staining, cells were incubated with anti-CD8 and anti-CD3 or anti-CD4 antibodies (FITC-CyChrome, PharMingen, San Diego, CA) in 200 μL PBS and 10% FCS for 15 minutes at 4°C. Cells were washed twice, resuspended in 200 μL PBS and 10% FCS, and analyzed in a FACSCalibur device.17
Lymphoproliferation assay
The proliferation assay was performed as described previously.8 HCMV antigen, phytohemagglutinin (Murex; Life Technology, Karlsruhe, Germany), and IL-2 (Biotest) were added at final concentrations of 1:400, 10 ng/mL, and 50 U/mL, respectively. A stimulation index of 3 was considered to indicate a positive lymphoproliferative response.
Detection of HCMV infection by PCR and assessment of viral load in the blood
All patients in the study were examined weekly by qualitative PCR assays using whole blood, beginning on day 0 and continuing until day 100 after SCT as described previously.2 4 Quantitative analysis of the HCMV DNA load was performed by using the standardized, commercially available COBAS Amplicor HCMV monitor test (Roche Diagnostics, Mannheim, Germany). Nucleic acid was extracted from 0.2 mL EDTA-anticoagulated plasma according to the protocol of the manufacturer. Specific primers amplified a 365-base-pair region of the HCMV DNA polymerase gene. The viral load was assessed by using an external standard and colorimetric amplicon detection. The lower detection limit of the assay was 400 genome equivalents (GE)/mL plasma, and linearity of the assay was achieved at between 400 and 2 × 105 GEs.
Results
Epitope prediction
Potential MHC class I–binding peptides from HCMV pp65 were predicted for the MHC class I restrictions HLA-A*0101, HLA-A*0201, HLA-A*0301, HLA-A*1101, HLA-B*0702, HLA-B*0801, and HLA-B*3501. Epitope prediction followed the rules reported previously and can be reproduced by following directions given at http://www.syfpeithi.de.18Fifty peptides predicted to be the best binders were synthesized. HLA-A*0201–restricted peptides were tested for HLA-A2 binding by using the reconstitution assay (data not shown). However, none of the peptides were excluded from analysis, since the peptide-binding assay is not sensitive enough to detect binding of all natural ligands.
Detection of HCMV-peptide–specific CTL responses in healthy HCMV-seropositive blood donors
The ELISPOT assay was used to screen HCMV-seropositive healthy blood donors for the presence in blood of CD8+ T cells directed against the predicted HCMV pp65 peptide epitopes. Peptide-pulsed mature DCs were found to be potent stimulator cells, demonstrating a T-cell stimulatory capacity comparable to that of peptide-loaded T2 (data not shown). Thus, to achieve an optimal T-cell stimulation, peptide-loaded autologous mature DCs were used as stimulator cells for detection of non–HLA-A*0201–restricted CTLs.
HLA-A*0201–positive donors had a dominant response to the NLVPMVATV peptide, as reported previously by others.6,13 16 In HLA-A*1101–positive donors, a strong release of IFN-γ was demonstrated by the ELISPOT assay after stimulation with GPISGHVLK-loaded mature DCs. This also was observed in HLA-B*0702–positive donors after stimulation with TPRVTGGGAM-loaded autologous DCs. In addition, after 3 in vitro stimulations of PBMNCs with peptide-loaded APCs from HLA-A*0101 HCMV-seropositive donors, HLA-A*0101–restricted, YSEHPTFTSQY-specific CD8+ T cells were expanded from below the detection level to about 25% of all CD8+ T cells. On intracellular IFN-γ staining, a low frequency of HLA-A*0101–restricted CD8+ T cells (0.08% to 0.2% of all CD8+ T cells) was documented after stimulation with YSEHPTFTSQY in 3 of 4 healthy individuals.
No HCMV-peptide–specific CTLs were detectable by intracellular IFN-γ staining in HCMV-seronegative healthy individuals expressing HLA-A*0101 (n = 4), HLA-A*0201 (n = 6), HLA-A*1101 (n = 5), or HLA-B*0702 (n = 4) after specific stimulation.
In 6 of 7 HCMV-seropositive individuals expressing HLA-A*0201 and HLA-B*0702, a dominant HLA-B*0702–restricted CTL response to HCMV was observed (frequency of HLA-B*0702–restricted CTLs of 0.7%-3.3% versus frequency of HLA-A*0201–restricted CTLs of 0-0.1%). The dominance of the HLA-B*0702–restricted CTL response was also confirmed in 3 healthy individuals expressing HLA-B*0702 and HLA-A*0101 (frequency of HLA-B*0702–restricted CTLs of 0.75% to 3.3% versus frequency of HLA-A*0101–restricted CTLs of 0-0.1%). Thus, CTL precursor frequencies specific for a single peptide seem to vary according to the genetic background of individual patients.
Frequency of HCMV-peptide–specific CD8+ T cells in leukapheresis products
The frequency of HCMV-peptide–specific CD8+ T cells in leukapheresis products was assessed by using MHC-peptide tetrameric complexes in 6 HCMV-seropositive donors. In 5 donors expressing HLA-A*0201, a median of 2.24% (range, 0.19%-14.21%) of all CD8+ T cells showed positive staining with the A2 tetramer. In one donor expressing HLA-A*0101, 0.11% of all CD8+ T cells showed positive staining with the A1 tetramer. Because of the limited number of donors studied, it was not possible to make any correlation between the percentage of HCMV-specific CTLs in the donor and subsequent reconstitution after SCT.
Reconstitution of CTLs specific for HCMV pp65 after allogeneic SCT
In 19 patients at risk of HCMV infection, blood samples were drawn every month after allogeneic SCT to assess the recovery of HCMV-specific T-cell responses. The following T-cell assays were performed: peptide-specific CTL responses evaluated by ELISPOT or intracellular INF-γ staining with flow cytometry (n = 19), HCMV-protein–specific CD4+ T-cell responses assessed by intracellular IFN-γ staining with flow cytometry (n = 18), and lymphoproliferative responses after stimulation with HCMV protein (n = 8).
HCMV-peptide–specific CD8+ T cells were observed in 14 of 19 patients at a median of 90 days after SCT (range, 35 to 234 days) and protein-specific CD4+ T cells in 11 of 18 patients at a median of 90 days after SCT (range, 35 to > 180 days). Peak counts of peptide-specific CD8+ T cells ranged from 0.14 to 60.6 cells/μL and peak counts of protein-specific CD4+ T cells from 0.64 to 18.97 cells/μL, as assessed by intracellular IFN-γ staining (Table 2). Early reconstitution of HCMV-specific T cells before day 100 was even observed in 2 of 3 patients who received CD34+-selected grafts from HCMV-seropositive mismatched family members, as well as in 4 of 6 patients given a transplant from an unrelated donor and 3 of 4 patients receiving a peak steroid dose of more than 5 mg/kg bw per day during the first weeks after SCT for early-onset acute GVHD. Even in 2 of 4 patients given a transplant from an HCMV-seronegative donor, HCMV-specific CD4+ and CD8+ T cells were observed on days 60 and 90, respectively, whereas in 2 patients, both of whom received a transplant from an unrelated donor, no HCMV-specific T cells were observed until day 120 and 150 after SCT (Table 2).
Correlation of human cytomegalovirus (HCMV) infection and HCMV-specific immune reconstitution in patients after allogeneic stem cell transplantation (SCT)
| Patient . | Donor . | HLA alleles . | Acute GVHD . | Steroids (mg/kg)* . | HCMV infection before day 100 after SCT† . | HCMV disease . | Peptide-specific CD8+‡ . | Protein-specific CD4+2-154 . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Duration, wk . | Day last positive PCR . | Peak viral load (GE/mL) . | Day first detected2-153 . | Peak count (n/μL) . | Day first detected2-153 . | Peak count (n/μL) . | ||||||
| Group 1 (donor positive for HCMV) | ||||||||||||
| 1 | Sibl | A2, B7 | II | 2 | 0 | — | — | — | + 35 (A2) | 1.01 | + 35 | SI positive |
| 2 | Sibl | A2 | 0 | 0 | 3 | + 35 | 3610 | — | + 38 | 23.2 | + 38 | 18.97 |
| 3 | Sibl | A3, B35 | II | 2 | 2 | + 84 | 2960 | — | > 120 | — | > 120 | — |
| 4 | Sibl | B35 | II | 10 | 0 | — | — | — | > 120 | — | > 120 | — |
| 52-159 | Sibl | A1, A2, B44 | II | 2 | 3 | + 221 | 3240 | — | + 234 (A2) | 9.7 | ND | — |
| 6 | Sibl | A11 | 0 | 0 | 6 | + 63 | 46 100 | — | > 180 | — | > 180 | — |
| 72-160 | Sibl | A1 | 0 | — | 6 | + 71 | ND | — | + 90 | 10.38 | + 90 | 3.42 |
| 82-160 | MM | A1 | III | 2 | 4 | > 100 | 5590 | Late retinitis | + 95 | 5.47 | > 120 | — |
| 92-160 | MM | B7 | I | 2 | 2 | + 57 | 2980 | — | + 66 | 7.95 | + 66 | 2.13 |
| 102-160 | MM | A2, A24 | II | 2 | 9 | + 139 | ND | — | + 120 (A2) | 60.6 | + 120 | 1.09 |
| 11 | MM | A2, B7 | III | 10 | 0 | — | — | — | + 61 (B7) | 0.14 | + 61 | SI positive |
| 12 | MM | A3, B35 | II | 7.5 | 3 | + 76 | 4580 | — | + 70 (B35) | 3.52 | + 140 | 0.64 |
| 13 | MUD | B44 | 0 | 0 | 2 | + 44 | 1950 | — | + 60 | 2.21 | + 60 | 9.22 |
| 142-159 | MUD | A1 | I | 2 | 3 | + 36 | 1520 | — | + 35 | 26.45 | + 63 | 5.6 |
| 152-159 | MUD | A2 | II | 10 | 2 | + 75 | ND | Late IP | + 85 | 37.96 | > 85 | — |
| Group 2 (donor negative for HCMV) | ||||||||||||
| 16 | Sibl | A1 | I | — | 1 | + 43 | ND | — | + 60 (A1) | 2.37 | + 60 | 5.77 |
| 17 | MUD | A1, A2 | 0 | 1 | 12 | + 230 | 210 000 | — | > 150 | — | > 150 | — |
| 18 | MUD | A3, A24 | II | 1 | 0 | — | — | — | > 120 | — | > 120 | — |
| 192-159 | MUD | A1, A3, B7 | 0 | 3 | 1 | + 92 | 2640 | — | + 90 (A1, B7) | 6.8 | + 90 | 1.02 |
| Patient . | Donor . | HLA alleles . | Acute GVHD . | Steroids (mg/kg)* . | HCMV infection before day 100 after SCT† . | HCMV disease . | Peptide-specific CD8+‡ . | Protein-specific CD4+2-154 . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Duration, wk . | Day last positive PCR . | Peak viral load (GE/mL) . | Day first detected2-153 . | Peak count (n/μL) . | Day first detected2-153 . | Peak count (n/μL) . | ||||||
| Group 1 (donor positive for HCMV) | ||||||||||||
| 1 | Sibl | A2, B7 | II | 2 | 0 | — | — | — | + 35 (A2) | 1.01 | + 35 | SI positive |
| 2 | Sibl | A2 | 0 | 0 | 3 | + 35 | 3610 | — | + 38 | 23.2 | + 38 | 18.97 |
| 3 | Sibl | A3, B35 | II | 2 | 2 | + 84 | 2960 | — | > 120 | — | > 120 | — |
| 4 | Sibl | B35 | II | 10 | 0 | — | — | — | > 120 | — | > 120 | — |
| 52-159 | Sibl | A1, A2, B44 | II | 2 | 3 | + 221 | 3240 | — | + 234 (A2) | 9.7 | ND | — |
| 6 | Sibl | A11 | 0 | 0 | 6 | + 63 | 46 100 | — | > 180 | — | > 180 | — |
| 72-160 | Sibl | A1 | 0 | — | 6 | + 71 | ND | — | + 90 | 10.38 | + 90 | 3.42 |
| 82-160 | MM | A1 | III | 2 | 4 | > 100 | 5590 | Late retinitis | + 95 | 5.47 | > 120 | — |
| 92-160 | MM | B7 | I | 2 | 2 | + 57 | 2980 | — | + 66 | 7.95 | + 66 | 2.13 |
| 102-160 | MM | A2, A24 | II | 2 | 9 | + 139 | ND | — | + 120 (A2) | 60.6 | + 120 | 1.09 |
| 11 | MM | A2, B7 | III | 10 | 0 | — | — | — | + 61 (B7) | 0.14 | + 61 | SI positive |
| 12 | MM | A3, B35 | II | 7.5 | 3 | + 76 | 4580 | — | + 70 (B35) | 3.52 | + 140 | 0.64 |
| 13 | MUD | B44 | 0 | 0 | 2 | + 44 | 1950 | — | + 60 | 2.21 | + 60 | 9.22 |
| 142-159 | MUD | A1 | I | 2 | 3 | + 36 | 1520 | — | + 35 | 26.45 | + 63 | 5.6 |
| 152-159 | MUD | A2 | II | 10 | 2 | + 75 | ND | Late IP | + 85 | 37.96 | > 85 | — |
| Group 2 (donor negative for HCMV) | ||||||||||||
| 16 | Sibl | A1 | I | — | 1 | + 43 | ND | — | + 60 (A1) | 2.37 | + 60 | 5.77 |
| 17 | MUD | A1, A2 | 0 | 1 | 12 | + 230 | 210 000 | — | > 150 | — | > 150 | — |
| 18 | MUD | A3, A24 | II | 1 | 0 | — | — | — | > 120 | — | > 120 | — |
| 192-159 | MUD | A1, A3, B7 | 0 | 3 | 1 | + 92 | 2640 | — | + 90 (A1, B7) | 6.8 | + 90 | 1.02 |
GVHD indicates acute graft-versus-host disease; PCR, polymerase chain reaction assay; GE, genome equivalents; Sibl, HLA-identical sibling; SI, stimulation index; MM, HLA-nonidentical family member; MUD, matched unrelated donor; IP, interstitial pneumonia; and ND, not done.
Peak steroid dose.
Duration indicates the weeks of viremia as assessed by the in-house PCR method before day 100 after SCT. Viral load was measured with a COBAS Amplicor HCMV monitor. The peak viral load before day 100 is indicated.
The HLA restriction of HCMV-peptide-specific cytotoxic T lymphocytes is indicated in parentheses.
Values are the first day after SCT that T-cell reactivities were observed.
T-cell responses were assessed by intracellular interferon-γ staining, lymphoproliferation (SI), or both.
Patients who received CD34+-selected stem cells obtained with use of the CliniMACS system.
Patients who received nonmyeloablative conditioning treatment.
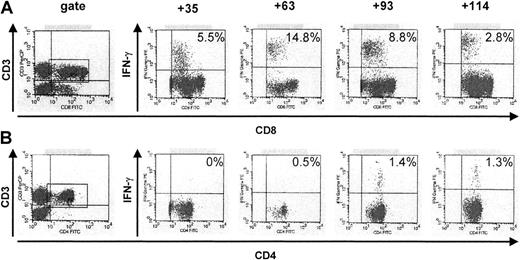
Patient 14 (Table 2), who received a transplant from a matched unrelated donor after dose-reduced conditioning therapy, had a massive expansion of HLA-A*0101–restricted, peptide-specific CTLs of up to 14.8% of all CD8+ T cells on day 63 and up to 1.4% of HCMV-reactive CD4+ T cells on day 93 after SCT (Figure1). This early HCMV-specific T-cell reconstitution correlated with a short duration of viremia (3 weeks) and a low peak viral load (1520 GE/mL). Patient 1 had a rapid reconstitution of HCMV-specific CD8+ and CD4+ T cells by day 35 after SCT and no detectable HCMV reactivation, even on PCR assay. In the early posttransplantation period, HLA-A*0201–restricted, CD8+ peptide-specific T cells dominated, whereas on day 120 after SCT, similar numbers of HLA-A*0201–restricted and HLA-B*0702–restricted, CD8+peptide-specific T cells were documented (Figure2A). Patient 9, who was given a transplant of highly purified CD34+-selected peripheral blood stem cells (2 × 105 CD3+ cells/kg bw) from a 1-Ag–mismatched family member (donor, HLA-A*0301; recipient, HLA-A*0201), showed early reconstitution of HLA-B*0702–restricted, peptide-specific T cells, whereas HLA-A*0201–restricted T cells were detectable only at background levels (Figure 2B).
Reconstitution of HCMV-specific CD8+ and CD4+ T cells in a patient after allogeneic transplantation of stem cells from a matched unrelated donor and reduced conditioning therapy.
Peptide-specific CD8+ and protein-specific CD4+T cells were analyzed by intracellular IFN-γ staining with flow cytometry after specific stimulation. The data show a massive initial expansion of HCMV-peptide–specific, HLA-A*0101–restricted CTLs, and later, of protein-specific CD4+ T cells in patient 14, who received a transplant from a matched unrelated donor after reduced conditioning treatment. A maximum of 14.8% of all CD8+ T cells were directed against a single HCMV peptide on day 63 after STC. Numbers represent the percentage of IFN-γ–producing CD3+CD8+ and CD3+CD4+cells in the lymphocyte gate.
Reconstitution of HCMV-specific CD8+ and CD4+ T cells in a patient after allogeneic transplantation of stem cells from a matched unrelated donor and reduced conditioning therapy.
Peptide-specific CD8+ and protein-specific CD4+T cells were analyzed by intracellular IFN-γ staining with flow cytometry after specific stimulation. The data show a massive initial expansion of HCMV-peptide–specific, HLA-A*0101–restricted CTLs, and later, of protein-specific CD4+ T cells in patient 14, who received a transplant from a matched unrelated donor after reduced conditioning treatment. A maximum of 14.8% of all CD8+ T cells were directed against a single HCMV peptide on day 63 after STC. Numbers represent the percentage of IFN-γ–producing CD3+CD8+ and CD3+CD4+cells in the lymphocyte gate.
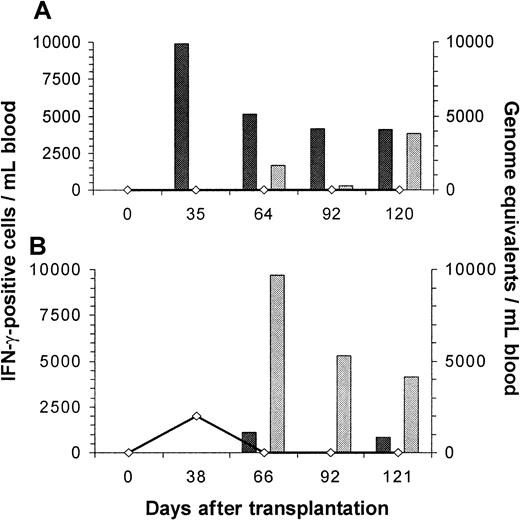
Correlation of viral load in the blood and reconstitution of HCMV-specific, HLA-A*0201– and HLA-B*0702–restricted CTLs in 2 patients after allogeneic SCT.
Peptide-specific CD8+ CTLs were analyzed by intracellular IFN-γ staining with flow cytometry after specific stimulation, and viral load was assessed by quantitative PCR. Black bars indicate HLA-A*0201–restricted CTLs and gray bars HLA-B*0702–restricted CTLs; the black line and open diamonds represent viral load. Early reconstitution of predominantly HLA-A*0201–restricted CTLs in patient 1 was associated with complete suppression of HCMV replication after allogeneic SCT (A). Patient 9, who was given a transplant of highly purified stem cells from a 1-Ag–mismatched family member (A3 A2), showed early reconstitution of HLA-B*0702–restricted CTLs associated with rapid clearance of the viral DNA from the blood (B). However, HLA-A*0201–restricted CTLs were detectable just above the lower detection limit on 2 occasions. Values are the absolute numbers of peptide-specific CTLs/mL blood.
Correlation of viral load in the blood and reconstitution of HCMV-specific, HLA-A*0201– and HLA-B*0702–restricted CTLs in 2 patients after allogeneic SCT.
Peptide-specific CD8+ CTLs were analyzed by intracellular IFN-γ staining with flow cytometry after specific stimulation, and viral load was assessed by quantitative PCR. Black bars indicate HLA-A*0201–restricted CTLs and gray bars HLA-B*0702–restricted CTLs; the black line and open diamonds represent viral load. Early reconstitution of predominantly HLA-A*0201–restricted CTLs in patient 1 was associated with complete suppression of HCMV replication after allogeneic SCT (A). Patient 9, who was given a transplant of highly purified stem cells from a 1-Ag–mismatched family member (A3 A2), showed early reconstitution of HLA-B*0702–restricted CTLs associated with rapid clearance of the viral DNA from the blood (B). However, HLA-A*0201–restricted CTLs were detectable just above the lower detection limit on 2 occasions. Values are the absolute numbers of peptide-specific CTLs/mL blood.
The number of HCMV-specific CD8+ T cells in the peripheral blood of patients expressing the HLA alleles A*0101, A*0201, or B*0702 was correlated with the viral load in the blood as assessed by quantitative PCR assay in 45 blood samples. In 11 of 13 blood samples with more than 5 peptide-specific CD8+ T cells/μL, no HCMV DNA was detectable, whereas HCMV DNA was detectable in 23 of 32 blood samples with a lower frequency of HCMV-specific CTLs (data not shown).
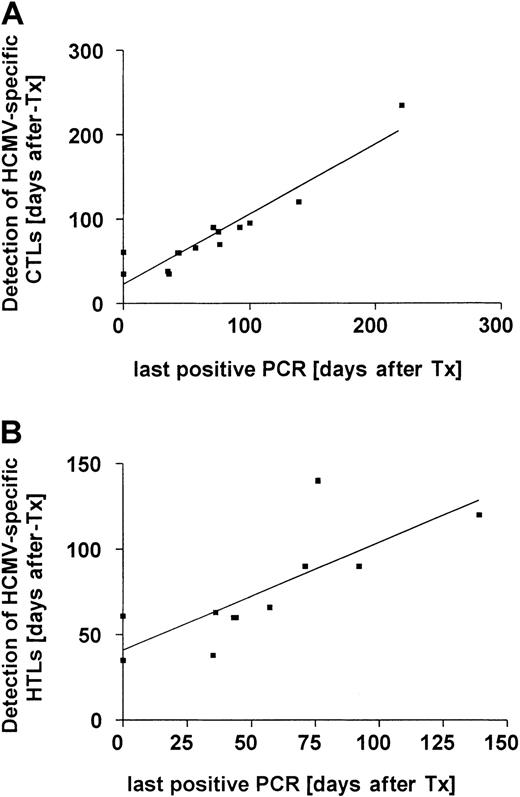
Reconstitution of HCMV-peptide–specific CD8+ T cells in 14 patients was found to be associated with rapid clearance of HCMV infection (r2 = 0.89, P < .0001 on linear regression analysis with GraphPad Prism, version 2.0 [GraphPad Software, San Diego, CA]; Figure 3A). A similar correlation was documented for reconstitution of HCMV-specific CD4+ T cells and clearance of viral DNA from the blood in 11 patients (r2 = 0.61, P = .0045 on linear regression analysis; Figure 3B). Thus, the duration of viremia was shorter in 12 patients who had HCMV-specific T-cell responses on day 100, with only 1 of 12 patients having viremia for more than 4 weeks compared with 3 of 7 patients lacking HCMV-specific T-cell responses on day 100 (P = .07 on χ2 testing). In 4 of the 14 patients with a documented HCMV peptide–specific CD8+T-cell response, recurrence of HCMV infection was documented later, after intensification of immunosuppression for GVHD. In 2 of these 4 patients, HCMV disease developed. Notably, both patients in whom late-onset HCMV disease developed had an HCMV-peptide–specific CD8+ T-cell response but no protein-specific CD4+ T-cell response 80 to 100 days after SCT (Table 2). In none of 5 HCMV-seronegative patients given a transplant from an HCMV-seronegative donor were HCMV-specific CTLs detectable.
Reconstitution of HCMV-peptide–specific CD8+ and protein-specific CD4+ T-cell responses associated with clearance of HCMV infection.
(A) Reconstitution of HCMV-peptide–specific CD8+ T cells in 14 patients was associated with rapid clearance of HCMV infection (r2 = 0.89, P < .0001 on linear regression analysis). The time of the last positive PCR result is shown on the x-axis and the time to reconstitution of peptide-specific CD8+ T cells on the y-axis. (B) A similar correlation was found for reconstitution of HCMV-protein–specific CD4+ T cells in 11 patients (r2 = 0.61, P = .0045 on linear regression analysis). The time of the last positive PCR result is shown on the x-axis and the time to reconstitution of protein-specific CD4+ T cells on the y-axis.
Reconstitution of HCMV-peptide–specific CD8+ and protein-specific CD4+ T-cell responses associated with clearance of HCMV infection.
(A) Reconstitution of HCMV-peptide–specific CD8+ T cells in 14 patients was associated with rapid clearance of HCMV infection (r2 = 0.89, P < .0001 on linear regression analysis). The time of the last positive PCR result is shown on the x-axis and the time to reconstitution of peptide-specific CD8+ T cells on the y-axis. (B) A similar correlation was found for reconstitution of HCMV-protein–specific CD4+ T cells in 11 patients (r2 = 0.61, P = .0045 on linear regression analysis). The time of the last positive PCR result is shown on the x-axis and the time to reconstitution of protein-specific CD4+ T cells on the y-axis.
HCMV-specific T-cell response in patients more than 150 days after allogeneic SCT
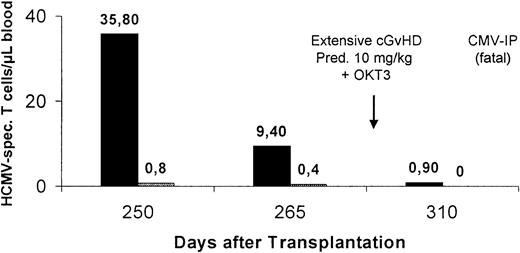
Eight patients were assessed for HCMV-specific T-cell responses late after allogeneic SCT. HCMV-peptide–specific CD8+ T cells were detectable in 6 of the 8, and HCMV-protein–specific CD4+ T cells were observed in 3 of 6 patients analyzed. Late-onset HCMV disease developed in 3 of the 8 patients, 2 of whom presented with prolonged viremia (8 weeks) and a high peak viral load (> 5 × 104 GE/mL blood) before day 100 (patients 21 and 23 on Table 3). In the third patient, HCMV-specific T cells were completely eliminated by high-dose corticosteroids and OKT3 (patient 15 on Table 3; Figure4). HCMV disease was fatal in 2 patients lacking HCMV-specific T-cell responses (patients 15 and 23 on Table 3; Figure 4). The only patient who survived HCMV pneumonia showed recovery of an HCMV-specific CTL response.
HCMV-specific immune reconstitution in 8 patients at more than 150 days after allogeneic SCT
| Patient . | Donor . | HCMV serostatus (D/R)3-150 . | Chronic GVHD . | Immune suppression . | HCMV infection3-151 . | HCMV disease . | Peptide-specific CD8+ . | Protein-specific CD4+ Count (n/μL) . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Duration, wk . | Peak viral load (GE/mL) . | HLA allele . | Count (n/μL) . | |||||||
| 53-152 | Sibl | +/+ | Limited | Pred 0.1 | 3 | 3240 | — | A2 | 9.6 | ND |
| 20 | Sibl | +/+ | No | CSA/pred 0.1 | 0 | < 400 | — | A2 | 7.6 | 8.3 |
| 213-153 | Sibl | +/+ | Limited | — | 8 | 56 900 | Late IP | A2 | 1.2 | — |
| 12 | MM | +/+ | Limited | — | 9 | ND | — | A2 | 52.5 | — |
| 153-152,3-154 | MUD | +/+ | Limited | CSA | 2 | < 400 | Late IP | A2 | 35.81 | 0.79 |
| 17 | MUD | −/+ | Limited | 0.1 | 12 | 210 000 | — | A1 | — | 1.51 |
| 22 | MUD | +/− | Limited | CSA/pred 0.2 | 8 | 11 300 | — | A2 | 5.45 | ND |
| 233-152 | MUD | −/+ | Limited | — | 8 | 50 400 | Late IP | A2 | — | — |
| Patient . | Donor . | HCMV serostatus (D/R)3-150 . | Chronic GVHD . | Immune suppression . | HCMV infection3-151 . | HCMV disease . | Peptide-specific CD8+ . | Protein-specific CD4+ Count (n/μL) . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Duration, wk . | Peak viral load (GE/mL) . | HLA allele . | Count (n/μL) . | |||||||
| 53-152 | Sibl | +/+ | Limited | Pred 0.1 | 3 | 3240 | — | A2 | 9.6 | ND |
| 20 | Sibl | +/+ | No | CSA/pred 0.1 | 0 | < 400 | — | A2 | 7.6 | 8.3 |
| 213-153 | Sibl | +/+ | Limited | — | 8 | 56 900 | Late IP | A2 | 1.2 | — |
| 12 | MM | +/+ | Limited | — | 9 | ND | — | A2 | 52.5 | — |
| 153-152,3-154 | MUD | +/+ | Limited | CSA | 2 | < 400 | Late IP | A2 | 35.81 | 0.79 |
| 17 | MUD | −/+ | Limited | 0.1 | 12 | 210 000 | — | A1 | — | 1.51 |
| 22 | MUD | +/− | Limited | CSA/pred 0.2 | 8 | 11 300 | — | A2 | 5.45 | ND |
| 233-152 | MUD | −/+ | Limited | — | 8 | 50 400 | Late IP | A2 | — | — |
HCMV indicates human cytomegalovirus; SCT, stem cell transplantation; GVHD, chronic graft-versus-host disease; GE, genome equivalents; Sibl, HLA-identical sibling; pred, prednisolone; ND, not done; CSA, cyclosporin A; IP, interstitial pneumonia; MM, HLA-nonidentical family member; and MUD, matched unrelated donor.
HCMV serostatus as assessed by enzyme-linked immunosorbent assay in stem cell donor (D) and recipient (R).
Duration indicates the weeks of viremia as assessed by the in-house polymerase chain reaction method before day 100 after SCT. Viral load was measured with a COBAS Amplicor HCMV monitor. The peak viral load before day 100 is indicated.
Patients who received nonmyeloablative conditioning treatment.
Patient who received CD34+-selected stem cells obtained with use of the CliniMACS system.
This patient received donor lymphocytes for relapse of the underlying malignant disease. Severe GVHD subsequently developed and was treated with corticosteroids and OKT3. The patient later died from HCMV-induced pneumonia (Figure 4).
Intensive immunosuppression associated with loss of HCMV-specific T-cell responses and late HCMV disease.
Peptide-specific CD8+ and protein-specific CD4+T cells were analyzed by intracellular IFN-γ staining with flow cytometry after specific stimulation. Black bars indicate HLA-A*0201–restricted CTLs and gray bars HCMV-specific CD4+ T cells. This patient (15 on Table 2) received donor lymphocytes for relapse of the underlying malignant disease, and severe GVHD subsequently developed. Intensive immunosuppression with prednisolone (Pred) and OKT3 was associated with complete ablation of HCMV-specific T-cell responses and subsequent onset of fatal HCMV disease.
Intensive immunosuppression associated with loss of HCMV-specific T-cell responses and late HCMV disease.
Peptide-specific CD8+ and protein-specific CD4+T cells were analyzed by intracellular IFN-γ staining with flow cytometry after specific stimulation. Black bars indicate HLA-A*0201–restricted CTLs and gray bars HCMV-specific CD4+ T cells. This patient (15 on Table 2) received donor lymphocytes for relapse of the underlying malignant disease, and severe GVHD subsequently developed. Intensive immunosuppression with prednisolone (Pred) and OKT3 was associated with complete ablation of HCMV-specific T-cell responses and subsequent onset of fatal HCMV disease.
Discussion
Despite major advances in the diagnosis and treatment of HCMV infection, HCMV disease remains a major cause of transplant-related morbidity and mortality after allogeneic SCT. Protective immunity toward HCMV infection is maintained by HCMV-specific CTLs. The viral structural matrix protein pp65 has been identified as a major target antigen for HCMV-specific, MHC class I–restricted CTLs derived from the blood of healthy individuals.14,21 With use of HLA class I–restricted T-cell clones, the HLA-A*0201–restricted peptide NLVPMVATV was defined,13,14 and this epitope was later also characterized by epitope prediction based on MHC class I motifs.16 17
However, only a few HCMV CTL epitopes have been characterized previously. In an attempt to define pp65-derived CTL epitopes that allow rapid and sensitive detection of HCMV-specific immune reconstitution after transplantation, a computer-based epitope prediction was performed. By screening HCMV-seropositive healthy individuals for IFN-γ secretion after peptide stimulation, we confirmed the existence of the HLA-B*0702–restricted peptide TPRVTGGGAM that was previously identified by cloning techniques.13,15 Moreover, the HLA-A*0101–restricted peptide YSEHPTFTSQY and the HLA-A*1101–restricted peptide GPISGHVLK were identified as target peptides of CTLs in HCMV-seropositive healthy individuals. More recently, the HLA-A*0101–restricted peptide was also identified by cloning techniques, whereas epitope prediction allowed identification of the optimized HLA-A*1101–restricted HCMV pp65 epitope.15
Because of the high frequency of CTLs specific for HCMV pp65 NLVPMVATV among peripheral blood lymphocytes in HCMV-seropositive and HLA-A*0201–expressing individuals, this peptide has been described as an immunodominant pp65 peptide antigen.14 16Interestingly, we found a dominance of the HLA-B*0702–restricted CTL response compared with the HLA-A*0101–restricted and the HLA-A*0201–restricted CTL response in HCMV-seropositive healthy individuals. In some HCMV-seropositive healthy individuals expressing these HLA restrictions, even a complete absence of HLA-A*0201–restricted CTLs was observed. Thus, CTL precursor frequencies specific for a single peptide seem to vary according to the genetic background of individual patients. Obviously, HLA-B*0702–restricted, HCMV-specific CTL responses seem to dominate the HLA−A*0101–restricted and HLA-A*0201–restricted CTL response in most HCMV-seropositive healthy individuals.
In addition, other HCMV proteins, such as the IE1 protein and pp150, have also been described as potential targets of HCMV-specific CTLs.10,15,27 The tegument protein pp65 was defined as the major target of HCMV-specific CTLs in studies using fibroblasts infected with the laboratory strain AD169.13,15 21However, this might have reflected a nonphysiologic overexpression of this protein in the specific culture system rather than a clear equal immunodominance in nature. Further characterization of HCMV-peptide epitopes for other HCMV proteins, such as pp150 and IE1, with the approach described here will help to further improve our understanding of protective HCMV immunity.
The introduction of novel techniques such as the ELISPOT assay and intracellular cytokine staining after specific stimulation has improved epitope mapping as well as the detection of peptide-specific CTLs present at a low frequency in peripheral blood.9,12,28 In this study, we used intracellular IFN-γ staining after specific stimulation to monitor reconstitution of HCMV-specific CD8+ and CD4+ T cells. In 19 patients, reconstitution of peptide-specific CTLs after allogeneic SCT was clearly associated with resolution of HCMV infection, and none of the patients with detectable HCMV-peptide–specific CTLs had onset of HCMV disease, suggesting the presence of protective immunity as shown previously by classic CTL assays.6,7,29 In patients given transplants from HCMV-seropositive donors, high frequencies of peptide-specific CTLs were observed until day 100 after transplantation, as was also reported by investigators using HLA tetramers.30,31 Early recovery of HCMV-specific CTLs correlated with rapid resolution of HCMV infection and thus a short duration of antiviral therapy in patients receiving PCR-based antiviral treatment. Late reconstitution of CTLs specific for HCMV pp65 peptide was associated with prolonged antiviral treatment, a major risk factor for development of late HCMV disease in patients receiving PCR-based treatment, as we demonstrated previously.4 Notably, patients who had late-onset HCMV disease lacked HCMV-protein–specific T cells at 3 months after SCT.
Ganciclovir prophylaxis is considered an effective strategy for preventing HCMV disease early after transplantation. However, ganciclovir has important toxic side effects, most importantly myelosuppressive and immunosuppressive effects,32 and it has been found to delay the development of HCMV-specific CTLs, thereby leading to late-stage interstitial pneumonia after cessation of drug treatment.29 A large prospective trial could not show a survival benefit in patients receiving ganciclovir prophylaxis, in spite of an impressive reduction in HCMV infection and disease most likely due to a delayed HCMV-specific immune reconstitution.33 Our analysis of HCMV-specific immune reconstitution in a cohort of patients receiving pre-emptive antiviral treatment based on sensitive detection methods shows that short courses of antiviral treatment for documented active HCMV infection allow rapid reconstitution of HCMV-peptide–specific CTLs in most patients, at least in those given a transplant from an HCMV-seropositive donor and even after transplantation of highly purified CD34+ stem cells or stem cells from unrelated donors.
Our data indicate that with the introduction of novel methods (such as intracellular cytokine staining or assessment of class I and class II tetrameric HLA–peptide complexes for well-defined peptides) that allow rapid and sensitive assessment of CTL precursor frequencies and a broader characterization of HCMV-peptide epitopes for many HLA restrictions, HCMV-specific immune reconstitution can be quantified easily. The potential use of these techniques for detection of HCMV-specific CTLs depends on the presence of frequent HLA alleles in the general population. If such frequency exists, identifying a limited number of CTL epitopes might allow assessment of HCMV-specific CTL precursor frequencies in most patients.15 In a recent analysis,34 definition of only 5 CTL epitopes from HCMV protein pp65 was considered sufficient to cover more than 97% of the Dutch population. In addition, loading of these peptides on monocyte-derived mature DCs allows in vitro generation of HCMV-specific T cells from a proportion of HCMV-seropositive and HCMV-seronegative donors.17 35
In this study, we demonstrated that epitope prediction by MHC peptide motifs and an IFN-γ ELISPOT assay allow identification of HLA class I ligands from viral proteins. These newly defined peptides can be used for rapid and quantitative assessment of HCMV-peptide–specific CTL responses after allogeneic SCT on a routine basis as well as for in vitro expansion of donor-derived, HCMV-peptide–specific CTLs for adoptive immunotherapy. Patients with a documented HCMV peptide–specific and protein-specific T-cell response are obviously at low risk of HCMV infection and disease. Although pre-emptive or prophylactic treatment of HCMV infection is mandatory early after transplantation, costly prolonged screening and antiviral treatment associated with life-threatening complications such as secondary bacterial and fungal infections and late HCMV disease can be restricted to patients with delayed HCMV-specific T-cell reconstitution. Thus, introduction of these highly sensitive and rapid techniques as diagnostic markers might help to further improve clinical management of HCMV infection after allogeneic SCT.
We thank M. Pirinen and F. Frank for excellent technical assistance.
Supported by grants from the Deutsche Forschungsgemeinschaft (SFB 510, project B3) and from the Federal Ministry of Education and Research (01KS9602) and the Interdisciplinary Center of Clinical Research Tübingen, project C2.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hermann Einsele, Medizinische Klinik II, Otfried-Müller-Straβe 10, D-72076 Tübingen, Germany; e-mail: hneinsel@med.uni-tuebingen.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal