We present a novel γ-chain dysfibrinogen that was discovered in a 32-year-old asymptomatic man admitted to the hospital after a car accident. He presented with a low fibrinogen concentration, 0.5 mg/mL, and a prolonged thrombin clotting time, 58 seconds. Analysis of purified fibrinogen by sodium dodecyl sulfate–polyacrylamide gel electrophoresis revealed a γ-chain variant with an apparently higher molecular weight. Isoelectric focusing (IEF) demonstrated an anodal shift in the banding pattern of the chains and electrospray ionization mass spectrometry (ESIMS) showed a 27-Da increase in the average mass of the unresolved variant and normal γ chains. DNA sequence analysis showed a heterozygous mutation of GGC (Gly)→GAC (Asp) at codon 309 of the γ chain gene. This Gly→ Asp substitution was consistent with the charge change shown by IEF as well as the mass change identified by ESIMS. Functional analysis revealed that thrombin-catalyzed polymerization occurred with a longer lag time, lower rate of lateral aggregation, and similar final turbidity compared to normal and that factor XIII cross-linking was normal. The polymerization results suggest that residue γ309 is necessary for proper alignment of fibrinogen molecules, specifically in protofibril formation and D:D interactions. γGly309 is highly conserved and x-ray structures support the conclusion that the lack of a side chain at this position helps facilitate the close contact between abutting γD domains of condensing fibrin monomers during polymerization.
Introduction
Fibrinogen is a plasma glycoprotein that consists of 2 sets of 3 polypeptide chains, Aα, Bβ and γ, which combine to form a symmetrical, trinodular molecule with 2 terminal D domains and a central E domain, denoted D-E-D. The E domain consists of the N-termini of all 6 chains, whereas the D domains consist of the C-termini of the Bβ and γ chains. Conversion of soluble fibrinogen into insoluble fibrin occurs when the serine protease thrombin removes 2 fibrinopeptides from the N-termini of the Aα and Bβ chains, in the E domain. The new N-terminal GPR sequences exposed by release of these peptides noncovalently interact with polymerization sites in the D domains of other fibrin molecules to form half-staggered, double-stranded protofibrils. These protofibrils grow linearly and laterally to form the final fibrin clot structure. During this polymerization process, activated factor XIII, a transglutaminase, cross-links the chains, thus stabilizing the overall clot structure.
The γ chain contains several sites that are crucial for normal fibrin polymerization. These sites include: (1) a high-affinity Ca2+ binding site, which is located between residues γ318 and 324,1 (2) the “a” polymerization site, which after fibrinopeptide release interacts with the newly exposed GPR sequence of the “A” site, to promote D:E interactions, (3) the D:D interface, where abutting D domains interact during protofibril formation, and (4) a cross-linking site, where activated factor XIII catalyzes the formation of covalent γ glutamyl-lysyl bonds between γGln398 or γGln399 of one γD domain and γLys406 in another γD domain.2,3 Recent x-ray crystallography studies have shown that the “a” polymerization pocket includes residues γGln329, γAsp330, γHis340, γAsp364 and γArg375,4,5 whereas the D:D interface includes residue γArg275, which forms asymmetric contacts with either γTyr280 or γSer300.5 6 Specifically, γArg275 of one D domain interacts with γTyr280 of a second D domain, whereas γArg275 in the second D domain interacts with γSer300 in the first D domain. Each of these γ chain sites plays a pivotal role in the normal functioning of fibrinogen during blood coagulation.
Over 30 different naturally occurring mutations have been reported in the γ chain and these have provided unique insights into residues involved in Ca2+ binding, D:E interactions, D:D interface contacts, and factor XIII cross-linking (for a review of these, see Cote et al7). In this report, we present a new dysfibrinogen, fibrinogen Hillsborough, which is associated with impaired fibrin polymerization but normal factor XIII cross-linking, due to a novel γ-chain mutation at the D:D interface.
Materials and methods
Coagulation tests
Citrated anticoagulated plasma was used in the routine coagulation assays. Functional fibrinogen levels were determined by the Clauss method and gravimetric levels were determined by reverse-phase quantitation of fibrinopeptides released from whole plasma.8
Fibrinogen purification
Fibrinogen was purified by precipitation with 22.5% ammonium sulfate.8 The pellet was washed twice with 25% saturated ammonium sulfate and redissolved in 20 mM Hepes, pH 7.4, 150 mM NaCl. The redissolved material was dialyzed against distilled water for an hour and then dialyzed extensively against 20 mM Hepes, pH 7.4, 150 mM NaCl. The protein concentration was determined from the absorbance at 280 nm using the extinction coefficient, ε = 1.51.9The purity of the normal and propositus reduced fibrinogen was assessed by separation on a 7.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel.10
Analysis of fibrinogen chains by mass spectrometry
Fibrinogen (5 mg/mL) was reduced in 8 M urea, 0.1 M Tris-HCl, pH 8.0, 15 mM dithiothreitol for 20 hours at 37°C and the chains separated on a Phenomenex C-4 column. The column was monitored at 215 nm and developed with a 0.05% trifluoroacetic acid/acetonitrile gradient system at a flow rate of 0.75 mL/min. Peak crests were collected and 20 μL was injected, at a flow rate of 5 μL/min, into the ion source of a VG Platform II electrospray ionization mass spectrometer (Micromass, Manchester, United Kingdom), as previously described.11 The probe was charged at +3500 V, the source maintained at 60°C, and the mass range of 700 to 1500 m/z scanned every 2 seconds with a 30- to 60-V cone voltage ramp over this range. Over 100 scans were averaged in acquiring the raw data. Data were acquired and processed using Mass-Lynx software and transformed onto a true molecular mass scale using maximum entropy (Max-Ent) software.11
Isoelectric focusing
Neuraminidase (type VI, Sigma, St Louis, MO) digestion was used to reduce the level of microheterogeneity prior to isoelectric focusing (IEF): 5 μg fibrinogen (5 mg/mL in 10 mM acetate buffer pH 5.6, 1 mM CaCl2) was incubated for 4 hours with 1 μg neuraminidase and the fibrinogen recovered by ammonium sulfate precipitation. The purified chains, collected from the peak crests above, were reduced in 8 M urea, 12.5 mM Tris-HCl, pH 8.0, 15 mM dithiothreitol for 2 hours at 37°C. These chains were focused over a pH range of 4 to 8 in a 7% acrylamide gel in 8 M urea, 0.5% Triton X-100.12
DNA sequence analysis
Genomic DNA was isolated from whole blood as previously described13 and all exons of the Aα, Bβ, and γ chains were amplified by the polymerase chain reaction (PCR) and sequenced. Exon 8 of the γ gene was amplified using the forward primer: 5′ TATCTATTGCCTCTTGCCAG and the reverse primer: 5′ TAGTGGTAGTATCGACCGTG. The products were amplified during 35 cycles at: 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 second, with a 5-minute extension at 72°C on a Bio-Rad I-Cycler (Hercules, CA). The PCR products were purified using the High Pure PCR Product Purification kit from Roche (Mannheim, Germany). Sequencing of exon 8 was performed with an internal primer, 5′ CCTACGAAAGAGGGAACTTC, using33P radiolabeled terminators and thermosequenase (Amersham, Amersham, United Kingdom). Sequencing parameters were 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds, for 40 cycles. The samples were denatured for 5 minutes at 94°C before loading onto a 6% sequencing gel.
Thrombin-catalyzed polymerization
Polymerization of purified fibrinogen (720 μL, 0.05 mg/mL) in 20 mM Hepes, pH 7.4, 150 mM NaCl was initiated by adding 80 μL thrombin (5.0 U/mL, Thrombostat, Parke-Davis, Morris Plains, NJ) and mixing. The absorbance at 350 nm was monitored for 15 minutes at room temperature. Lag time and Vmax were calculated as previously described.14 Lag time represents the time when the slope of the steepest part of the polymerization curve (Vmax) crossed the x-axis and final turbidity represents the absorbance at 350 nm after 15 minutes. Statistical values were determined by unpaired t tests using StatView (Berkeley, CA). A significant difference is evident atP < .05.
Factor XIII cross-linking
Factor XIII cross-linking of normal and propositus fibrinogen was initiated by adding human α-thrombin (12 μL, final concentration 0.1 U/mL, Enzyme Research Labs, South Bend, IN) to the reaction mixture (108 μL), containing 0.3 mg/mL fibrinogen, 7.6 U/mL factor XIII in 20 mM Hepes, pH 7.4, 150 mM NaCl, 5 mM CaCl2. The reaction was mixed by pipetting up and down and 12 μL was aliquoted into tubes labeled 0, 1, 2, 3, 4, 5, and 15 minutes. At each appropriate time point, 20 μL Laemmli SDS-reducing buffer was added to the corresponding tube to stop the reaction. On completion of the experiment, all samples were boiled at 100°C for 10 minutes before loading onto a 10% SDS-PAGE gel. The gels were stained with Coomassie blue R-250.
BLAST analysis
Sequence homology of the γ, β, and αE chains of fibrinogen from human, bovine, rat, Xenopus, and lamprey were determined using BLAST (Basic LocalAlignment Search Tool). BLAST can be found at: http://www.ncbi.nlm.nih.gov/BLAST/. Accession numbers are presented in Figure 4. SwissPdbViewer was used to model the γGly309Asp substitution in the DD dimer structure.15
Results
A 32-year-old asymptomatic man was hospitalized as a result of an automobile accident. Routine coagulation tests indicated a low functional fibrinogen level of 0.5 mg/mL (normal range, 1.5-4.0 mg/mL) and a prolonged thrombin clotting time (TCT) of 58 seconds (normal range, 18-24 seconds). Prothrombin time (PT) and activated partial thromboplastin time (aPTT) were normal. On gravimetric analysis of fibrinopeptide release, the circulating fibrinogen level was determined to be 1.5 mg/mL. This 3-fold difference between functional and actual fibrinogen levels, as well as the prolonged clotting time, suggested the presence of an abnormal fibrinogen. Examination of the parents revealed a similar coagulation defect in the father, who had a Clauss fibrinogen of 0.6 mg/mL and gravimetric concentration of 3.0 mg/mL together with a TCT of 57 seconds. The mother was normal with values of 2.7 mg/mL, 2.1 mg/mL, and 21 seconds, respectively.
Analysis of the propositus' fibrinogen by SDS-PAGE revealed 2 γ chain bands: a normal band and a band of apparently higher molecular weight that migrated more slowly through the gel (Figure1A). The equal intensity of the 2 bands suggested a heterozygous mutation in the γ chain and heterozygosity was confirmed by the finding of a similar doublet γ band in the father but not the mother. Further examination of the propositus' γ chains by isoelectric focusing revealed a shift of the individual elements of the banding pattern toward the positive electrode suggesting an increase in net negative charge in some 50% of its chains (Figure 1B). Reverse-phase high-performance liquid chromatography profiles of the chains were normal; however, electrospray ionization mass spectrometry (ESIMS) of the unresolved normal and variant γ chains from the propositus indicated an average mass of 48 416 Da for the dominant monosialylated form, which is 27 Da higher than the normal control, 48 389 Da (data not shown). Together, these results suggest a heterozygous γ chain substitution that results in a negative charge change and a 54-Da increase in mass.
SDS-PAGE and IEF demonstrate an abnormal chain in fibrinogen Hillsborough.
(A) SDS-PAGE of normal (N) and propositus (P) fibrinogens under reduced conditions. The normal Aα, Bβ, and γ chains are labeled on the left and the abnormal γ chain is indicated on the right (arrow). (B) IEF of purified γ chains over the pH range of 4 to 8.
SDS-PAGE and IEF demonstrate an abnormal chain in fibrinogen Hillsborough.
(A) SDS-PAGE of normal (N) and propositus (P) fibrinogens under reduced conditions. The normal Aα, Bβ, and γ chains are labeled on the left and the abnormal γ chain is indicated on the right (arrow). (B) IEF of purified γ chains over the pH range of 4 to 8.
DNA sequencing of the γ chain gene showed a heterozygous GGC→ GAC mutation (Figure 2) at codon γ309. This mutation results in a substitution of aspartic acid for glycine at position 309 in the γ chain and is consistent with the negative charge change indicated by IEF. The known mass difference of 58/2 for the heterozygous Gly→Asp substitution is consistent with the experimental 27-Da increase determined by ESIMS. We have named this novel variant fibrinogen Hillsborough.
DNA sequence of exon 8 of the fibrinogen γ chain reveals a heterozygous mutation in the propositus (P) as compared to normal (N).
The propositus is heterozygous for a GGC→ GA*C mutation, indicated by arrow on left, which causes a Gly→ Asp amino acid change at residue γ309.
DNA sequence of exon 8 of the fibrinogen γ chain reveals a heterozygous mutation in the propositus (P) as compared to normal (N).
The propositus is heterozygous for a GGC→ GA*C mutation, indicated by arrow on left, which causes a Gly→ Asp amino acid change at residue γ309.
Decreased functional levels and a prolonged TCT indicated functional differences between the normal and variant fibrinogens. In vitro functional studies of the propositus' purified fibrinogen revealed that this fibrinogen polymerized with a significantly longer lag time, a slightly lower rate of lateral aggregation (Vmax, Table1) and a similar final turbidity to normal (Figure 3). Factor XIII cross-linking of the propositus' fibrinogen occurred at a similar rate to normal (data not shown). These results indicate that the heterozygous substitution of aspartic acid for glycine at position 309 in the γ chain impairs fibrinogen's polymerization function without altering its ability to be cross-linked by factor XIII.
Polymerization parameters for control and propositus fibrinogen polymerization
| . | Lag time (s) . | Vmax (× 10−4 U/s) . |
|---|---|---|
| Control | 161 ± 9 | 3.0 ± 0.4 |
| Propositus | 278 ± 27 | 2.6 ± 0.2 |
| P | .0002 | .16 |
| . | Lag time (s) . | Vmax (× 10−4 U/s) . |
|---|---|---|
| Control | 161 ± 9 | 3.0 ± 0.4 |
| Propositus | 278 ± 27 | 2.6 ± 0.2 |
| P | .0002 | .16 |
Results are expressed as an average of 4 runs ± SD.
Thrombin-catalyzed polymerization of fibrinogen Hillsborough (dashed) is impaired compared to normal (solid).
Purified fibrinogen (0.05 mg/mL) was mixed with thrombin (5 U/mL) and absorbance monitored at 350 nm as a function of time. Each curve represents an average of 4 runs.
Thrombin-catalyzed polymerization of fibrinogen Hillsborough (dashed) is impaired compared to normal (solid).
Purified fibrinogen (0.05 mg/mL) was mixed with thrombin (5 U/mL) and absorbance monitored at 350 nm as a function of time. Each curve represents an average of 4 runs.
Discussion
Here we identify a novel dysfibrinogen with the mutation, γGly309Asp, in the functionally important C-terminal region of the γ chain. This variant, named fibrinogen Hillsborough, has characteristics similar to several known dysfibrinogens with mutations located in the C-terminal end of the γ chain.7Like fibrinogens Baltimore III (γAsn308Ile) and Kyoto I, Matsumoto II, and Bicêtre II (γAsn308Lys),16-22 fibrinogen Hillsborough presents with a normal prothrombin time and aPTT but a prolonged TCT. Similarly, each of these mutations is associated with low functional but normal immunologic/gravimetric levels. The fibrin polymerization profiles of these purified fibrinogens, when monitored spectrophotometrically, are remarkably similar, with each having a longer lag time and lower rate of lateral aggregation than normal but reaching a similar final turbidity to normal. Additionally, the rates of factor XIII cross-linking of the abnormal γ chains in fibrinogens Kyoto I, Baltimore III, and Hillsborough are all normal. Clinically, however, these dysfibrinogens present differently with fibrinogens Hillsborough, Baltimore III, and Kyoto I being asymptomatic, fibrinogen Bicêtre II being thrombotic, and fibrinogen Matsumoto II having a bleeding tendency. This difference remains unexplained, although it may relate to ascertainment or clinical selection bias or could be a result of different genetic environments. It is clear that abnormal fibrin clot structures can result in either bleeding or clotting tendencies and it is possible that expression of these variants within different coagulation environments could cause the different clinical presentations.
The similar functional abnormalities between the aforementioned dysfibrinogens suggest that the region around residues γ308 and γ309 is important for normal fibrin polymerization. Specifically, the significantly delayed lag times suggest that these residues play a crucial role in the interactions that dictate protofibril formation. Because both D:E and D:D interactions are involved in protofibril formation and these interactions involve sites that are spatially close to residue γ309, the mutation in fibrinogen Hillsborough could impair protofibril formation by disrupting one or both of these interactions. Additionally, the normal cross-linking seen in fibrinogens Hillsborough, Kyoto I, and Baltimore III suggests that mutations in this region do not upset the distal C-terminal region, where γ-γ dimer formation occurs. This result is consistent with the structural data that show the C-terminus of the γ chain as a flexible attachment separate from the globular component of the D domain.1Thus, mutations at γ308 and γ309 likely impair the initial stage of fibrin polymerization, protofibril formation, rather than the final stages characterized by factor XIII cross-linking and γ-γ dimer formation.
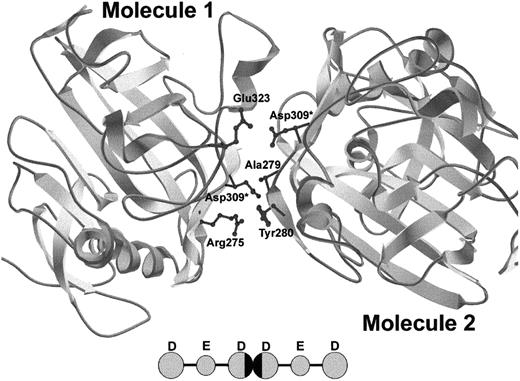
Recent crystal structure determinations provide insight into the possible mechanism of protofibril formation disruption by the γGly309Asp substitution. The fragment D structure5 shows that γ309 is solvent exposed and in the vicinity of the D:D interface, along with residues γ308, γ310, γ268, and γ275. It is likely that replacement of glycine by a bulky, charged aspartic acid residue disrupts the normal interactions at the D:D interface. Figure 4 simulates the D:D interactions of 2 D domains containing the γAsp309 substitution. It is evident that the bulky, charged side chains of the aspartic acid residues extend significantly into the D:D interface. More specifically, when Asp is introduced into molecule 1 (left), the Asp sterically clashes with γAla279 and the side chains of γTyr280 of molecule 2 (right) and when the Asp is placed in molecule 2, it electrostatically clashes with γGlu323 of molecule 1. Although this is only one of the conformations that these aspartic acid residues can take relative to the rest of the D domain, simulations of this type support the hypothesis that a bulky, charged aspartic acid residue can upset the D:D interaction that dictates protofibril formation by causing both steric and electrostatic repulsions. Because the D:D interactions do not dictate lateral aggregation, this mutation does not affect lateral aggregation of protofibrils, as evidenced by the similar maximal turbidity reached between normal and γGly309Asp fibrinogen (Figure 3).
Crystal structure of DD dimer interface showing simulated γGly309Asp mutation.
Because of the asymmetric nature of the D:D interface, the simulated γAsp309 residue has 2 different sets of potential interactions. γAsp309 in molecule 1 causes steric repulsion of γAla279 and γTyr280 in molecule 2 and γAsp309 in molecule 2 causes electrostatic repulsion of γGlu323 in molecule 1. Asterisks indicate new Asp side chains. The model was created using Swiss-PdbViewer. The inset shows 2 fibrin monomers interacting, with the D:D interface blackened.
Crystal structure of DD dimer interface showing simulated γGly309Asp mutation.
Because of the asymmetric nature of the D:D interface, the simulated γAsp309 residue has 2 different sets of potential interactions. γAsp309 in molecule 1 causes steric repulsion of γAla279 and γTyr280 in molecule 2 and γAsp309 in molecule 2 causes electrostatic repulsion of γGlu323 in molecule 1. Asterisks indicate new Asp side chains. The model was created using Swiss-PdbViewer. The inset shows 2 fibrin monomers interacting, with the D:D interface blackened.
Examining the sequence homology of fibrinogen from various species supports a functional importance for residue γ309, which is conserved as Gly in the γ chain of all species that have been sequenced (Figure5). The conservation seems to be driven by functional rather than structural constraints, because the corresponding position in the nonfunctional Bβ and AαEchains shows considerable variation with bulky polar residues of Ser and Asn being allowed. We suggest that the absence of a side chain at γ309 may be necessary to facilitate the D:D interactions that occur between the chains of docking fibrin monomers. This conservation further supports the idea that the γGly309Asp substitution imposes its functional abnormality by altering the normal interactions that occur at the D:D interface, which are crucial for protofibril formation and thus the subsequent steps in fibrin polymerization.
Sequence homology of γ, β, and αEchains of fibrinogen from 5 different species as determined by BLAST analysis.
Sequence homology of γ, β, and αEchains of fibrinogen from 5 different species as determined by BLAST analysis.
We gratefully acknowledge the donation of purified factor XIII by Dr Kevin Siebenlist.
Supported in part by National Institutes of Health grant F32 HL10436-01 to J.L.M.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jennifer L. Mullin, Molecular Pathology Laboratory, Canterbury Health Laboratories, PO Box 151, Christchurch, New Zealand; e-mail: jennifer.mullin@chmeds.ac.nz.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal