FLT3 receptor tyrosine kinase is expressed on lymphoid and myeloid progenitors in the hematopoietic system. Activating mutations in FLT3 have been identified in approximately 30% of patients with acute myelogenous leukemia, making it one of the most common mutations observed in this disease. Frequently, the mutation is an in-frame internal tandem duplication (ITD) in the juxtamembrane region that results in constitutive activation of FLT3, and confers interleukin-3 (IL-3)–independent growth to Ba/F3 and 32D cells. FLT3-ITD mutants were cloned from primary human leukemia samples and assayed for transformation of primary hematopoietic cells using a murine bone marrow transplantation assay. FLT3-ITDs induced an oligoclonal myeloproliferative disorder in mice, characterized by splenomegaly and leukocytosis. The myeloproliferative phenotype, which was associated with extramedullary hematopoiesis in the spleen and liver, was confirmed by histopathologic and flow cytometric analysis. The disease latency of 40 to 60 days with FLT3-ITDs contrasted with wild-type FLT3 and enhanced green fluorescent protein (EGFP) controls, which did not develop hematologic disease (> 200 days). These results demonstrate that FLT3-ITD mutant proteins are sufficient to induce a myeloproliferative disorder, but are insufficient to recapitulate the AML phenotype observed in humans. Additional mutations that impair hematopoietic differentiation may be required for the development of FLT3-ITD–associated acute myeloid leukemias. This model system should be useful to assess the contribution of additional cooperating mutations and to evaluate specific FLT3 inhibitors in vivo.
Introduction
FLT3 (FMS-like receptor tyrosine kinase) also referred to as fetal liver kinase 2 (FLK-2) or stem cell tyrosine kinase 1 (STK-1) was originally described as a tyrosine kinase receptor with strong sequence similarity to FMS, KIT, and platelet-derived growth factor receptor (PDGFR).1,2 These proteins are all members of the receptor tyrosine kinase type (RTK) III subfamily based on a common domain structure including an interrupted kinase domain (for review, see Rosnet and Birnbaum3). FLT3 is expressed by cells found in the hematopoietic stem cell compartment (Sca1+, Lin−)1 and in more restricted progenitors of the lymphoid and myeloid series4,5 (for review, see Shurin et al6). Like the other RTK III family members, the stimulation of FLT3 by its ligand (FLT ligand, FL) has been proposed to play a role in cell proliferation.7 8
It is therefore interesting that activating mutations in FLT3 have been found in approximately 25% to 30% of patients with acute myelogenous leukemia (AML).9,10 The majority of FLT3 mutations are internal tandem duplications (ITDs) in the juxtamembrane domain encoded by exon 11, and were first reported in patients with AML in 1996.11 This mutation appears to confer a poor prognosis in several studies reported to date.12-14 All ITD repeats were in-frame, but varied significantly in length. No such ITD abnormalities were detected in remission samples, indicating that these were acquired mutations.11 Subsequent studies have confirmed the presence of the juxtamembrane ITDs in human leukemias, with an overall incidence of approximately 20% in AML. In addition, substitution mutations in the FLT3 kinase domain at Asp835 have recently been reported in approximately 7% of patients with AML.10 Both the FLT3-ITD and FLT3-Asp835 mutations result in constitutive activation of the receptor and are associated with FLT3 autophosphorylation and phosphorylation of downstream targets such as STAT5 and mitogen-activated protein (MAP) kinase.15-18 FLT3-ITD mutations are also found but at much lower frequencies in related diseases such as myelodysplastic syndrome (< 5%)9,10,19 and have rarely been identified in acute lymphocytic leukemia (ALL) unless they are biphenotypic.20
Constitutive activation of RTKs is not unprecedented in leukemias; however, the vast proportion of activated RTKs studied thus far are associated with chronic myelogenous leukemia (CML) phenotypes and occur as a result of a chromosomal translocation. Examples include the BCR/ABL, TEL/PDGFβR, TEL/JAK2, HIP1/PDGFβR, H4/PDGFβR, and TEL/ABL fusion proteins that are expressed as a consequence of the t(9;22)(q34;q22),21-25 t(5;12)(q33;p13),26-30t(9;12)p(24;p13),31-33t(5;7)(q33;q11.2),34,35t(5;10)(q33;q11.2),36,37 and t(9;12)(q34;p13)38 translocations, respectively. Each of these fusion proteins contains a carboxy-terminal tyrosine kinase domain and an amino terminal oligomerization domain that constitutively activates the respective tyrosine kinase (for a review, see Sawyers39). These fusion proteins are all localized to the cytoplasm, and constitutive activation of the respective tyrosine kinase confers factor-independent growth to cultured hematopoietic cell lines.21-38,40,41 Furthermore, each of these fusions is capable of conferring a myeloproliferative phenotype in murine bone marrow transplant (BMT) assays for leukemia,21-38,40,41 indicating that tyrosine kinase fusions are sufficient to induce a myeloproliferative phenotype. Mutational analysis has demonstrated that transformation in cell culture, or in the murine BMT assay, is absolutely dependent on kinase activity.21-38,40 41
Activating mutations in FLT3 differ from tyrosine kinase fusions associated with the CML phenotypes. FLT3-ITD and activating loop mutants are localized to the plasma membrane as opposed to the cytoplasmic localization of tyrosine kinase fusions. The residual tyrosine kinase allele in the tyrosine kinase fusions associated with chromosomal translocations is typically either expressed at low levels or is localized to a different subcellular compartment than the tyrosine kinase fusion. In contrast, both the wild-type and FLT3-ITD alleles are expressed at comparable levels and are localized to the plasma membrane where they may interact. Perhaps most importantly, activating mutations in FLT3 are most frequently associated with AML rather than CML phenotypes.9Furthermore, FLT3-ITDs are most often observed in AML patients negative for other cytogenetic abnormalities.14 Taken together, these data suggest that FLT3-ITDs may play a critical role in the development of AML.
We have investigated the transforming properties of the FLT3-ITD in primary hematopoietic cells using a murine BMT assay. We report that the FLT3-ITDs cloned from primary human myeloid leukemias confer factor-independent growth to Ba/F3 cells and induce a myeloproliferative phenotype similar to that observed with the translocation-associated tyrosine kinase fusions. These data have important implications for the contribution of the FLT3-ITD to the pathogenesis of human AML. Furthermore, this model should provide a useful system for analysis of cooperativity of FLT3 with other mutations and gene rearrangements associated with human leukemia and for testing of FLT3-specific low molecular weight inhibitors.
Materials and methods
Cloning and vector construction
The complementary DNA (cDNA) for human FLT3 was kindly provided by H. E. Broxmeyer (Indiana University School of Medicine) and amplified by polymerase chain reaction (PCR) using primers containing an additional HpaI site, flt5′HpaI: GTTAACCATGCCGGCGTTGGCGC and FLT3′HpaI: GTTAACTACGAATCTTCGACC and cloned into pGEM-T easy (Stratagene, La Jolla, CA) according to the manufacturer's instructions and sequenced. After informed consent was obtained, DNA taken from leukemic blasts from 12 patients was screened by PCR strategy for length mutations in exon 11 of the FLT3 gene using the primers, F1: GGTGTTTGTCTCCTCTTCATTGTCA and B1: AAAGCACCTGATCCTAGTACCTTCC, to give a 223-base pair (bp) PCR product following amplification of the wild-type exon. The PCR was performed with Taq polymerase (Perkin Elmer, Foster City, CA) in the manufacturer's buffer including 10% glycerol for 35 cycles at an annealing temperature of 50°C using 3 μM of each primer. Higher molecular weight PCR products, indicative of a length mutation, were subcloned into “pGEM-T easy” and sequenced. The ITD mutations were recreated in the FLT3 cDNA by amplifying 2 PCR products that overlap in the duplicated region within exon 11. To generate the W51 mutation, a PCR product from nucleotides 603 to 1801 was generated using primers FLT3/604F: CAGGGGGAAAGCTGTAAAG and W51B2: GATCATATTCATATTCTCTG. The product was digested withXhoI, which cleaves at nucleotide 874. The second PCR product spanned nucleotides 1781 to 2982 at the end of the cDNA using primers W51F2 (phosphorylated): TCAGAGAATATGAATATGAT and SP6: TATTTAGGTGACACTATAG (binds in pGEM). The second product was digested with SacI, which cleaves in the pGEM polylinker 3′ of the FLT3 cDNA. These 2 PCR fragments were ligated resulting in a fragment that contained the appropriate duplication and could be cloned into theXhoI and SacI sites in the FLT3 cDNA to replace the wild-type sequence. The same strategy was used to generate each of the mutants where the primers used to generate the W73, W78, T6, and POS mutant were W73B2: CCATATTCATATTCTCTGAAA and W73F2: GCTCTACGTTGATTTCAGAG; W78B2: CCTTCATATTCTCTGAAATCAACG and W78F2: GTTGGTACAGGTGACCGGCTC; T6B2: CCAAACTCTAAATTTTCTCTTGG and T6F2: TTACGTTGATTTCAGAGAATATG; POSB2: TAAATTTTCTCTTGGAAACTCCCATTTG and POSF2: TGCTCCTCAGATAATGAGTACTTC, respectively.
Cell culture and virus production
The Ba/F3 cells were maintained in RPMI, 10% fetal calf serum (FCS), and interleukin-3 (IL-3) (0.5 ng/mL, R & D Systems, Minneapolis, MN). The 293T cells were maintained in Dulbecco modified Eagle medium (DMEM) with 10% FCS. Retroviral stocks were generated as described previously.32 Briefly, transient cotransfection of equimolar amounts of MSCV constructs and a packaging construct, pIK6 (Cell Genesys, Redwood City, CA), was performed in 293T cells using Superfect (Qiagen, Valencia, CA) according to the manufacturer's instructions. Supernatants were harvested 48 hours after transfection, filtered (0.45 μM), and aliquoted for storage at −80°C. Viral titers were estimated by transduction of Ba/F3 cells in the presence of Polybrene (10 μg/mL) as described previously.44 Briefly, cells (1 × 106) were infected for 48 hours and analyzed by flow cytometry to determine the percentage of enhanced green fluorescent protein-positive (EGFP+) cells. Viral titers for all constructs used were normalized to the lowest titer and were used at titers of 1 × 105 infectious units/mL. To test for transforming ability, Ba/F3 cells were transduced as above with MSCV-neo constructs, cultured for 48 hours, and selected with G418 (Life Technologies, Carlsbad, CA) for 14 days. Factor-independent growth was determined by washing resistant cells 3 times in phosphate-buffered saline (PBS) followed by plating in medium without IL-3. Live cells, determined by trypan blue dye exclusion, were counted daily for 7 days.
Mouse strains and BMT
BALB/c mice were purchased from Taconic (Germantown, NY). BMT assays were carried out as described previously.32 44Briefly, 4- to 6-week-old male donor mice were primed with intraperitoneal injection of 5′-fluorouracil (150 mg/kg, Sigma, St Louis, MO) and subsequently killed after 6 days by CO2 asphyxiation. Bone marrow was flushed from femurs and tibias, and red blood cells were lysed (Red Blood Cell Lysis, RBCL buffer, Sigma). Cells were cultured overnight with IL-3 (6 ng/mL, R & D Systems), IL-6 (10 ng/mL, R & D systems), and stem cell factor (10 ng/mL, Peprotech, Rocky Hill, NJ) in RPMI with 10% FCS (transplant medium). Cells were transduced by 2 rounds of spin-infection, at 24 hours and 48 hours after harvesting. Centrifugation of 1 mL viral supernatant and 4 × 106cells in 3 mL transplant media containing 5 μg/mL Polybrene and 7.5 mM HEPES buffer was carried out for 90 minutes at 1800g. Cells were washed in PBS, resuspended in Hanks balanced salt solution (Life Technologies) and injected (5 × 105 cells/0.5 mL) into the lateral tail vein of lethally irradiated (2 × 450 cGy) female recipient mice. Mice were housed in microisolator cages with autoclaved chow and acidified water.
Histopathology
Murine tissues were fixed for at least 72 hours in 10% neutral buffered formalin (Sigma), dehydrated in alcohol, cleared in xylene, and infiltrated with paraffin on an automated processor (Leica, Bannockburn, IL). The tissue sections (4 μm thick) from paraffin-embedded tissue blocks were placed on charged slides and deparaffinized in xylene, rehydrated through graded alcohol solutions, and stained with hematoxylin and eosin. To visualize the reticulin fibers, rehydrated sections were sequentially treated with potassium permanganate (0.5%), potassium metabisulfite (2%), ferric ammonium sulfate (2%), ammonical silver solution, formalin (10%), gold chloride (0.2%), potassium metasulfite (2%), sodium thiosulfate (2%), and counterstained with methyl green dye according to the published method.45
Flow cytometric immunophenotyping and fluorescence-activated cell sorting
Single-cell suspensions of spleen, bone marrow, and blood were prepared as described previously.32 44 Briefly, red blood cells were lysed in RBCL buffer (Sigma) for 5 minutes at room temperature and frozen in 90% fetal bovine serum and 10% dimethyl sulfoxide. Prior to analysis, cells were washed in PBS/0.1% NaN3/0.1% bovine serum albumin (BSA) and preincubated for 20 minutes on ice with supernatant from the 2.4G2 hybridoma cell line (anti-CD16/CD32; American Type Culture Collection, Rockville, MD) to block nonspecific Fc receptor-mediated binding. Cells were stained for 20 minutes on ice with monoclonal antibodies, washed in staining buffer, and stained with secondary antibodies where necessary. Antibodies used were allophycocyanin (APC)–conjugated Gr-1 and CD4; phycoerythrin (PE)–conjugated Mac 1, Thy 1.2, and CD8; biotin-conjugated CD19; and APC-conjugated streptavidin. All antibodies were purchased from Pharmingen, San Diego, CA apart from APC-CD4 and APC streptavidin, which were purchased from Caltag, Burlingame, CA. Flow cytometric analysis was carried out using a 4-color FACSort cytometer (Becton Dickinson, Mountain View, CA) and analyzed using CellQuest software.
Fluorescence-activated cell sorting (FACS) was performed on a Coulter Epics Altra at approximately 15 000 cells/s. A single cell suspension from the spleen was resuspended in 1% BSA in PBS at 107cells/mL and sorted into EGFP+ and EGFP−fractions. Gates were set around the clearly negative and positive cell populations and weakly EGFP+ cells not included in the gates were discarded (Figure 5C). The EGFP− fraction was 98.3% pure and the EGFP+ fraction was 86.5% pure after sorting. DNA was prepared as described below.
Southern analysis
DNA was prepared using a PUREGENE DNA isolation kit according to the manufacturer's protocol (Gentra Systems, Minneapolis, MN). After enzymatic digestion of 20 μg DNA with either EcoRI orXhoI and electrophoretic separation, the genomic DNA was blotted to Hybond-N+ nylon membranes (Amersham, Arlington Heights, IL) by capillary transfer.46 The green fluorescent protein (GFP) probe was a 750-bp fragment isolated using aNcoI/SalI digest from MSCV-EGFP. The 500-bp granzyme A probe was a kind gift from J. Pollock and T. Ley, Washington University. Probes were labeled with 32P by random priming using Radprime (Life Technologies), and Southern hybridization was performed as described.32
Western analysis
The Ba/F3 cells were washed 3 times in PBS and lysed in 10 mM Tris pH 7.5, 130 mM NaCl, 1% Triton X-100, 10 mM NaF, 10 mM sodium phosphate, 10 mM sodium pyrophosphate, 10 mM EDTA, 1 mM sodium vanadate, and protease inhibitor cocktail tablets (Roche-Boehringer, Indianapolis, IN). Total cell lysate (150 μg) was separated on a 7.5% gel and Western analysis was performed as described previously44 using a 1:200 dilution of anti-FLT3 SC-479 (Santa Cruz, Santa Cruz, CA) as the primary antibody, followed by horseradish peroxidase–conjugated secondary antibody (Amersham Life Science) and visualization by enhanced chemoluminescence (Amersham Life Science).
Results
Cloning of FLT3-ITD mutations from primary human AML cells
The DNA samples from peripheral blood of 12 patients with AML were screened by PCR (see “Materials and methods”) for length mutations in the juxtamembrane region, encoded by exon 11. Four independent FLT3-ITD mutants were cloned, which had a duplicated sequence of between 7 and 25 amino acids (Figure1). The length and position of the duplication varied between samples and differed from other published FLT3-ITD mutants.9,11,13,15 No abnormalities were observed in samples from 54 healthy individuals, consistent with published data.9 Each FLT3-ITD mutant, including a previously reported FLT3-ITD mutant (Npos is Mut117) positive control, and the wild-type FLT3 gene were subcloned into a MSCV murine ecotropic retrovirus that expressed either the neomycin selectable marker or the EGFP under the control of an internal ribosome entry site (IRES).
A schematic diagram of the FLT3 receptor tyrosine kinase.
The extracellular domain is labeled and shaded light gray, arrows point to the transmembrane (hatched box) and juxtamembrane domains, and the 2 protein kinase domains (PTK) are shaded dark gray and labeled. The amino acid sequence of exon 11, which encodes the juxtamembrane sequence, is shown below. WT is the wild-type sequence and W78, W73, W51, Npos, and T6 are the FLT3-ITD mutations where the duplicated residues are shown, together with an arrow indicating the duplication position with respect to wild-type exon 11. The amino terminus is labeled NH2 and the carboxy terminus is labeled COOH.
A schematic diagram of the FLT3 receptor tyrosine kinase.
The extracellular domain is labeled and shaded light gray, arrows point to the transmembrane (hatched box) and juxtamembrane domains, and the 2 protein kinase domains (PTK) are shaded dark gray and labeled. The amino acid sequence of exon 11, which encodes the juxtamembrane sequence, is shown below. WT is the wild-type sequence and W78, W73, W51, Npos, and T6 are the FLT3-ITD mutations where the duplicated residues are shown, together with an arrow indicating the duplication position with respect to wild-type exon 11. The amino terminus is labeled NH2 and the carboxy terminus is labeled COOH.
Expression and transforming properties of FLT3 and FLT3 mutants in Ba/F3 cells
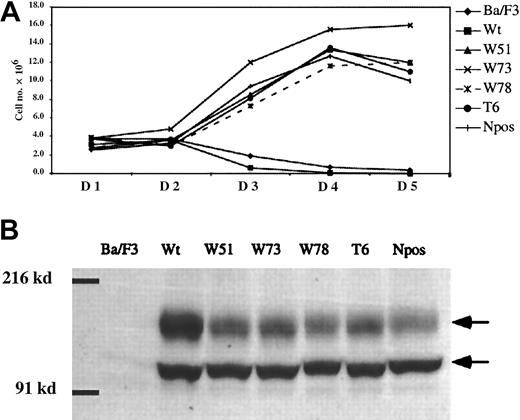
To confirm that the FLT3-ITD mutants we cloned had similar properties to those previously reported, Ba/F3 cells were stably transduced with MSCV-neo retrovirus containing either the wild-type FLT3 or FLT-ITD cDNAs. Pools of transduced cells were maintained in IL-3 and selected with G418 for 2 weeks prior to analysis of factor-independent growth. Each of the FLT3-ITD mutants including the previously reported positive control Npos,17 conferred IL-3 factor–independent growth to Ba/F3 cells. Parental cells or cells stably transduced with the wild-type FLT3 died after IL-3 deprivation (Figure 2A). There were no significant differences in growth rates in replicate experiments over the time course of this assay. A point mutation in the activation loop that inhibits kinase activity of wild-type FLT347 abrogated transforming activity of FLT3-ITD when mutated in the context of W51 (data not shown). Western blot analysis confirmed that the cell pools expressed comparable levels of FLT3 and each FLT3-ITD mutant at the expected molecular weights of approximately 130 and 155 kd (Figure 2B), and that parental Ba/F3 cells did not express FLT3 (Figure 2B). The FLT3 wild-type protein was identified by immunoprecipitation analysis as 2 distinct bands of approximately 130 and 155 kd, respectively. It has been previously demonstrated by expression in COS-1 and COS-7 cells that these 2 bands are generated from a predicted 110-kd protein via posttranslational glycosylation.15 47
Growth rate of Ba/F3 cells expressing wild-type and ITD mutant FLT3 receptor.
(A) Cell number is indicated on the y-axis, and time in days on the x-axis. Ba/F3 cells transduced with MSCV-neo retroviruses containing FLT3 wild-type and ITD mutant cDNAs were selected for 14 days with G418 in the presence of IL-3. The cell numbers plotted represent the viable cells in each population over 5 days when cells were cultured in the absence of IL-3. The legend on the right indicates the symbols used for the curves of Ba/F3 cells, cells transduced with wild-type Flt3 (Wt) and each of the FLT3-ITD mutants, W51, W73, W78, T6, and Npos. (B) Western analysis of Ba/F3 cells expressing either wild-type (Wt) or ITD mutant FLT3 using an anti-FLT3 antibody. Two FLT3-specific bands are detected in cells transduced with the FLT3 constructs as indicated by arrows, where the upper band is a more glycosylated form. The position of molecular weight markers of 216 and 91 kd is indicated.
Growth rate of Ba/F3 cells expressing wild-type and ITD mutant FLT3 receptor.
(A) Cell number is indicated on the y-axis, and time in days on the x-axis. Ba/F3 cells transduced with MSCV-neo retroviruses containing FLT3 wild-type and ITD mutant cDNAs were selected for 14 days with G418 in the presence of IL-3. The cell numbers plotted represent the viable cells in each population over 5 days when cells were cultured in the absence of IL-3. The legend on the right indicates the symbols used for the curves of Ba/F3 cells, cells transduced with wild-type Flt3 (Wt) and each of the FLT3-ITD mutants, W51, W73, W78, T6, and Npos. (B) Western analysis of Ba/F3 cells expressing either wild-type (Wt) or ITD mutant FLT3 using an anti-FLT3 antibody. Two FLT3-specific bands are detected in cells transduced with the FLT3 constructs as indicated by arrows, where the upper band is a more glycosylated form. The position of molecular weight markers of 216 and 91 kd is indicated.
Two independent FLT3-ITD mutants, but not wild-type FLT3, induce a lethal myeloproliferative disease in a murine BMT assay
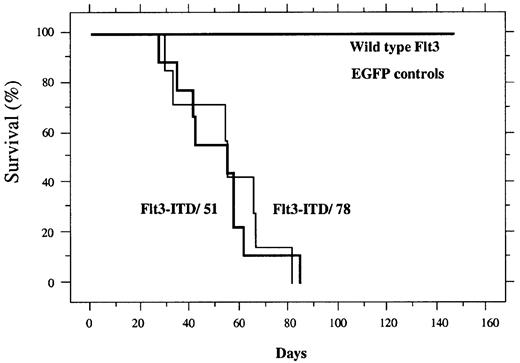
Donor mice were treated with 5-fluorouracil to increase the number of cycling stem cells for retroviral transduction, as described in “Materials and methods.” Equivalent titers of FLT3-ITD or FLT3 were then transduced into primary mouse bone marrow cells using the MSCV-EGFP retrovirus. Transduced bone marrow for each construct was also analyzed by flow cytometry to confirm EGFP expression in a comparable fraction of the transplanted cells. Transduced bone marrow cells were transferred by injection into the lateral tail vein of lethally irradiated syngeneic recipient mice. Mice receiving transplants of cells transduced with retrovirus containing the wild-type FLT3 or the MSCV vector expressing EGFP engrafted normally and had a normal survival with a follow-up of more than 200 days (Figure 3). In contrast, animals transduced with cells containing either the FLT3-ITD/51 or FLT3-ITD/78 developed a lethal hematopoietic disease with a median latency of approximately 40 to 50 days (Figure 3 and Table1).
Kaplan Meier plot of survival.
Mice receiving transplants of bone marrow transduced with wild-type FLT3 (n = 8), EGFP (n = 4), FLT3-ITD/51 (n = 9), or FLT3-ITD/78 (n = 8). The percentage of surviving mice (y-axis) is plotted with respect to time in days (x-axis).
Kaplan Meier plot of survival.
Mice receiving transplants of bone marrow transduced with wild-type FLT3 (n = 8), EGFP (n = 4), FLT3-ITD/51 (n = 9), or FLT3-ITD/78 (n = 8). The percentage of surviving mice (y-axis) is plotted with respect to time in days (x-axis).
Analysis of mice that received transplants of wild-type FLT3 or FLT3-ITD-transduced bone marrow
| Construct . | No. of mice . | WBC count, per μL (median) . | Spleen weight, mg (median) . | Phenotype . |
|---|---|---|---|---|
| Wt-FLT3 | 6 | 3.5-5.5 × 103 (5 × 103) | 100-171 (113) | Normal |
| FLT3-ITD/51 | 8 | 28-145 × 103 (36 × 103) | 300-1000 (800) | Myeloproliferation |
| FLT3-ITD/78 | 6 | 6.5-129 × 103 (75 × 103) | 250-900 (670) | Myeloproliferation |
| Construct . | No. of mice . | WBC count, per μL (median) . | Spleen weight, mg (median) . | Phenotype . |
|---|---|---|---|---|
| Wt-FLT3 | 6 | 3.5-5.5 × 103 (5 × 103) | 100-171 (113) | Normal |
| FLT3-ITD/51 | 8 | 28-145 × 103 (36 × 103) | 300-1000 (800) | Myeloproliferation |
| FLT3-ITD/78 | 6 | 6.5-129 × 103 (75 × 103) | 250-900 (670) | Myeloproliferation |
The construct used is listed in column 1, followed by the number of mice analyzed in column 2. The phenotype is listed as normal or myeloproliferation based on the histologic and immunologic phenotype.
Animals receiving transplants of FLT3-ITD developed marked leukocytosis, with a white blood cell (WBC) count ranging from 28 to 145 × 103/μL (Table 1). Differential counts analyzed by the scatter property of peripheral blood of several animals indicated that the increase in WBCs was due almost entirely to neutrophils, which were increased from between 13.7% to 21.8% in 3 controls to 75.7% to 87% in 3 FLT3-ITD–diseased animals analyzed. Animals with FLT3-ITD–induced disease appeared to have a higher proportion of mature neutrophils with fewer immature myeloid cells observed than in animals with myeloproliferative disease associated with other tyrosine kinase fusions.28 The absolute numbers of other blood cells were unchanged and no abnormality in erythrocyte count or composition was observed. Wright-Giemsa stains of peripheral blood demonstrated leukocytosis with myeloid lineage cells in all stages of maturation, predominantly terminally differentiated neutrophils (Figure 4A).
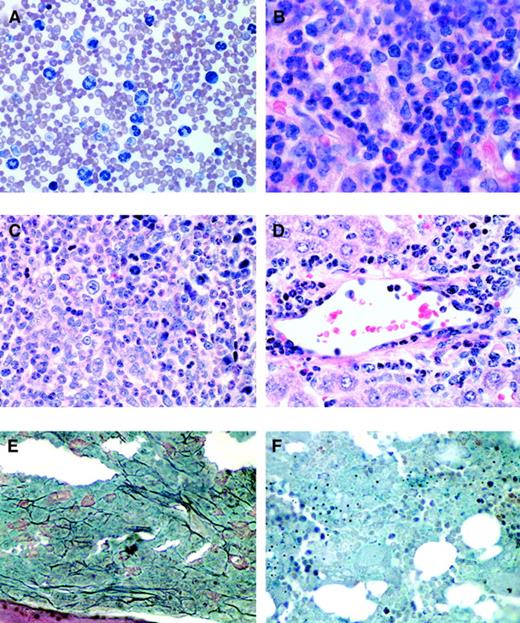
Histopathology of mice receiving transplants of FLT3-ITD–transduced bone marrow.
(A) Peripheral blood smear (Wright-Giemsa stain, original magnification × 400) from a representative FLT3-ITD mouse (mouse 51.1) reveals marked leukocytosis comprised predominantly of maturing myeloid elements. (B) Bone marrow from the femur of the same mouse (hematoxylin and eosin, original magnification × 500) reveals features of a myeloproliferative disorder with marked hypercellularity and myeloid hyperplasia consisting predominantly of mature granulocytic elements. Spleen (C) and liver (D) (hematoxylin and eosin, original magnification, × 500) also reveal an identical myeloid infiltrate in the Flt3-ITD mouse (mouse 51.1). (E) Bone marrow of a Flt3-ITD mouse (mouse 78.5) and (F) bone marrow from a wild-type FLT3 mouse. Both were stained to highlight reticulin fibers (black) (original magnification, × 500). There is a marked increase in reticulin fibrosis in the FLT3-ITD mice compared to wild-type FLT3 mice.
Histopathology of mice receiving transplants of FLT3-ITD–transduced bone marrow.
(A) Peripheral blood smear (Wright-Giemsa stain, original magnification × 400) from a representative FLT3-ITD mouse (mouse 51.1) reveals marked leukocytosis comprised predominantly of maturing myeloid elements. (B) Bone marrow from the femur of the same mouse (hematoxylin and eosin, original magnification × 500) reveals features of a myeloproliferative disorder with marked hypercellularity and myeloid hyperplasia consisting predominantly of mature granulocytic elements. Spleen (C) and liver (D) (hematoxylin and eosin, original magnification, × 500) also reveal an identical myeloid infiltrate in the Flt3-ITD mouse (mouse 51.1). (E) Bone marrow of a Flt3-ITD mouse (mouse 78.5) and (F) bone marrow from a wild-type FLT3 mouse. Both were stained to highlight reticulin fibers (black) (original magnification, × 500). There is a marked increase in reticulin fibrosis in the FLT3-ITD mice compared to wild-type FLT3 mice.
Gross pathologic examination demonstrated emaciation, ruffled fur, and marked splenomegaly, with spleen weights ranging from 300 to 1000 mg in FLT3-ITD/51 and 250 to 900 mg in FLT3-ITD/78 compared to 100 to 171 mg in wild-type FLT3 or EGFP controls (Table 1). The slight increase in spleen weight in the controls was typical for animals after irradiation and BM transplantation. Bone marrow stained with hematoxylin-eosin demonstrated hypercellularity with a marked predominance of maturing myeloid lineage cells, consistent with a myeloproliferative disease (Figure 4B). In addition, the bone marrow, as in other mouse models of myeloproliferative disease,48 displayed a significant degree of reticulin fibrosis as demonstrated by silver stain when compared to control FLT3 mice (Figure 4E,F). A reduced yield of cells was obtained from the marrow of FLT3-ITD animals compared to EGFP and wild-type FLT3 controls. Splenic architecture was effaced by a marked expansion of red pulp comprised of maturing myeloid cells (Figure 4C) and scattered admixed megakaryocytes (not shown). Analysis of the liver demonstrated a perivascular parenchymal infiltration comprised predominantly of maturing myeloid elements, with admixed erythroid and megakaryocytic elements similar to that seen in the spleen (Figure 4D). The lungs showed evidence of pulmonary hemorrhage, as observed in BMT assays with other tyrosine kinase fusions such as with TEL/PDGFβR28 and TEL/JAK2.48 FLT3-ITD disease was not transplantable into lethally irradiated secondary recipient mice (n = 16) when 1 × 106 spleen cells from 4 independent mice with myeloproliferative disease were used as donors with a follow-up of more than 80 days. This is consistent with observations made in other tyrosine kinase fusion models of myeloproliferative disease.29 32
Flow cytometric analysis of blood and spleen cells from FLT3-ITD mice further confirmed the myeloproliferative phenotype. Several FLT3-ITD animals were examined in detail showing 57% to 70% of cells in the spleen (n = 3) and 70% to 90% of cells in the blood (n = 5) were positive for the late myeloid markers Gr-1 (Ly 6-G) and Mac-1, indicative of mature neutrophils, compared to 5% to 9% and 16% to 24% Gr-1, Mac-1 double-positive cells in spleen and blood of controls, respectively. A representative set of samples of one EGFP control, one wild-type FLT3, and one FLT3-ITD are presented in Figure5. Gr-1+ cells were also EGFP+, consistent with proviral integration in these cells. In addition, significant proportions of Gr-1+ cells were EGFP− (Figure 5). Potential explanations for this observation include cell nonautonomous contributions to myeloid proliferation,49 lower undetectable levels of EGFP in some clones due to lower expression levels depending on integration site of the provirus, or loss of EGFP by leaching from the short-lived neutrophils, which are highly overrepresented in this disease.50 To address this issue, we sorted cells from the spleen into EGFP− and EGFP+ fractions and carried out Southern analysis for the presence of the provirus in both populations. Hybridization with a probe for EGFP shows that cells both positive and negative for EGFP by flow cytometry carry the provirus (Figure 5C). The blot was hybridized with a second probe (granzyme A) as a control for DNA loading and relative copy number. This indicates that the expansion in the Gr-1+, EGFP− population observed (Figure 5A, B) is probably due to a cell autonomous effect as a direct result of FLT3-ITD expression.
Immunophenotype of cells from the peripheral blood and spleen of FLT3-ITD mice.
Spleen (A) and blood (B) cells from mice that received bone marrow transduced with retroviruses encoding EGFP only, wild-type FLT3 and EGFP, or FLT3-ITD (mouse N78.5) and EGFP. Cells were stained with APC-conjugated anti-Gr-1 alone, APC-anti-Gr-1 and PE-conjugated Mac-1, APC-conjugated CD4 and PE-conjugated CD8 or the combination of biotinylated anti-CD19 and PE-conjugated anti-Thy-1.2 followed by APC-conjugated streptavidin. The dot plots are gated for live cells based on forward and side scatter profiles. (C) Retroviral integration. EGFP− and EGFP+ fractions of spleen cells were sorted and the purity of each population is shown using EGFP fluorescence and forward scatter. Southern analysis of these samples, where an EGFP probe demonstrates viral integration or a granzyme A probe demonstrates DNA loading, is shown on the right.
Immunophenotype of cells from the peripheral blood and spleen of FLT3-ITD mice.
Spleen (A) and blood (B) cells from mice that received bone marrow transduced with retroviruses encoding EGFP only, wild-type FLT3 and EGFP, or FLT3-ITD (mouse N78.5) and EGFP. Cells were stained with APC-conjugated anti-Gr-1 alone, APC-anti-Gr-1 and PE-conjugated Mac-1, APC-conjugated CD4 and PE-conjugated CD8 or the combination of biotinylated anti-CD19 and PE-conjugated anti-Thy-1.2 followed by APC-conjugated streptavidin. The dot plots are gated for live cells based on forward and side scatter profiles. (C) Retroviral integration. EGFP− and EGFP+ fractions of spleen cells were sorted and the purity of each population is shown using EGFP fluorescence and forward scatter. Southern analysis of these samples, where an EGFP probe demonstrates viral integration or a granzyme A probe demonstrates DNA loading, is shown on the right.
No abnormalities were observed in T-cell (Figure 5A) or B-cell (Figure5A, B) populations. Comparable results were obtained from immunophenotypic analysis of cells from the bone marrow (n = 4), with an increase in Gr-1+ cells from 54% on average in controls to 87% average in FLT3-ITD mice.
Southern blot analysis of spleen, bone marrow, and blood demonstrated proviral integration in all affected tissues. Data from the spleen are shown in Figure 6, where the DNA was digested to excise the proviral DNA as a single band (3 kb for MSCV-EGFP or 6 kb for the FLT3 series in MSCV-EGFP) or cleaved once within the construct and randomly in the flanking DNA, according to the insertion position, which demonstrated the clonality. DNA integrity and equal loading was demonstrated by reprobing the blot for an endogenous gene, granzyme A (Figure 6, lower panel). Southern blot analysis of control mice sacrificed at the same time confirmed transduction of bone marrow cells with FLT3-ITD and MSCV-EGFP, although these animals did not develop disease. Restriction digests designed to determine clonality indicated that the myeloproliferative disease induced by the FLT3-ITD was oligoclonal (Figure 6, upper panel).
Southern analysis of proviral integration in the spleen cells of animals that received transplants of wild-type FLT3 and FLT3-ITD.
For each animal analyzed 2 digests were performed; AnXhoI digest (X) cleaves in the retroviral long terminal repeats (LTRs) releasing a fragment corresponding to the intact retrovirus and an EcoRI (E) digest cleaves within the proviral sequence and in the flanking genomic sequence allowing the clonality of the disease to be estimated. The mouse identity number for each construct is shown above the enzyme digest. The upper panel shows hybridization with a probe for EGFP. In the lower panel hybridization of the same blot with a probe for granzyme A exon I and II. The size of molecular weight marker bands is indicated on the left of each panel.
Southern analysis of proviral integration in the spleen cells of animals that received transplants of wild-type FLT3 and FLT3-ITD.
For each animal analyzed 2 digests were performed; AnXhoI digest (X) cleaves in the retroviral long terminal repeats (LTRs) releasing a fragment corresponding to the intact retrovirus and an EcoRI (E) digest cleaves within the proviral sequence and in the flanking genomic sequence allowing the clonality of the disease to be estimated. The mouse identity number for each construct is shown above the enzyme digest. The upper panel shows hybridization with a probe for EGFP. In the lower panel hybridization of the same blot with a probe for granzyme A exon I and II. The size of molecular weight marker bands is indicated on the left of each panel.
Discussion
This is the first demonstration that expression of FLT3-ITD, but not wild-type FLT3, has transforming properties in primary bone marrow cells. FLT3-ITD expression causes a myeloproliferative phenotype characterized by leukocytosis and comprised mainly of mature neutrophils. Splenomegaly with extramedullary hematopoiesis in the spleen and the liver are also consistently observed. Myeloproliferative disease induced by FLT3-ITD has a latency of 40 to 60 days and is oligoclonal.
These findings demonstrate that FLT3-ITD is sufficient to induce a myeloproliferative disease. However, although the FLT3-ITD is primarily associated with AML in humans, it is not sufficient to induce an AML phenotype in the murine BMT assay. These data suggest that FLT3-ITDs may require additional cooperating mutations to generate the AML phenotype. Evidence supporting this hypothesis includes clinical observations that FLT3-ITD may coexist with virtually any known chromosomal translocation associated with AML, including the t(8;21), inv(16), and t(15;17) associated with the AML1/ETO, CBFβ/MHY11, and PML/RARα fusion proteins, respectively14 (S. Schnittger, abstract 3569, American Society of Hematology, 2000). Furthermore, several laboratories have convincingly demonstrated that each of these latter fusions alone is not sufficient to cause AML, but require secondary mutations.51-55 Taken together, these data support a 2-hit model of AML in which one mutation, such as FLT3-ITD, serves primarily to provide proliferative and survival signals to cells, with minimal effects on differentiation. A second mutation, such as theAML1/ETO gene rearrangement, which may confer subtle proliferative or survival advantage, serves primarily to impair hematopoietic differentiation. The combination of these mutations may generate the AML phenotype due to the unregulated proliferation of hematopoietic cells with impaired differentiation.
Initial analysis suggests that FLT3 and FLT3-ITD use the same signal transduction pathways. On stimulation with FL, FLT3 activates signal transduction pathways through engagement of SH2-containing proteins, including PLCγ, RAS-GTPase activating protein, the p85 subunit of PI3K, SHC, GRB2, STAT5, VAV, FYN, and SRC, among others.17,18,56,57 The consequence of the ITD, regardless of length, appears to be a constitutive increase in FLT3 tyrosine autophosphorylation. Expression of various FLT3-ITD mutants in the murine 32D and Ba/F3 hematopoietic cell lines demonstrates this autophosphorylation, activation of known downstream effectors of FLT3 activation, including PI3K, STAT5, and transformation to growth factor independence.16-18,58 Wild-type FLT3 is expressed at high levels in blast cells of more than 75% of AML patients,59and is also expressed in a significant proportion of patients with ALL and in leukemia-lymphoma cell lines.11,16,60 The physiologic consequences of FLT3 activation by the ITD differ from activation of wild-type FLT3 by its ligand. The wild-type receptor, when overexpressed in Ba/F3 and 32D cells, may be weakly tyrosine phosphorylated and may activate downstream effectors, but it does not confer factor independent growth. Furthermore, stimulation of FLT3-overexpressing cells with FL is also not sufficient to confer IL-3–independent growth.17,61 In contrast each of the FLT3 mutants that we and others have tested confer factor-independent growth to Ba/F3 cells. It has also been suggested that FLT3-ITD but not wild-type FLT3 activates STAT5 and could be responsible for its constitutive activation in AML samples analyzed.17 62Furthermore, in the murine BMT assay, FLT3-ITD but not wild-type FLT3, induces a myeloproliferative phenotype. These data indicate that there are qualitative or quantitative differences in signal transduction mediated by FLT-ITD compared to wild-type FLT3.
The mechanism by which the ITD activates the FLT3 tyrosine kinase is not understood. However, insertion of the duplicated residues may disrupt a kinase inhibitory domain, resulting in kinase activation. Several lines of evidence support this hypothesis. First, FLT3-ITD in-frame mutations in patients with AML may range in size from several to more than 40 amino acids and vary in position within the juxtamembrane domain. In addition, it has recently been demonstrated that deletion of several amino acid residues in the juxtamembrane domain of FLT3 also results in constitutive activation of FLT3.58 The observation that a spectrum of insertions or deletions in the JM domain serve to activate the kinase is most consistent with loss of function of a kinase inhibitory domain through disruption of the tertiary structure. Second, in-frame deletions in the juxtamembrane domain of c-KIT, a related type III receptor tyrosine kinase, results in constitutive activation of c-KIT in gastrointestinal stromal cell tumors and mastocytoses.63,64 Third, regulation of the FLT1 receptor tyrosine kinase through an autoinhibitory function of the JM domain has recently been reported.65 Collectively, these data support the hypothesis that disruption of an autoinhibitory motif in the JM domain, either by deletion or insertion, results constitutive activation of the FLT3 kinase. Structural analysis of FLT3 and related mutants will be required to confirm this hypothesis. An additional question regarding the mechanism of FLT3-ITD transformation relates to the involvement of the wild-type FLT3 in modulating the signal transduction by the mutant. It is plausible that wild-type FLT3 could potentiate or impair signal transduction by FLT3-ITD.
The data from the FLT3-ITD murine BMT assay are consistent with previously reported data on the role of FL and FLT3 receptor in hematopoiesis. FL synergizes with other growth factors in colony assays to stimulate proliferation of hematopoietic progenitors and myeloid precursors but not erythroid precursors.66,67 Furthermore, mice that do not express the FLT3 receptor have reduced B lymphopoiesis.68 On transplantation, reduced T lymphopoiesis and myelopoiesis are also observed.68 Mice that lack FL have reduced numbers of myeloid and B-lymphoid progenitors and a deficiency in natural killer and dendritic cells.69In addition, mice treated with exogenous FL accumulate progenitor cells in the peripheral blood.70 Overexpression of FL in a BMT assay induces a proliferative phenotype71 and can predispose animals to leukemia after a 5-month latency period where the malignant cells were all shown to express FLT3.72 Taken together these data suggest that the FLT3-FL signaling pathway has a unique role in expansion of stem and progenitor cells and sustained activation of FLT3 can lead to hyperproliferation of cells expressing the FLT3 receptor.
Our findings confirm the role of FLT3 in cellular proliferation and support the hypothesis that FLT3-ITD mutations contribute to the pathogenesis of human leukemias. Each independent FLT3-ITD mutant tested was sufficient to induce a myeloproliferative disease in the murine BMT assay, a phenotype similar to that generated by the activated tyrosine kinases that result from chromosomal translocations.21-38,40 41 These data suggest that it may be worthwhile to investigate other myeloproliferative syndromes, such as myeloid metaplasia with myelofibrosis, polycythemia vera, and essential thrombocythemia, for the presence of activating mutations in FLT3.
Finally, the recently reported successes in the therapy of chronic myelogenous leukemias associated with the BCR/ABL gene rearrangement with the ABL-specific kinase inhibitor STI57173,74 suggest that small molecule inhibitors of FLT3 may have therapeutic efficacy in AML.75 This murine model should also allow pharmacologic and therapeutic properties of small molecule inhibitors to be tested in vivo.
We thank L. Seaton for administrative assistance. We thank J. Daley and S. Lazo for expert cell sorting, and D. Cain and S. Amaral for technical support. We also thank E. Anastasiadou, A. Dash, D. Sternberg, and other members of the Gilliland and Tenen laboratories for valuable discussions.
Supported in part by National Institutes of Health grants CA66996 and DK50654. L.M.K. is an associate and D.G.G. is an associate investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
D. Gary Gilliland, Harvard Institutes of Medicine, 4 Blackfan Cir, Rm 418, Boston, MA 02115; e-mail:gilliland@calvin.bwh.harvard.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal