The AF10 gene encodes a putative transcription factor containing an N-terminal LAP/PHD zinc finger motif, a functional nuclear localization signal, an AT-hook domain, and a leucine zipper toward the C-terminus. AF10 is involved in 2 distinct chromosomal translocations associated with hematologic malignancy. The chimeric fusion proteins MLL/AF10 and CALM/AF10, resulting from the t(10;11)(p12;q23) and the t(10;11)(p12;q14), respectively, consistently retain the leucine zipper motif of AF10. This part of the C-terminal region was used as bait in a yeast 2 hybrid screening of a testis complementary DNA library. The leucine zipper interacted with GAS41, a protein previously identified as the product of an amplified gene in a glioblastoma. GAS41 shows significant homology to theSaccharomyces cerevisiae protein ANC1 and to the human MLL fusion partners AF9 and ENL. The interaction was confirmed in vivo. Furthermore, the study showed by coimmunoprecipitation that GAS41 interacts with INI1 (Integrase Interactor 1) and that INI1 was present in the AF10 immunoprecipitate. INI1 is the human homologue of the yeast SNF5 protein, a component of the SWI/SNF complex, which acts to remodel chromatin and to modulate transcription. The retention of the leucine zipper in the MLL and CALM fusions suggests that a key feature of these chimeric proteins may be their ability to interfere in normal gene regulation through interaction with the adenosine triphosphate–dependent chromatinremodeling complexes.
Introduction
The disruption of the human homologue of theDrosophila Trithorax (trx) gene, MLL on 11q231-4 by chromosomal translocations is a frequent event in human acute leukemia. These translocations, leading to the juxtaposition of genetic elements and formation of MLLfusion genes, occur in approximately 5% to 10% of human acute leukemias, but with a higher frequency in infant leukemias5 and secondary leukemias.6 Although the full biological function of MLL is uncertain, it is known to act as a positive regulator of HOX gene expression in development.7,8 Currently, 20 different translocations affecting the MLL gene have been molecularly cloned and the partner genes identified.9 Significantly, all such translocations result in in-frame fusions at the messenger RNA level, and, therefore, the rearrangements result in the production of chimeric proteins in which the N-terminus of MLL is consistently fused to the C-terminus encoded by the partner gene.10-12Because such a diverse group of proteins can be fused to MLL, the role of the fusion proteins in leukemogenesis remains obscure.13 Although most of the fusion partners are structurally and functionally unrelated to each other,14 a number are involved in transcriptional regulation. For example, ENL, AF9, and AF4 activate transcription from synthetic reporter genes in vivo.15-20 The AF10 gene is one of the fewMLL partner genes to be independently rearranged with a third gene in leukemia, the CALM gene in the t(10;11)(p12;q14) translocation.21,AF10complementary DNA (cDNA) encodes a 1084-aa protein, a member of a family of proteins including MLL, all carrying a conserved LAP/PHD finger domain.22,23 There is also a putative AT-hook motif,24 a bipartite nuclear localization signal, a leucine zipper domain, and a glutamine-rich region at the C-terminus. The latter is not present in all isoforms, as a result of alternative splicing.25,26 Although different breakpoints have been described for AF10, the resultant fusion protein in both products consistently loses the LAP/PHD finger but retains the putative leucine zipper region. The leucine zipper motif of AF10 along with its immediate upstream region was found to be homologous to the equivalent region in AF1727,28 and to be conserved among other species (Figure 1A). To gain an insight into the potential role of this motif in leukemogenesis, we have investigated its potential protein interactions. We have established that the leucine zipper interacts both in vitro and in vivo with GAS41, previously identified as the product of an amplified gene in a glioblastoma.29 GAS41 shows significant homology to the human AF9 and ENL proteins and to the ANC1 protein in yeast.30 Furthermore, we have shown that GAS41 interacts with INI1 (Integrase Interactor 1) the human homologue of the yeast SNF5, a component of the SWI/SNF complex,31-33 and INI1 was detected in the AF10 immunoprecipitate. The evolutionarily conserved SWI/SNF complex is one of several multiprotein complexes that modulate transcription by remodeling chromatin in an adenosine triphosphate–dependent manner.
Cluster analysis of regions of homology.
Amino acids matching the consensus are shaded black. (A) Comparison of the leucine zipper regions of human AF10 (Accession No. P55197), human AF17 (Accession No. P55198), dAF10 (Drosophila)(Accession No. AAF54065), and CEZF (C elegans)(Accession No. AAK26137). (B) Comparison of the conserved N-terminal region of GAS41 with other related proteins. The following sequences were used: GAS41 (10-121 of AAD121188), AF9 (1-107 of Accession No.P42568), ENL (1-107 of Accession No. Q03111), M04B2.3 (1-113 of Accession No. T23696), ANC1 (1-112 of Accession No. P35189), YNK7 (1-120 of Accession No. P53930), YD67 (1-118 of Accession No. Q10319), and SPAC22H12.02 (1-115 of Accession No. AL034565).
Cluster analysis of regions of homology.
Amino acids matching the consensus are shaded black. (A) Comparison of the leucine zipper regions of human AF10 (Accession No. P55197), human AF17 (Accession No. P55198), dAF10 (Drosophila)(Accession No. AAF54065), and CEZF (C elegans)(Accession No. AAK26137). (B) Comparison of the conserved N-terminal region of GAS41 with other related proteins. The following sequences were used: GAS41 (10-121 of AAD121188), AF9 (1-107 of Accession No.P42568), ENL (1-107 of Accession No. Q03111), M04B2.3 (1-113 of Accession No. T23696), ANC1 (1-112 of Accession No. P35189), YNK7 (1-120 of Accession No. P53930), YD67 (1-118 of Accession No. Q10319), and SPAC22H12.02 (1-115 of Accession No. AL034565).
Materials and methods
Yeast 2 hybrid screening
The AF10 bait (aa 683-972),34 was obtained by reverse transcriptase-polymerase chain reaction (RT-PCR) from placental messenger RNA and cloned in-frame with the Gal4 DNA-binding domain into the pBD-Gal4-Cam vector (Stratagene, La Jolla, CA). The PJ69-4A yeast strain was simultaneously transformed with the bait plasmid pBD-Gal4-AF10 and a human testis cDNA library cloned into pAD-Gal4 vector, according to the manufacturers' instructions (Hybri-ZAP 2-Hybrid cDNA Gigapack Cloning Kit; Stratagene). Positive interactions were tested for their Lac Z activity. DNA from positive clones was extracted and sequenced with 2 pAD-Gal4–specific oligonucleotides, AD-for (CTGTCACCTGGTTGGACGGACCAA) and pGAD-rev (GTGAACTTGCGGGGTTTTTCAG). DNA and protein databases were searched by using the BLAST package of search algorithms (http://ncbi.nlm.nih.gov/cgi.bin/BLAST). The interaction strength between the potential partner protein GAS41 and multiple AF10 constructs was assessed for β-gal activity and for growth on fully selective medium. As negative control, the pBD-Gal4 vector fused to Lamin C was used.
In vitro glutathione-S-transferase pull-down assay
Cloned glutathione-S-transferase (GST) fusion cDNAs (pGEX-2T vector; Pharmacia, Uppsala, Sweden) were expressed in a 200-mL culture of the Escherichia coli strain Dh5α as GST fusion proteins after 3 hours of induction with 0.1 mM isopropyl thiogalactoside. Cells were harvested, resuspended in 1× phosphate-buffered saline (PBS; 80 mM Na2HPO4, 20 mM NaH2PO4, 100 mM NaCl, pH 7.5), and lysed by mild sonication. The cleared lysate was immobilized on affinity Glutathione-Sepharose beads. The cDNAs for the partner proteins were cloned in the pCR 2.1 vector (Invitrogen, Carlsbad, CA), and the proteins were synthesized and radiolabeled with 20 μCi (0.74 MBq) 35S-Met (TNT Quick Coupled Transcription/Translation System Kit; Promega, Madison, WI) in a coupled transcription-translation system. About 25 μg of each immobilized GST chimeric fusion protein was incubated at 4°C for 2 hours with one fifth of the labeled polypeptides in binding buffer (20 mM Tris-HCl pH 8.0, 0.2% Triton X-100, 2 mM EDTA pH 8.0, 300 mM NaCl, 1 μg/mL aprotinin, 1 μg/mL leupeptin, and 1 mM phenyl-methyl sulfonyl fluoride [PMSF]). After extensive washing in RIPA buffer (10 mM Tris-HCl pH 7.5, 0.2% NP-40, 1 mM EDTA pH 8.0, 400 mM NaCl, 1 μg/mL aprotinin, 1 μg/mL leupeptin, and 1 mM PMSF) bound proteins were eluted by boiling in 2× sodium dodecyl sulfate (SDS) sample buffer (125 mM Tris-HCl pH 6.8, 20% glycerol, 4% SDS, 0.05% bromophenol blue, and 10% β-mercaptoethanol) and analyzed on a 10% or 12% SDS polyacrylamide gel. The gels were stained with Coomassie blue to ensure the loading of equal amounts of GST fusion protein. Dried gels were scanned on a Phospho-Imager (STORM840; Molecular Dynamics, Sunnyvale, CA).
Cell culture
KG1a cells35 were grown in RPMI 1640 medium containing 1% penicillin/streptomycin and 1% glutamine and were supplemented with 10% heat-inactivated fetal calf serum. Cells were grown at 37°C in a humidified incubator (MK11 Leec, Nottingham, United Kingdom) supplemented with 5% carbon dioxide.
Protein extraction
Approximately 5 million cells were pelleted at 1400 rpm for 10 minutes and washed 3 times in ice-cold 1× PBS. The proteins were extracted with ice-cold lysis buffer (60 mM Tris-HCl pH 7.5, 5 mM EDTA pH 8.0, 150 mM NaCl, 1.0% Triton X-100, 10% glycerol, 5 μg/mL aprotinin, 5 μg/mL leupeptin, 1 mM Na3VO4, 1 μg/mL pepstatin, and 1 mM PMSF). The lysate was kept on ice for 20 minutes, then microfuged at 14 000 rpm for 5 minutes. Protein concentrations were measured against a calibration curve by using bovine serum albumin (BSA) as a reference in a Bradford assay (Bio-Rad, Hercules, CA). Aliquots containing 30 μg protein lysate were mixed with an equal volume of 2× SDS sample buffer, boiled for 4 minutes, and stored at −20°C.
Immunoprecipitation
Immunoprecipitation assays were performed with KG1a cells. Thirty million cells were lysed in 1.3 mL lysis buffer (60 mM Tris-HCl pH 7.5, 5 mM EDTA pH 8.0, 150 mM NaCl, 1% Triton X-100, 10% glycerol, 5 μg/mL aprotinin, 5 μg/mL leupeptin, 1 mM Na3VO4, 1 μg/mL pepstatin, and 1 mM PMSF) for 20 minutes in ice. Whole cell extracts were clarified by centrifugation and quantified. For each immunoprecipitation, 1 mg total lysate was diluted in 1 mL lysis buffer and incubated for 2 hours at 4°C with 50 μL precleared Protein A Sepharose CL-4B (Pharmacia). The supernatant was then incubated at 4°C overnight either with 10 μg primary antibody or preimmune serum and further incubated with 50 μL fresh Protein A for 1 hour at 4°C with agitation. The precipitate was washed 3 times in buffer A (1× PBS, 1% NP-40, 100 μM Na3VO4), twice in buffer B (100 mM Tris-HCl pH 7.4, 5 mM LiCl, 100 μM Na3VO4), and twice in buffer C (100 mM Tris-HCl pH 7.4, 5 mM EDTA pH 8.0, 5 mM NaCl, 100 μM Na3VO4). The pellet was resuspended in 2× SDS sample buffer, boiled, and analyzed on SDS polyacrylamide gel for coimmunoprecipitated proteins.
Immunoblot analysis
Samples were electrophoresed on an SDS polyacrylamide gel in a Mighty Small miniature slab gel unit (Hoefer, Uppsala, Sweden) in running buffer (25 mM Tris-HCl, 192 mM glycine, and 0.1% SDS) and blotted onto a polyvinylidene diflouride membrane (Immobilon-P; Millipore, Bedford, MA) at 300 mA for 2.5 hours at 4°C by using a Bio-Rad Mini Trans-Blot Cell, containing transfer buffer (25 mM Tris/HCl, 192 mM glycine, and 20% methanol). The filters were blocked (3% nonfat dried milk, 0.1% Tween 20 in 1× Tris-buffered saline: 20 mM Tris-HCl pH 7.6, 137 mM NaCl) for 1 hour at room temperature and then incubated with the primary antibodies diluted in blocking solution, with gentle agitation for 2 hours at room temperature. Membranes were washed 5 times with 0.1% Tween 20 in 1× Tris-buffered saline and then incubated with the diluted horseradish peroxidase (HRP)–conjugated secondary antibodies for 1 hour at room temperature. The blots were washed and proteins detected by using the Super Signal West Dura Substrate Working Solution (Pierce, Rockford, IL) according to the manufacturers' instructions.
Immunofluorescence
KG1a cells were washed twice in 1× PBS and finally resuspended at a concentration of 106 cells/mL. A total of 100 mL of this suspension was pipetted onto a cytospin column and spun at 400 rpm for 5 minutes (Cytospin3; Thermo-Shandon, Runcorn, United Kingdom) on poly-L-lysine–coated glass slides. Cells were immediately fixed in 2% paraformaldehyde for 20 minutes and then permeabilized in 0.1% saponin for 10 minutes. For AF10/GAS41 colocalization cells were rinsed in 1× PBS and covered for 1 hour with 200 μL chicken anti-AF10 antibody diluted 100 times in 1× PBS, 1% BSA, and 0.02 M sodium azide. Slides were washed 3 times in 1× PBS and 200 μL rabbit anti-GAS41 antibody, diluted 50 times, and was applied at room temperature for 1 hour. After 3 washes, cells were first incubated with fluorescein isothiocyanate (FITC)–conjugated rabbit antichicken secondary antibody for 45 minutes at room temperature and then with the Cy3-labeled goat antirabbit antibody again for 45 minutes. Slides were washed 3 times and then mounted in 70% glycerol in PBS containing antifade. Staining was visualized by using the Bio-Rad Laser sharp MRC-600 confocal imaging system.
Antibodies
Primary antibodies for immunoblot and immunoprecipitation of AF10, a rabbit polyclonal antibody obtained from rabbit immunized with the first 331 amino acids produced as GST fusion protein (Clare Hall, ICRF)25 was used. The rabbit polyclonal antibody, GAS41-N, against GAS41 was a kind gift of Dr A. Munnia (Department of Human Genetics, University Hospital, Homburg, Germany). Anti INI1, C-18, is a goat polyclonal antibody (Santa Cruz Biotechnologies, Santa Cruz, CA). For the immunofluorescence of AF10, a chicken antibody was used (BMA, Biomedicals, AG, Augst, Switzerland). Secondary antibodies for immunoblot were donkey antirabbit immunoglobulin g (IgG) HRP-conjugated (Amersham, Uppsala, Sweden) and antigoat IgG HRP-conjugated (Santa Cruz, Santa Cruz, CA). Secondary antibodies for immunofluorescence were FITC-conjugated rabbit antichicken IgG (Sigma, St Louis, MO) and fluoroLinkTM CyTM3-labeled goat antirabbit IgG (Amersham).
Results
AF10 interacts with GAS41 in a yeast 2 hybrid screening
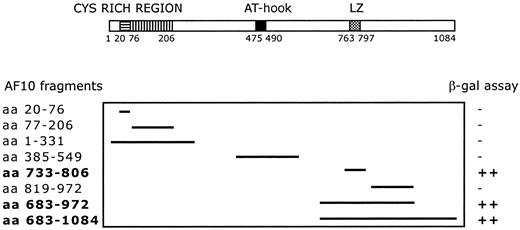
A C-terminal region of AF10 containing the leucine zipper motif (aa 683-972) was used as bait for yeast 2 hybrid screening. The screening of a human testis cDNA library led to the identification of 17 positive clones. Sequence analysis identified 4 independent clones containing the complete open-reading frame (227 amino acids) of theGAS41 gene, linked in-frame to the Gal4 transactivation domain. The GAS41 gene was previously cloned (Glioma Amplified Sequence) from a glioblastoma cell line.29 The first N-terminal 100 amino acids of GAS41 are homologous to AF9 and ENL in humans and to ANC1 and YNK7 in Saccharomyces cerevisiae, YD67 and SPAC22H12.02 in Schizosaccharomyces pombe, and M04B2.3 in Caenorhabditis elegans (Figure 1B). The remainder of GAS41 consists of a C-terminal coiled-coil domain. To determine the specificity of the AF10/GAS41 interaction, we generated a series of deletions encoding portions of AF10, in-frame with the binding domain of Gal4. Yeast plated on selective media was able to grow only when the leucine zipper region was present in the expressed proteins, following cotransformation with the AF10 constructs and the pAD-Gal4 vector expressing GAS41. The shortest active segment identified contained the leucine zipper motif alone (aa 733-806) (Figure2).
Deletion mapping of interacting fragments.
Diagram shows the 8 AF10 deletion fragments tested with the yeast 2 hybrid system. β-Galactosidase assay was used to determine the minimal interaction domain of AF10 with the potential partner protein GAS41. ++, strong interaction; −, no interaction.
Deletion mapping of interacting fragments.
Diagram shows the 8 AF10 deletion fragments tested with the yeast 2 hybrid system. β-Galactosidase assay was used to determine the minimal interaction domain of AF10 with the potential partner protein GAS41. ++, strong interaction; −, no interaction.
The leucine zipper motif of AF10 interacts with GAS41 in a GST pull-down experiment
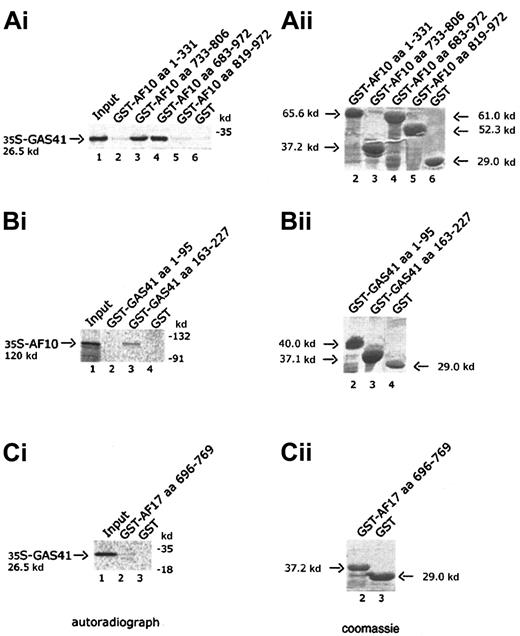
Four AF10 constructs, one containing the cysteine-rich region (aa 1-331), 2 containing the leucine zipper motif (aa 733-806 and aa 683-972), and the fourth containing the C-terminal portion of AF10 (aa 819-972), were expressed in bacteria as GST fusion products and bound to Glutathione-Sepharose beads. Binding of radiolabeled GAS41 protein was examined against the 4 GST chimeric polypeptides and GST alone. Only the constructs containing the leucine zipper motif were capable of binding to GAS41 protein (Figure 3A, lanes 3 and 4). This result demonstrated that the leucine zipper motif of AF10 was essential for the in vitro binding of AF10 to GAS41. Two deleted constructs of GAS41 (aa 1-95 and aa 163-227) were fused to GST and used to map the region of interaction with AF10. In vitro–translated and –radiolabeled AF10 bound the C-terminal coiled-coil region of GAS41 fused to GST (Figure 3B, lane 3) but not the N-terminal ENL/AF9-like domain (Figure 3B, lane 2). The leucine zipper of AF17 (aa 696-769) was expressed as a GST fusion protein and incubated with the radiolabeled in vitro–translated GAS41 protein. AF17 leucine zipper was unable to mediate the binding to GAS41 (Figure3C, lane 2), suggesting that GAS41 may have specificity for AF10.
In vitro interaction of AF10 and GAS41.
(Ai) Autoradiograph of a 12% polyacrylamide gel with in vitro–translated GAS41 protein pulled down with 4 different GST-AF10 fusion polypeptides. Lane 1, input of in vitro–translated GAS41 protein (one fifth of total amount); lane 2, GST-AF10–cysteine-rich region (aa 1-331); lane 3, GST-AF10–leucine zipper motif (aa 733-806); GST-AF10–leucine zipper region (aa 683-972); lane 5, GST-AF10–C-terminal portion (aa 819-972); and lane 6, GST as negative control. (Bi) Autoradiograph of a 10% polyacrylamide gel with in vitro–translated AF10 protein precipitated with 2 different GST-GAS41 fusion polypeptides. Lane 1, input of in vitro–translated AF10 protein (one fifth of total amount); lane 2, GST-GAS41–AF9-like region (aa 1-95); lane 3, GST-GAS41–coil-coiled region (aa 163-227); and lane 4, GST as negative control. (Ci) Autoradiograph of a 12% polyacrylamide gel with in vitro–translated GAS41 (lane 1, input of in vitro–translated protein), precipitated with the leucine zipper motif of AF17 (aa 696-769), lane 2; and with GST in lane 3 as negative control. (Aii,Bii,Cii) The right panels represent the equivalent Coomassie blue–stained gel of the pull-down experiment.
In vitro interaction of AF10 and GAS41.
(Ai) Autoradiograph of a 12% polyacrylamide gel with in vitro–translated GAS41 protein pulled down with 4 different GST-AF10 fusion polypeptides. Lane 1, input of in vitro–translated GAS41 protein (one fifth of total amount); lane 2, GST-AF10–cysteine-rich region (aa 1-331); lane 3, GST-AF10–leucine zipper motif (aa 733-806); GST-AF10–leucine zipper region (aa 683-972); lane 5, GST-AF10–C-terminal portion (aa 819-972); and lane 6, GST as negative control. (Bi) Autoradiograph of a 10% polyacrylamide gel with in vitro–translated AF10 protein precipitated with 2 different GST-GAS41 fusion polypeptides. Lane 1, input of in vitro–translated AF10 protein (one fifth of total amount); lane 2, GST-GAS41–AF9-like region (aa 1-95); lane 3, GST-GAS41–coil-coiled region (aa 163-227); and lane 4, GST as negative control. (Ci) Autoradiograph of a 12% polyacrylamide gel with in vitro–translated GAS41 (lane 1, input of in vitro–translated protein), precipitated with the leucine zipper motif of AF17 (aa 696-769), lane 2; and with GST in lane 3 as negative control. (Aii,Bii,Cii) The right panels represent the equivalent Coomassie blue–stained gel of the pull-down experiment.
AF10 and GAS41 interact in vivo: coimmunoprecipitation of the endogenous proteins
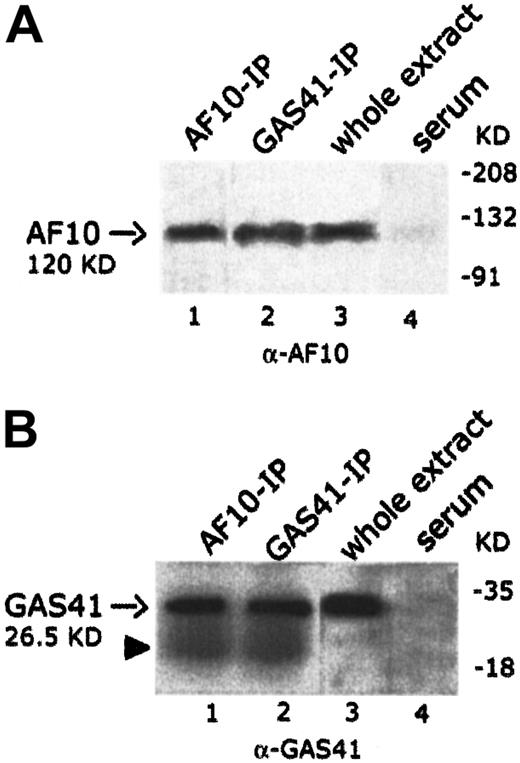
To establish whether endogenous AF10 and GAS41 interacted in vivo, the 2 proteins were independently immunoprecipitated from KG1a cell line with polyclonal antibodies to AF10 and to GAS41 proteins, respectively. Both immunoprecipitates were loaded on separate denaturing polyacrylamide gels and immunoblotted with the 2 antibodies (rabbit polyclonal anti-AF10 and rabbit polyclonal anti-GAS41). An immunoblot on total lysate was performed to test the antibodies (Figure4A, lane 3, and 4B, lane 3). In the AF10 immunoprecipitate, the rabbit antibody to GAS41 identified a single band of the size expected for the endogenous GAS41 (26.5 KDa) (Figure4B, lane 1), whereas the rabbit antibody to AF10 detected, in the GAS41 immunoprecipitate, a band of about 120 KDa, the size predicted for the AF10 protein (Figure 4A, lane 2). These experiments suggest a direct physical interaction in vivo between GAS41 and AF10.
In vivo interaction of AF10 and GAS41.
(A) Immunoblot analysis with a rabbit polyclonal anti-AF10 antibody of the endogenous AF10 and GAS41 immunoprecipitates from whole extract of KG1a cells (lanes 1 and 2, respectively); 30 μg protein extract (3% of that used for immunoprecipitations) was used as antibody positive control (lane 3). Immunoprecipitates with rabbit preimmune serum was used as negative control (lane 4). Samples were run on an 8% polyacrylamide gel. (B) Immunoblot analysis with a rabbit polyclonal anti-GAS41 antibody of AF10 and GAS41 immunoprecipitates from whole extract of KG1a cells (lanes 1 and 2, respectively); 30 μg protein extract was used as antibody positive control (lane 3). Immunoprecipitates with rabbit preimmune serum were used as negative control (lane 4). The closed arrowhead indicates the presence of the light chain (22 KD) of the rabbit antibody. Samples were run on a 15% polyacrylamide gel.
In vivo interaction of AF10 and GAS41.
(A) Immunoblot analysis with a rabbit polyclonal anti-AF10 antibody of the endogenous AF10 and GAS41 immunoprecipitates from whole extract of KG1a cells (lanes 1 and 2, respectively); 30 μg protein extract (3% of that used for immunoprecipitations) was used as antibody positive control (lane 3). Immunoprecipitates with rabbit preimmune serum was used as negative control (lane 4). Samples were run on an 8% polyacrylamide gel. (B) Immunoblot analysis with a rabbit polyclonal anti-GAS41 antibody of AF10 and GAS41 immunoprecipitates from whole extract of KG1a cells (lanes 1 and 2, respectively); 30 μg protein extract was used as antibody positive control (lane 3). Immunoprecipitates with rabbit preimmune serum were used as negative control (lane 4). The closed arrowhead indicates the presence of the light chain (22 KD) of the rabbit antibody. Samples were run on a 15% polyacrylamide gel.
Cytoplasmic and nuclear colocalization of AF10 and GAS41
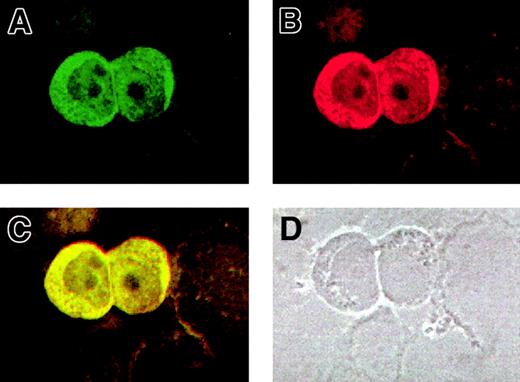
For a further validation of the AF10/GAS41 interaction, we determined the subcellular localization of both proteins by using immunofluorescence assays in KG1a cells. The cells were first alternatively incubated with the 2 primary antibodies and then with a FITC-conjugated secondary antibody to detect AF10 (Figure5A) and with a Cy3-labeled secondary antibody for GAS41 (Figure 5B). The images obtained at the confocal microscope showed for both the proteins a cytoplasmic localization, although a lighter signal was detected in the nucleus. After overlay, a mixed (yellow) color was observed for all signals (Figure 5C), which is indicative of colocalization. A phase contrast image of the same slides (Figure 5D) indicated the subcellular compartments. Debris of cells lightly stained was visible in the background.
Colocalization of AF10 and GAS 41 endogenous proteins in KG1a cells by immunofluorescence.
(A) Subcellular localization of AF10, detected with a FITC-conjugated secondary antibody. (B) Subcellular localization of GAS41, detected with a Cy3-labeled secondary antibody. (C). Overlay of the previous images showing colocalization of the 2 proteins, both in the cytoplasm and in the nucleus. (D) Phase contrast image of the same field, showing the presence of cell debris.
Colocalization of AF10 and GAS 41 endogenous proteins in KG1a cells by immunofluorescence.
(A) Subcellular localization of AF10, detected with a FITC-conjugated secondary antibody. (B) Subcellular localization of GAS41, detected with a Cy3-labeled secondary antibody. (C). Overlay of the previous images showing colocalization of the 2 proteins, both in the cytoplasm and in the nucleus. (D) Phase contrast image of the same field, showing the presence of cell debris.
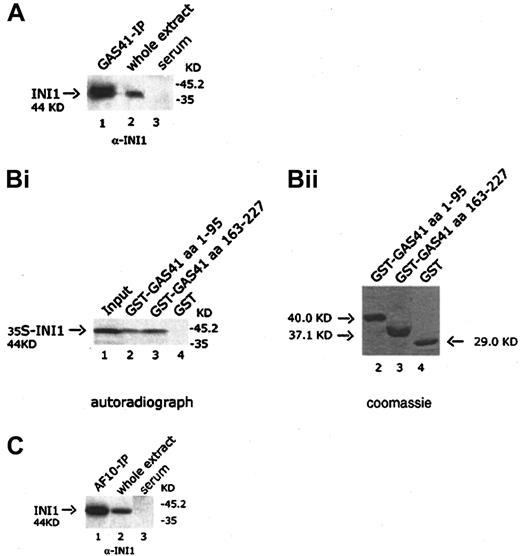
GAS41 interacts with INI1 both in vivo and in vitro
It has been previously shown in S cerevisiae that the ANC1 protein interacted with SNF5, a protein component of the SWI/SNF complex.30 Because GAS41 appears to be the human homologue of ANC1, we investigated the possible interaction with INI1, the human homologue of SNF5. The GAS41 immunoprecipitate was immunoblotted with a goat anti-INI1 antibody that detected a 44-KDa band corresponding to the expected size of INI1, suggesting an association of GAS41 with INI1 in vivo (Figure 6A, lane 1). The interaction was mapped by using 2 GST-deleted constructs of GAS41 (aa 1-95 and aa 163-227) expressed in bacteria, both of which pulled down in vitro–translated INI1 (Figure 6B, lanes 2 and 3). The results of the experiments shown above prompted us to investigate whether a similar association could be shown between AF10 and INI1. An AF10 immunoprecipitate was immunoblotted with the anti-INI1 antibody. The antibody identified a protein of 44 KDa, the expected size for INI1 (Figure 6C, lane 1). Further experiments were performed to investigate whether this interaction was direct. In a yeast 2 hybrid experiment, yeast plated on selective media was not able to grow when cotransformed with the leucine zipper region (aa 683-972) of AF10 fused in-frame with the DNA-binding domain of Gal4 and the pAD-Gal4 vector expressing INI1 (data not shown). We therefore concluded that AF10, GAS41, and INI1 may be present in the same complex but without a direct interaction between AF10 and INI1.
In vivo and in vitro interaction of GAS41 and INI1.
(A) Immunoblot analysis with a goat polyclonal anti-INI1 antibody of GAS41 protein immunoprecipitated from whole extract of KG1a cells (lane 1); 30 μg protein extract was used as antibody positive control (lane 2). Immunoprecipitates with rabbit preimmune serum were used as negative control (lane 3). Samples were run on a 12% polyacrylamide gel. (Bi) Autoradiograph of a 12% polyacrylamide gel with in vitro–translated INI1 protein precipitated with 2 different GST-GAS41 fusion polypeptides. Lane 1, input of in vitro–translated INI1 protein (one fifth of total amount); lane 2, GST-GAS41–AF9-like region (aa 1-95); lane 3, GST-GAS41–coil-coiled region (aa 163-227); and lane 4, GST as negative control. (Bii) The equivalent Coomassie blue–stained gel of the pull down experiment. (C) Immunoblot analysis with a goat polyclonal anti-INI1 antibody of AF10 protein immunoprecipitated from whole extract of KG1a cells (lane 1). Protein extract (30 μg) was used as antibody positive control (lane 2). Immunoprecipitates with rabbit preimmune serum were used as negative control (lane 3). Samples were run on a 12% polyacrylamide gel.
In vivo and in vitro interaction of GAS41 and INI1.
(A) Immunoblot analysis with a goat polyclonal anti-INI1 antibody of GAS41 protein immunoprecipitated from whole extract of KG1a cells (lane 1); 30 μg protein extract was used as antibody positive control (lane 2). Immunoprecipitates with rabbit preimmune serum were used as negative control (lane 3). Samples were run on a 12% polyacrylamide gel. (Bi) Autoradiograph of a 12% polyacrylamide gel with in vitro–translated INI1 protein precipitated with 2 different GST-GAS41 fusion polypeptides. Lane 1, input of in vitro–translated INI1 protein (one fifth of total amount); lane 2, GST-GAS41–AF9-like region (aa 1-95); lane 3, GST-GAS41–coil-coiled region (aa 163-227); and lane 4, GST as negative control. (Bii) The equivalent Coomassie blue–stained gel of the pull down experiment. (C) Immunoblot analysis with a goat polyclonal anti-INI1 antibody of AF10 protein immunoprecipitated from whole extract of KG1a cells (lane 1). Protein extract (30 μg) was used as antibody positive control (lane 2). Immunoprecipitates with rabbit preimmune serum were used as negative control (lane 3). Samples were run on a 12% polyacrylamide gel.
Discussion
The AF10 gene was first identified as a translocated partner of MLL.34 It has been subsequently found that the AF10 gene is one of the few MLLpartner genes to be independently rearranged, with a third gene,CALM, encoding a clathrin assembly protein.21Although the MLL/AF10 fusions were initially thought to be restricted to the AML M4/M5 subtype, recent evidence36suggests that they are more widely distributed among the AML subtypes. The fusion of AF10 with CALM occurs in a wider spectrum of hematologic malignancies, including acute myeloid and lymphoid leukemias and lymphomas.37-39 A consistent feature of both the MLL/AF10 and CALM/AF10 fusion proteins appears to be the juxtaposition of the leucine zipper motif of AF10 onto the N-terminal region of MLL or CALM.27,40 The fact that in all these cases the leucine zipper region is retained underlines its relevance in leukemic transformation, as has been already demonstrated in murine stem cells retrovirally transduced with MLL/AF10 fusion cDNAs. Constructs containing the leucine zipper were capable of mediating transformation, whereas those lacking this motif were not.41 Because of this observation, a C-terminal fragment of AF10 containing the leucine zipper motif was used as bait in a yeast 2 hybrid screening to identify candidate interacting proteins. Four independent clones were isolated, each containing the full open-reading frame of GAS41, a previously known protein that was identified as the product of a gene amplified in a glioblastoma.29 Deletion constructs of AF10 indicated that the region of interaction with GAS41 in yeast was confined to the leucine zipper motif. We have confirmed this interaction by using in vitro GST pull-down experiments and have furthermore demonstrated that the endogenous AF10 and GAS41 proteins can be coimmunoprecipitated from cell lines.
AF17, together with AF10 and MLL, is a member of the LAP/PHD zinc finger–containing family and is itself involved in a t(11;17)(q23;q21) translocation with MLL.28 The leucine zipper of AF17 has 77% (27 of 35 aa) identity with the leucine zipper motif of AF10 (Figure 1A). The published data suggest that, as in the case of AF10, the leucine zipper of AF17 is consistently retained in the fusion product of the translocation. Thus, there has been selective pressure to maintain the motif, suggesting that it has an important functional role. However, the leucine zipper motif of AF17, when tested in vitro, did not pull down GAS41.
GAS41 appears to be a human homologue of ANC1, a 244-amino acid protein in S cerevisiae known to be an integral member of 2 basal transcription factor complexes, TFIID and TFIIF, and to be an interacting component of the SWI/SNF chromatin-remodeling complex.30 GAS41, ANC1, and the other yeast homologues have a significant similarity (Figure 1B) in their N-terminal region to 2 human proteins involved in fusion with MLL, AF942 and ENL.43 Both AF9 and ENL, when fused to MLL, are known to mediate transformation in mice.15,17,18,44 The biochemical role ANC1 plays in the transcription complexes is not yet clear, but ANC1 is known to bind to the SWI/SNF complex through its interaction with SNF5. Homologues of SNF5 have been isolated in both humans andDrosophila, INI1, and Snr1, respectively. They have been shown to be associated in large complexes equivalent to the yeast SWI/SNF45,46 and to bind the SET domains ofMLL and Trx genes.47 To extend these findings and investigate a putative relation of AF10 with the chromatin-remodeling complexes, we have further shown that GAS41 interacts in vitro and in vivo with INI1. Although a yeast 2 hybrid experiment excluded the direct interaction between AF10 and INI1, the presence of the latter in an AF10 immunoprecipitated, suggested the possibility that the 3 proteins exist in a protein complex.
It has been shown that the AT-hook motif of AF10 mediated the binding to synthetic cruciform DNA.26 The published data suggested that proteins containing AT-hooks, like HMG-I (Y), play an important role in chromatin structure and transcriptional regulation by acting as accessory factors that influence the association of transcription factors with chromatin.48,49 It is plausible that AF10 and GAS41 when bound together recruit the SWI/SNF complex through the interaction with INI1. The AT-hook motif of AF10 could use AT-rich tracks to target specific regions on chromatin. When AF10 is fused to MLL, the AT-hook region of AF10 will not be consistently present in the fusion proteins, whereas the N-terminal AT-hooks of MLL will be retained. Fused MLL lacks the C-terminal SET domain that mediates the interaction with INI1.47 The MLL/AF10 fusion would create a protein, which could still interact with GAS41 and which could still bind DNA, but the interaction with INI1 would be mediated by GAS41, if still possible, leading to the loss of regulation ofMLL target genes, thus compromising its role in development and affecting the expression of AF10 target genes. The interaction described here may therefore play an important role in neoplastic transformation, and further work is required to determine the role of the AF10 fusion proteins in leukemogenesis.
We thank Professor Ad Geurts van Kessel (Department of Human Genetics, University Medical Center St Radbound, Nijmegen, The Netherlands) for gifts of the human testis cDNA library, all the expressing vectors, the oligonucleotides, as well as the yeast strain used in the yeast 2 hybrid screening. We also thank Dr A. Munnia (Department of Human Genetics, Medical School, University of Saar, Homburg/Saar, Germany) for the gift of the rabbit polyclonal antibody GAS41-N. Finally, we thank Dr V. Saha for proofreading the paper.
Supported by the Kay Kendall Leukaemia Fund (S.D.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Bryan D. Young, ICRF Med Oncology Unit, St Bartholomew's Hospital Medical School, Charterhouse Square, London EC1M 6BQ, United Kingdom; e-mail: b.young@icrf.icnet.uk.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal