The effects of colony-stimulating factor 1 (CSF-1), the primary regulator of mononuclear phagocyte production, are thought to be mediated by the CSF-1 receptor (CSF-1R), encoded by the c-fms proto-oncogene. To investigate the in vivo specificity of CSF-1 for the CSF-1R, the mouse Csf1r gene was inactivated. The phenotype ofCsf1−/Csf1r− mice closely resembled the phenotype of CSF-1-nullizygous(Csf1op/Csf1op) mice, including the osteopetrotic, hematopoietic, tissue macrophage, and reproductive phenotypes. Compared with their wild-type littermates, splenic erythroid burst-forming unit and high-proliferative potential colony-forming cell levels in bothCsf1op/Csf1op andCsf1−/Csf1r− mice were significantly elevated, consistent with a negative regulatory role of CSF-1 in erythropoiesis and the maintenance of primitive hematopoietic progenitor cells. The circulating CSF-1 concentration inCsf1r−/Csf1r− mice was elevated 20-fold, in agreement with the previously reported clearance of circulating CSF-1 by CSF-1R–mediated endocytosis and intracellular destruction. Despite their overall similarity, several phenotypic characteristics of theCsf1r−/Csf1r− mice were more severe than those of theCsf1op/Csf1op mice. The results indicate that all of the effects of CSF-1 are mediated via the CSF-1R, but that subtle effects of the CSF-1R could result from its CSF-1–independent activation.
Introduction
Colony-stimulating factor 1 (CSF-1) regulates the survival, proliferation, and differentiation of mononuclear phagocytic cells and is the primary regulator of mononuclear phagocyte production in vivo.1,2 However, CSF-1 also regulates cells of the female reproductive tract and plays an important role in fertility.3,4 The effects of CSF-1 are mediated by a high-affinity receptor tyrosine kinase (CSF-1R)5-8 encoded by the c-fms proto-oncogene.9 The CSF-1R is expressed on primitive multipotent hematopoietic cells,10,11 mononuclear phagocyte progenitor cells,12 monoblasts, promonocytes, monocytes,5,6 tissue macrophages,6,13-15osteoclasts,16 B cells,17,18 smooth muscle cells,19 and neurons.20,21 CSF-1R messenger RNA (mRNA) is expressed in Langerhans cells,22 in the female reproductive tract, in oocytes and embryonic cells of the inner cell mass and trophectoderm,23 in decidual cells,24-26 and in cells of the trophoblast.24,25 The expression of the CSF-1R on primitive hematopoietic cells that are unable to proliferate in vitro in response to CSF-1 alone10,11 but are able to proliferate and differentiate if stimulated with combinations of CSF-1 and other hematopoietic growth factors10,11 27 suggests that CSF-1R is involved in the regulation of more primitive hematopoietic cells than those that form macrophage colonies in vitro in response to CSF-1 alone.
Mice homozygous for the mutationosteopetrotic28 possess an inactivating mutation in the coding region of the CSF-1 gene and are devoid of detectable CSF-1.29,30 TheseCsf1op/Csf1op mice are osteopetrotic because of an early and marked deficiency of osteoclasts28 that spontaneously recovers with age,31,32 probably because of the action of vascular endothelial growth factor.33 However, the phenotype of these mice is pleiotropic.3 They are toothless; have low body weight, low growth rate, and skeletal abnormalities; and are deficient in tissue macrophages.2,28,30,34,35 They have defects in both male and female fertility, neural development, the dermis, and synovial membranes.3 The pleiotropic phenotype of the Csf1op/Csf1op mouse may be due to a reduction in trophic and/or scavenger functions of the tissue macrophages regulated by CSF-1, secondary to the reduction of their concentration in tissues,2 because outside the female reproductive tract the CSF-1R is primarily expressed in mononuclear phagocytes.1,3 However, it is possible that some of these effects may also be due to loss of function of other cells such as neuronal cells and muscle precursors, which have also been reported to express the CSF-1R.20 36
To address the questions of whether CSF-1 activates other receptors besides the CSF-1R and, conversely, whether the CSF-1R mediates the response to ligands other than CSF-1, we have carried out the targeted inactivation of the CSF-1R gene. The present study describes the phenotype of mice homozygous for a targeted CSF-1R–null mutation and compares it with the phenotype ofCsf1op/Csf1op mice. The slightly more severe phenotype ofCsf1r−/Csf1r− overCsf1op/Csf1op mice indicates that the effects of CSF-1 are uniquely mediated through the CSF-1R and that subtle effects of the CSF-1R could result from its CSF-1–independent activation.
Materials and methods
Construction of the Csf1r gene-targeting vector
Mouse Csf1r 5′ promoter region and exon 2 region primers included cfms-75F, 5′-GGC ACG GGG CTC CCA GCT GCT AGT TCT GTG-3′, and cfms-497R, 5′-AAG GGC AGA TGA GAA AGG TAT GAA GAA TGT-3′; and mouse Csf1r 3′ terminal region and exon 22 sequence primers included cfms-3024F, 5′-CCT CAG CTT GGC CCG ACT CTG ACA ATT CAG-3′, and cfms-3360R, 5′-AGT GAA GGT CAA GAG TGG TGG CCA ATA ATG-3′. These primers were synthesized and submitted to Genome Systems (St Louis, MO) to screen a 129Sv mouse-derived embryonic stem (ES) cell genomic P1 library. Three genomic clones containing theCsf1r gene were obtained. One of these clones was mapped by a combination of restriction enzyme digestion, field inversion gel electrophoresis, Southern analyses, polymerase chain reaction (PCR), and sequencing. A 12-kb AseI fragment spanning exons 2 to 6 was subcloned into the SmaI site in the multiple cloning site of pGEM7Z in which the ApaI site had been mutated, and the subclone was designated pGEM-Ase-Csf1r. The AseI fragment was subjected to restriction enzyme mapping, PCR, and DNA sequencing to generate the restriction map shown in Figure1A. A humanized green fluorescent protein (hGFP) sequence and the neomycin resistance (PGKneo) cassette was cloned in-frame into the ApaI site in the exon 3 of pGEM-Ase-Csf1r, and a PGK-tk (thymidine kinase) cassette was cloned into the ClaI site in the multiple cloning site of the pGEM-Ase-Csf1r, yielding the final targeting vector (Figure 1A). A unique MluI site in the vector was used for linearization.
Targeted disruption of the mouse
Csf1r gene: decreased growth rate and increased circulating CSF-1 inCsf1r−/Csf1r−mice. (A) The targeted region of the Csf1r gene, theCsf1r gene-targeting vector, and the correctly targeted allele, showing exons 1 to 7, restriction enzyme sites (As,AseI; H, HindIII; A, ApaI; C,ClaI), PCR primers used (P1-P6, see text), the in-frame humanized green fluorescent protein (hGFP) sequence and the neomycin resistance (PGKneo) and thymidine kinase (PGKtk) cassettes above the flanking intron 7 probe were used to identify the 7-kb wild-type allele and 8-kb targeted allele HindIII fragments depicted below it. (B) Southern blot analyses of the DNA from individual G418 and gancyclovir-resistant ES cell clones using the probe shown in A. Asterisks mark clones possessing the correctly targeted allele. (C) Anti–CSF-1R Western blot analysis of BMM (upper panel). The molecular masses of the mature CSF-1R (165 kDa) and its precursor (130 kDa) are indicated. RT-PCR of BMM RNA with primers P5, P6 (A) specific for the targeted allele (middle panel) and control primers for β-actin (lower panel) are shown. (D) Growth curves of progeny of double heterozygote (Csf1r+/Csf1r−;Csf1+/Csf1op×Csf1r+/Csf1r−;Csf1+/Csf1op) crosses (n ≥ 5 for each genotype). (E) Serum CSF-1 concentration determined by a radioimmunoassay that selectively detects biologically active CSF-1 (± SD; n ≥ 5 for each genotype).
Targeted disruption of the mouse
Csf1r gene: decreased growth rate and increased circulating CSF-1 inCsf1r−/Csf1r−mice. (A) The targeted region of the Csf1r gene, theCsf1r gene-targeting vector, and the correctly targeted allele, showing exons 1 to 7, restriction enzyme sites (As,AseI; H, HindIII; A, ApaI; C,ClaI), PCR primers used (P1-P6, see text), the in-frame humanized green fluorescent protein (hGFP) sequence and the neomycin resistance (PGKneo) and thymidine kinase (PGKtk) cassettes above the flanking intron 7 probe were used to identify the 7-kb wild-type allele and 8-kb targeted allele HindIII fragments depicted below it. (B) Southern blot analyses of the DNA from individual G418 and gancyclovir-resistant ES cell clones using the probe shown in A. Asterisks mark clones possessing the correctly targeted allele. (C) Anti–CSF-1R Western blot analysis of BMM (upper panel). The molecular masses of the mature CSF-1R (165 kDa) and its precursor (130 kDa) are indicated. RT-PCR of BMM RNA with primers P5, P6 (A) specific for the targeted allele (middle panel) and control primers for β-actin (lower panel) are shown. (D) Growth curves of progeny of double heterozygote (Csf1r+/Csf1r−;Csf1+/Csf1op×Csf1r+/Csf1r−;Csf1+/Csf1op) crosses (n ≥ 5 for each genotype). (E) Serum CSF-1 concentration determined by a radioimmunoassay that selectively detects biologically active CSF-1 (± SD; n ≥ 5 for each genotype).
ES cell culture and chimeric mouse production
Mouse ES cells (Go Germline; Genome System) (ESVJ-1182) derived from 129SvJ strain were cultured on feeder layers of mitomycin C–treated mouse embryonic fibroblasts (MEFs) in ES cell medium (Dulbecco modified Eagle medium containing leukemia inhibitory factor [LIF]; Gibco BRL, Rockville, MD) at 37°C and 7.5% CO2. MEF preparation, ES cell propagation, electroporation, and selection of recombinants with G418 and gancyclovir (Ganc) were carried out exactly as described in the Genome Systems manual. Briefly, ES cells were expanded on MEF feeder layers; after trypsinization and preplating to remove MEF, 1 × 107 ES cells were electroporated with 20 μg of the MluI-linearized targeting vector DNA. Twenty-four hours later, G418 and Ganc were added into the LIF-containing ES cell medium for selection over 6 days. G418r/Gancr ES clones were then picked and replated onto gelatinized 24-well plates in ES cell medium. After a further 2 days, the ES cells were trypsinized and split in 2, one half being used to expand cells for DNA extraction and the other half cultured for 2 further days before freezing. Genomic DNA was extracted from the expanded cells, digested with HindIII, and subjected to Southern blotting with an intron 7 probe to flanking sequence not included in the targeting vector to identify the targeted clones. The correctly targeted allele yielded an 8-kbHindIII band that was clearly resolved from a 7-kb band derived from the wild-type allele (Figure 1B). In addition, PCR products of the correctly targeted gene were obtained with forward and reverse primers corresponding to 5′ and 3′ sequences flanking the targeting vector sequence that were used respectively with reverse and forward primers within the 5′ and 3′ regions of the targeting vector. Selected clones bearing the correctly targeted allele were thawed, expanded, and injected into C57BL/6-derived blastocysts, which were transplanted into CD1 pseudopregnant females to generate chimeric founder mice. Subsequent genotyping of mice was carried out by PCR of tail DNA using the primers P1, P2 (wild-type allele) and P3, P4 (targeted allele) (Figure 1A). Primers P1 (5′-TCT CCT GGG ATG GGA AAC GAT CCC AAA GGC-3′) and P2 (5′-GAT TCA GGG TCC AAG GTC CAG ATG GGA GAG-3′) yielded a 536–base pair (bp) product, exclusively from the wild type allele, whereas P3 (5′-GCC AGC CAC GAT AGC CGC GCT GCC TCG TC-3′) and P4 (5′-CTT CCT GGC CCT CAA CCA CTG TCA C-3′) gave rise to a 1.6-kb product exclusively from the targeted allele.
Mice
To obtain germline transmission, chimeras were mated with C57BL/6J × C3Heb/FeJ-a/a strain mice on which theCsf1op allele was maintained. All mice were maintained on this segregating background, behind a barrier at the Albert Einstein College of Medicine animal facility.Csf1r−/Csf1− andCsf1op/Csf1op mice were distinguished from normal siblings at 10 days of age by the absence of incisor eruption and were fed ad libitum a powdered mixture of mouse food and infant milk formula (Enfamil) daily to improve their nutritional status. Control mice received mouse chow ad libitum. Genotyping of the Csf1op allele was carried out as described.37
Radiographic analysis of mouse skeletal structure and CSF-1 radioimmunoassay
Radiographs were produced by exposing euthanized or anesthetized mice in a Faxitron pathology specimen x-ray cabinet (Faxitron X-Ray, Buffalo Grove, IL). The animals were posed immediately above a fine-grained Polaroid 665 instant negative film package. Exposure was set at 50 kV for 1.5 minutes. The negatives were developed and printed according to the manufacturer's instructions (Polaroid, Cambridge, MA). CSF-1 in mouse sera was determined by a radioimmunoassay that selectively detects biologically active CSF-1.38 39
Immunohistochemistry and histochemistry
Rat monoclonal antibody to F4/80 was a gift from Dr David Hume (Department of Microbiology, University of Queensland). For immunostaining with F4/80 antibodies and histochemical localization of tartrate-resistant acid phosphatase (TRAP), siblings of the different genotypes were perfused and tissues were fixed, decalcified (knee joint only), embedded, sectioned, and immunostained as described.2,40 TRAP staining was carried out as described.40 Quantitative histomorphometric analyses of the hematoxylin and eosin–stained sections were performed using a digital camera to capture images. Image analysis was performed by using Image-Pro Plus (Media Cybernetics, Silver Spring, MD). F4/80+ cells in tissue sections of at least 2 mice of a particular genotype at each age were quantitated as described.2 Whole-mount preparations of the 4th inguinal mammary gland were stained with alum carmine as described.41 For the estrus cycle analyses, daily vaginal smears were stained with hematoxylin and eosin. Mice were assessed as being in one of the 4 stages: proestrus (100% intact live epithelial cells), estrus (100% cornified epithelial cells), metestrus (50% cornified epithelial cells and 50% leukocytes), or diestrus (80%-100% leukocytes).42
Hematologic analysis and hematopoietic progenitor cell assays
Mice (6- to 8-week-old) were euthanized by exposure to a high concentration of CO2. Blood was collected in heparinized tubes. Total white blood cells were counted in 6% CH3COOH by using a hemocytometer. Red cell parameters were analyzed by using an automatic blood cell counter. Monocytes, granulocytes, and lymphocytes were resolved by differential flow cytometry analysis as described.43 Spleen and bone marrow cell suspensions were assayed for high-proliferative potential colony-forming cells (HPP-CFCs) and CSF-1–dependent colony-forming units (CFU-Cs) in agar cultures as previously described.31 Erythroid burst-forming unit (BFU-E) and granulocyte, erythroid, megakaryocyte, and macrophage colony-forming unit (CFU-GEMM) assays were performed by using reagents supplied by Stem Cell Technology (Vancouver, BC, Canada) in methylcellulose cultures as described by the manufacturers. The growth factors used for HPP-CFC agar cultures were stem cell factor, interleukin-6 (IL-6), IL-3, and granulocyte-macrophage CSF (GM-CSF). CFU-GEMM assays were performed in methylcellulose culture medium containing stem cell factor, IL-6, IL-3, and EPO.
Reverse transcriptase–PCR and Western blot
To assess Csf1r gene expression, bone marrow–derived macrophages (BMMs) were prepared from siblings of the different genotypes as previously described44 but with GM-CSF replacing CSF-1. BMM cultured in mouse GM-CSF (R & D Systems, MN) was solubilized in sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) running buffer,45 subjected to SDS-PAGE, and Western blotted with a 1:1 mixture of 2 affinity purified goat antimouse CSF-1R cytoplasmic domain peptide antibodies.46 Total RNA samples were prepared from BMM using the Trizol reagent (Gibco) and assayed by reverse transcriptase (RT)–PCR with primers that bind to exon 2 of the Csf1r gene and 5′ coding sequence of the GFP gene (P5 and P6, Figure 1A; P5 [5′-CTAGCAGCTGGGAGCCCCGTGCCCAGCCGACTC-3′]; P6 [5′-GGGTAGCGGCTGAAGCACTGCACGCCGTAGGTC-3′]). RT-PCR was carried out with an Advantage one-step RT-PCR kit (Clontech, Palo Alto, CA).
Statistical analyses
The mean, SD, or SEM of all numeric data was calculated. Data were analyzed by either Student t test or chi-square test, where appropriate. Comparisons of data sets yieldingP > .05 were considered as not statistically significant.
Results
Targeted disruption of the mouse CSF-1R gene
A P1 clone containing the entire coding region and flanking DNA of CSF-1R gene was used to prepare the targeting construct (Figure 1A) as described in “Materials and methods.” This vector was linearized and electroporated into 129SvJ strain ES cells, and clones of transfected cells resistant to both G418 and Ganc were screened for homologous recombinants by Southern blotting and PCR (Figure 1B). Of the 164 G418/Ganc-resistant clones obtained, 20 were identified as homologous recombinants and 2 of these (2C5 and 6D3) were injected into blastocysts. Of the 5 chimeras obtained, 3 (2 derived from 6D3 and one from 2C5) were transmitted to the germline. These lines were maintained on the same C57BL/6J × C3Heb/FeJ-a/a background as theCsf1op/Csf1op mice. Western blot analysis of whole cell lysates of BMM prepared fromCsf1r−/Csf1r− and littermate control mice indicated that theCsf1r−/Csf1r− cells were devoid of CSF-1R (Figure 1C, top panel). Consistent with these results, RT-PCR analysis using primers P5 and P6 (Figure 1A) indicated that bothCsf1r−/Csf1r− andCsf1r+/Csf1r− cells expressed mRNA encoding the CSF-1R–hGFP fusion protein (Figure 1C, lower panels). However, no significant expression of GFP could be detected inCsf1r−/Csf1r− orCsf1r+/Csf1r− cells by fluorescence microscopy or flow cytometry (data not shown).
Gross phenotype ofCsf1r−/Csf1r−mice
Csf1r−/Csf1r− mice were identical in appearance toCsf1op/Csf1op mice. They were small (see below) and toothless and possessed truncated limbs, a domed skull, and, occasionally, a kinked tail. Studies withCsf1op/Csf1op mice indicate that they have a low body weight and a low growth rate.28,37 Mice homozygous for theCsf1op or Csf1r−mutations possessed a significantly lower growth rate and lower adult weight than double-positive control mice or mice heterozygous for the mutations and were indistinguishable from each other in this respect (Figure 1D). Furthermore, the growth curves of the double mutants were indistinguishable from those of mice homozygous for either mutation. The Csf1r−/Csf1r− mice, likeCsf1op/Csf1opmice,47 were also deaf (data not shown).
A comparison of the genotypic frequencies for single heterozygous crosses (Csf1+/Csf1op ×Csf1+/Csf1op andCsf1r+/Csf1r− ×Csf1r+/Csf1r−) is presented in Table 1. At birth, the frequency of the progeny of Csf1op heterozygote andCsf1r− heterozygote crosses was as expected for a nondeleterious gene inherited in a Mendelian fashion. By weaning, however, the survival ofCsf1r−/Csf1r− mice approximated the survival ofCsf1op/Csf1op mice, and the survival of both mutant mice was significantly lower than the survival of wild-type mice, as previously reported forCsf1op/Csf1opmice.28 Analysis of the data from double heterozygote crosses at weaning (Table 2) revealed that survival ofCsf1op/Csf1op mice was not significantly different from the survival ofCsf1r−/Csf1r− mice or of double mutantCsf1op/Csf1op;Csf1r−/Csf1r−, except for the survival ofCsf1op/Csf1op;Csf1r+/Csf1r−mice, which unexpectedly exhibited normal survival, suggesting that theCsf1op/Csf1op phenotype might be slightly less severe than theCsf1r−/Csf1r− phenotype.
Frequency of genotypes of progeny of single heterozygote crosses of Csf1op andCsf1r− mutations
| Age of progeny . | Cross . | Total progeny . | Homozygous wild type . | Heterozygotes . | Homozygous mutant . | χ2 test probability* . |
|---|---|---|---|---|---|---|
| 1 d | Csf1+/Csf1op × Csf1+/Csf1op | 40 | 12 (1.2)† | 17 (1.7) | 11 (1.1) | 0.62 |
| Csf1r+/Csf1r− × Csf1r+/Csf1r− | 72 | 21 (1.17) | 38 (2.11) | 13 (0.72) | 0.37 | |
| 3 wk | Csf1+/Csf1op × Csf1+/Csf1op | 214 | 71 (1.32) | 109 (2.04) | 34 (0.64) | 0.002 |
| Csf1r+/Csf1r− × Csf1r+/Csf1r− | 393 | 107 (1.09) | 221 (2.24) | 65 (0.66) | 0.005 |
| Age of progeny . | Cross . | Total progeny . | Homozygous wild type . | Heterozygotes . | Homozygous mutant . | χ2 test probability* . |
|---|---|---|---|---|---|---|
| 1 d | Csf1+/Csf1op × Csf1+/Csf1op | 40 | 12 (1.2)† | 17 (1.7) | 11 (1.1) | 0.62 |
| Csf1r+/Csf1r− × Csf1r+/Csf1r− | 72 | 21 (1.17) | 38 (2.11) | 13 (0.72) | 0.37 | |
| 3 wk | Csf1+/Csf1op × Csf1+/Csf1op | 214 | 71 (1.32) | 109 (2.04) | 34 (0.64) | 0.002 |
| Csf1r+/Csf1r− × Csf1r+/Csf1r− | 393 | 107 (1.09) | 221 (2.24) | 65 (0.66) | 0.005 |
Probability that results do not differ significantly from the 1:2:1 ratio.
Ratio compared with expected 1:2:1 ratio of an independently segregating gene.
Genotypic frequencies as a percentage of total 3-week-old progeny from double heterozygote(Csf1r+/Csf1r−; Csf1+/Csf1op × Csf1r+/Csf1r−; Csf1+/Csf1op) crosses
| Csf1 genotype . | Csf1r genotype . | ||
|---|---|---|---|
| Csf1r+/Csf1r+ . | Csf1r+/Csf1r− . | Csf1r−/Csf1r− . | |
| Csf1+/Csf1+ | 6.25/7.7* | 12.5/17.6 | 6.25/3.8 |
| Csf1+/Csf1op | 12.5/15.3 | 25.0/31.0 | 12.5/4.2 |
| Csf1op/Csf1op | 6.25/4.2 | 12.5/12.6 | 6.25/3.4 |
| Csf1 genotype . | Csf1r genotype . | ||
|---|---|---|---|
| Csf1r+/Csf1r+ . | Csf1r+/Csf1r− . | Csf1r−/Csf1r− . | |
| Csf1+/Csf1+ | 6.25/7.7* | 12.5/17.6 | 6.25/3.8 |
| Csf1+/Csf1op | 12.5/15.3 | 25.0/31.0 | 12.5/4.2 |
| Csf1op/Csf1op | 6.25/4.2 | 12.5/12.6 | 6.25/3.4 |
Percentage expected for independently segregating alleles/percentage observed, n = 261.
Previous studies from our laboratory indicated that 95% of the circulating CSF-1 is cleared by CSF-1R–mediated endocytosis and intracellular destruction by sinusoidally located macrophages and that circulating CSF-1 has a half-life of only 10 minutes.48 We therefore determined the CSF-1 concentration in the sera ofCsf1r−/Csf1r−,Csf1r+/Csf1r+, andCsf1r+/Csf1r− mice (Figure 1E). Consistent with these previous observations, levels of serum CSF-1 inCsf1r−/Csf1r− mice were elevated approximately 20-fold compared with the levels inCsf1r+/Csf1r+ littermate control mice. Interestingly, the levels of circulating CSF-1 in theCsf1r+/Csf1r− mice (19.0 ± 5.4 ng/mL) were within normal range (19.9 ± 6.1 ng/mL).
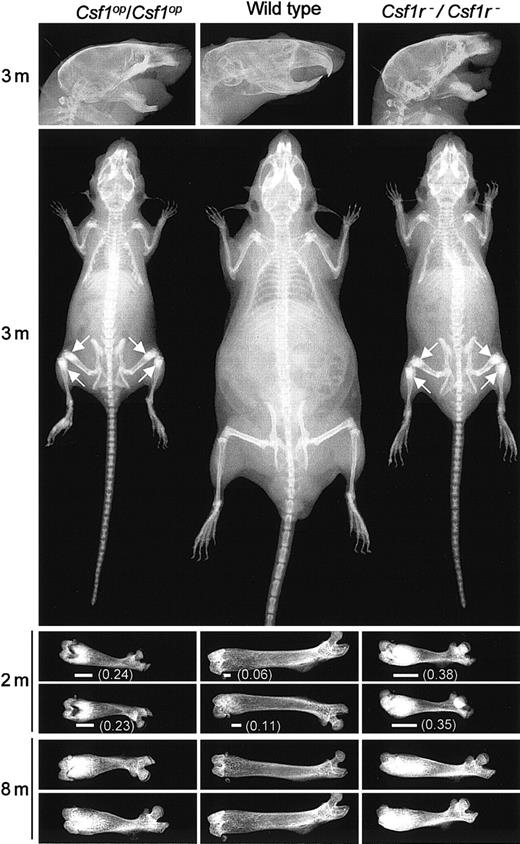
Comparison of the osteopetrotic phenotypes ofCsf1r−/Csf1r−andCsf1op/Csf1opmice
Csf1op/Csf1op mice exhibit impaired bone resorption associated with a reduction in the number of osteoclasts.28 Their inability to remodel bone resulted in general skeletal deformities that, as shown in Figure2, were shared with theCsf1r−/Csf1r− mice. Compared with wild-type littermates, the long bones of their limbs were short (Figure2) with increased bone density at the metaphyses (Figure 2), deformities in the flat bony plates result in a domed skull (Figure 2), and the increased bone density in the mandible presumably leads to the failure of tooth eruption (Figure 2). Interestingly, a radiograph analysis of the temporal changes in the femurs of wild-type and mutant mice with age (Figure 2) indicates that there is significantly more radiopacity in the distal metaphysis of the femurs ofCsf1r−/Csf1r− than ofCsf1op/Csf1op mice. Similar results were obtained withCsf1r−/Csf1r− mice derived from both 2C5 and 6D3 ES cell lines. All further experiments were carried out with Csf1r−/Csf1r− mice derived from 2C5 ES cells.
Comparison of the skeletal development of wild-type,
Csf1op/Csf1op, and Csf1r−/Csf1r−mice. Radiograms of the heads, bodies, and femurs of mice of the indicated genotypes at different ages (m, months). Each femur is from a different mouse. Arrows indicate regions of increased bone density most easily visualized at this magnification. Also shown is the extent (horizontal line) and fraction (in parenthesis) of the total femur length that is of high radiopacity.
Comparison of the skeletal development of wild-type,
Csf1op/Csf1op, and Csf1r−/Csf1r−mice. Radiograms of the heads, bodies, and femurs of mice of the indicated genotypes at different ages (m, months). Each femur is from a different mouse. Arrows indicate regions of increased bone density most easily visualized at this magnification. Also shown is the extent (horizontal line) and fraction (in parenthesis) of the total femur length that is of high radiopacity.
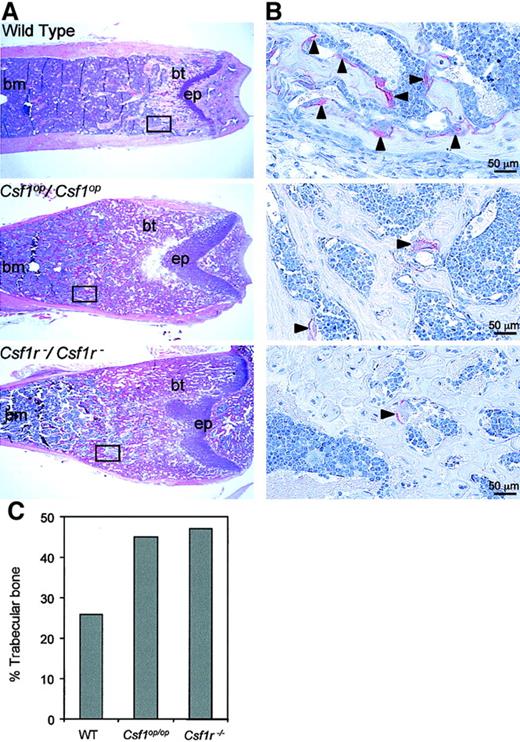
The metaphyseal radiopacity of the long bones ofCsf1op/Csf1op andCsf1r−/Csf1r− mice is the result of the failure of osteoclast development at this site. Hematoxylin and eosin staining of longitudinal sections of the distal metaphyseal regions of the femurs of these mice revealed that bothCsf1r−/Csf1r− andCsf1op/Csf1op mice possessed higher amounts of bony trabeculae than littermate control mice, consistent with impaired bone remodeling (Figure3A). In addition, TRAP+ cells (osteoclasts) were present in low numbers in the bony trabecula regions of Csf1op/Csf1op femurs compared with littermate control femurs and even fewer in number inCsf1r−/Csf1r− femurs (Figure 3B). Histomorphometric analyses of the entire bone marrow cavity of the femurs were consistent with these observations, the percentage of trabecular bone in Csf1r−/Csf1r−and Csf1op/Csf1op femurs being greater than in wild-type femurs (Figure 3C). In the metaphyseal region, the retention of trabecular bone was greater forCsf1r−/Csf1r− (92%) than forCsf1op/Csf1op (83%) mice (compared with wild-type mice [43%]), reflecting the greater radiopacity of the Csf1r−/Csf1r−over Csf1op/Csf1op femurs in this region (Figure 2).
Histology of bone marrow of wild type,
Csf1op/Csf1op, and Csf1r−/Csf1r−mice. (A) Low-power photomicrographs of hematoxylin and eosin–stained midsagittal 5-μm sections of the distal femoral metaphyses of 8-week-old mice (bm, bone marrow; bt, bony trabeculae; ep, epiphyseal plate). Boxes indicate comparable areas of TRAP-stained sections photographed in B. (B) High-power photomicrographs of TRAP staining for osteoclasts in midsagittal 5-μm sections of femurs of 4-week-old mice in areas comparable to those boxed in A. Counterstained with hematoxylin. Arrowheads indicate TRAP+ cells. (C) Percentage of trabecular bone in the entire bone marrow cavity determined from the sections used in A. Original magnification A, × 25; B, × 400.A.
Histology of bone marrow of wild type,
Csf1op/Csf1op, and Csf1r−/Csf1r−mice. (A) Low-power photomicrographs of hematoxylin and eosin–stained midsagittal 5-μm sections of the distal femoral metaphyses of 8-week-old mice (bm, bone marrow; bt, bony trabeculae; ep, epiphyseal plate). Boxes indicate comparable areas of TRAP-stained sections photographed in B. (B) High-power photomicrographs of TRAP staining for osteoclasts in midsagittal 5-μm sections of femurs of 4-week-old mice in areas comparable to those boxed in A. Counterstained with hematoxylin. Arrowheads indicate TRAP+ cells. (C) Percentage of trabecular bone in the entire bone marrow cavity determined from the sections used in A. Original magnification A, × 25; B, × 400.A.
Consequent to their decreased volume of femoral bone marrow, the total femoral marrow cellularity ofCsf1op/Csf1op andCsf1r−/Csf1r− mice was significantly lower than the bone marrow cellularity of wild-type mice (Table 3). As previously reported forCsf1op/Csf1opmice,31 32 the bone marrow cellularityCsf1r−/Csf1r− mice recovered to the levels observed in control wild type mice by 8 months of age (Table 3), despite evidence of some residual osteopetrosis in bothCsf1op/Csf1op andCsf1r−/Csf1r− mice (Figure 2).
Bone marrow cellularity of 7-week-old and 8-month-old mice
| Age . | Total femur cellularity/body weight (×10−6/g) . | ||
|---|---|---|---|
| Wild type . | Csf1op/Csf1op . | Csf1r−/Csf1r− . | |
| 7 wk | 0.50 ± 0.10 | 0.25 ± 0.043-150 | 0.23 ± 0.103-150 |
| 8 mo | 1.57 ± 0.28 | 1.90 ± 0.14 | 1.53 ± 0.82 |
| Age . | Total femur cellularity/body weight (×10−6/g) . | ||
|---|---|---|---|
| Wild type . | Csf1op/Csf1op . | Csf1r−/Csf1r− . | |
| 7 wk | 0.50 ± 0.10 | 0.25 ± 0.043-150 | 0.23 ± 0.103-150 |
| 8 mo | 1.57 ± 0.28 | 1.90 ± 0.14 | 1.53 ± 0.82 |
Significantly different from wild type (Studentt test P ≤ .01).
Reduced mononuclear phagocyte production inCsf1r−/Csf1r−mice
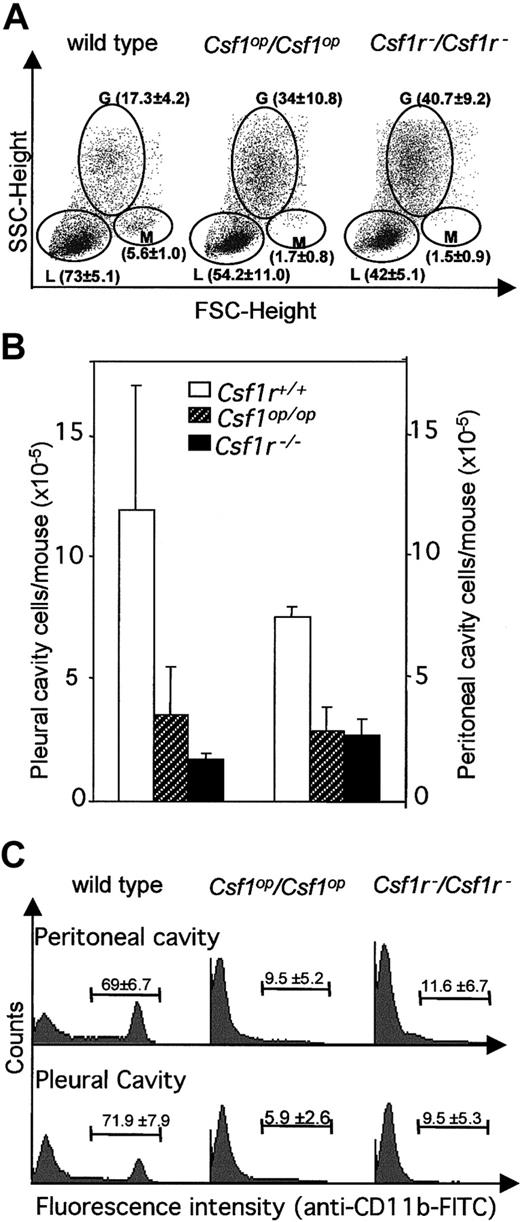
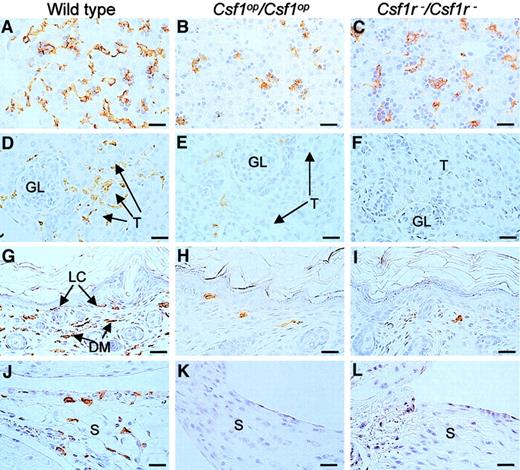
Previous reports34,49 have shown that blood monocyte and lymphocyte percentages are reduced inCsf1op/Csf1op mice. Fluorescence-activated cell sorter (FACS) analysis of these populations by forward and side-light scatter revealed that there was a decrease in monocytes and lymphocytes and an increase in granulocytes in the circulation of Csf1r−/Csf1r− mice and that they were not significantly different fromCsf1op/Csf1op mice in this respect (Figure 4A). Similarly, the total cellularities of the pleural and peritoneal cavities ofCsf1r−/Csf1r− mice were reduced to the same extent as inCsf1op/Csf1op mice (Figure 4B) as were the frequencies of Mac1(CD11b)+ cells in these cavities (Figure 4C). Tissue macrophages expressing macrophage-specific cell-surface protein, F4/80, are significantly decreased in many tissues ofCsf1op/Csf1opmice.2 The F4/80+ cell densities of several such tissues were determined in wild-type,Csf1op/Csf1op, andCsf1r−/Csf1r− mice at ages in which the F4/80+ macrophage density had previously been shown2 to be maximum for the particular wild-type tissue (Figure 5, Table4).2,50,51 As previously shown,2 the F4/80+ cell densities of the tissues of Csf1op/Csf1opmice were significantly lower than those of wild-type control mice (including the Langerhans cells of the epidermis, previously reported to be normal inCsf1op/Csf1opmice2). There was no difference between the lowered F4/80+ cell densities of these tissues and those ofCsf1r−/Csf1r− mice, except that the F4/80+ cell density in the kidney was significantly lower inCsf1r−/Csf1r− mice than inCsf1op/Csf1op mice (Table4). Thus, apart from this difference, the mononuclear phagocyte numbers were equivalently reduced inCsf1r−/Csf1r− andCsf1op/Csf1opmice.
FACS analysis of blood leukocytes, peritoneal cavity, and pleural cavity cells.
(A) Typical FACS analyses of monocytes, granulocytes, and lymphocytes by forward and side light scatter. Separate regions encompassing the monocyte (M), granulocyte (G), and lymphocyte (L) subpopulations are indicated. The means of results of such analyses for 3 mice of each genotype are shown in brackets (± SD). (B) Total pleural cavity and peritoneal cavity cells (n = 3). (C) Typical FACS analyses of CD11b+ peritoneal and pleural cavity cells. Percentage of positive cells for 3 mice of each genotype (± SD) is shown within each FACS distribution. FITC indicates fluorescein isothiocyanate; FSC, forward scatter; SSC, side scatter.
FACS analysis of blood leukocytes, peritoneal cavity, and pleural cavity cells.
(A) Typical FACS analyses of monocytes, granulocytes, and lymphocytes by forward and side light scatter. Separate regions encompassing the monocyte (M), granulocyte (G), and lymphocyte (L) subpopulations are indicated. The means of results of such analyses for 3 mice of each genotype are shown in brackets (± SD). (B) Total pleural cavity and peritoneal cavity cells (n = 3). (C) Typical FACS analyses of CD11b+ peritoneal and pleural cavity cells. Percentage of positive cells for 3 mice of each genotype (± SD) is shown within each FACS distribution. FITC indicates fluorescein isothiocyanate; FSC, forward scatter; SSC, side scatter.
F4/80+ cells in liver, kidney, skin, and synovial membrane.
Tissues from wild-type,Csf1op/Csf1op, andCsf1r−/Csf1r− mice were immunostained with a monoclonal antibody to F4/80 that selectively stains macrophages and were counterstained with hematoxylin. Sections of (A-C) 2-day-old livers, (D-E) 2-week-old kidneys showing macrophages surrounding the glomeruli (GL) and tubules (T) of wild-type mice (D), (F-I) 2-day-old skin showing immunostaining of both Langerhans cells (LC) and dermal macrophages (DM) from wild- type mice, and (J-L) longitudinal sections of 2-week-old knee joints in the region of the synovial membrane (S) showing immunostaining of cells in the wild-type synovial membranes. Note the more rounded and less dendritic appearance of the F4/80+ cells in the tissues of theCsf1op/Csf1op andCsf1r−/Csf1r− mice, previously reported forCsf1op/Csf1op mice. Bar, 50 μm. Original magnification A-L, × 400.
F4/80+ cells in liver, kidney, skin, and synovial membrane.
Tissues from wild-type,Csf1op/Csf1op, andCsf1r−/Csf1r− mice were immunostained with a monoclonal antibody to F4/80 that selectively stains macrophages and were counterstained with hematoxylin. Sections of (A-C) 2-day-old livers, (D-E) 2-week-old kidneys showing macrophages surrounding the glomeruli (GL) and tubules (T) of wild-type mice (D), (F-I) 2-day-old skin showing immunostaining of both Langerhans cells (LC) and dermal macrophages (DM) from wild- type mice, and (J-L) longitudinal sections of 2-week-old knee joints in the region of the synovial membrane (S) showing immunostaining of cells in the wild-type synovial membranes. Note the more rounded and less dendritic appearance of the F4/80+ cells in the tissues of theCsf1op/Csf1op andCsf1r−/Csf1r− mice, previously reported forCsf1op/Csf1op mice. Bar, 50 μm. Original magnification A-L, × 400.
Tissue F4/80+ cell densities
| Tissue . | Age of mice . | Wild type . | Csf1op/Csf1op . | Csf1r−/Csf1r− . |
|---|---|---|---|---|
| Bone marrow4-150,4-151 | 2 wk | 697.7 ± 11.7 | 210.5 ± 14.8 | 173.7 ± 41.0 |
| Liver4-150,4-151 | 2 d | 678.1 ± 108.4 | 413.4 ± 13.0 | 463.7 ± 45.3 |
| Liver4-150,4-151 | 4 mo | 372.9 ± 44.2 | 124.3 ± 7.2 | 150.0 ± 68.2 |
| Kidney4-150,4-151,‡ | 2 wk | 184.3 ± 83.0 | 34.5 ± 1.6 | 12.3 ± 0.4 |
| Thymus4-150,4-151 | 2 wk | 376.4 ± 40.1 | 110.6 ± 18.1 | 168.3 ± 15.2 |
| Langerhans cells4-150,4-151 | 2 d | 15.8 ± 1.34-153 | 3.0 ± 1.44-153 | 2.0 ± 1.44-153 |
| Dermis4-150,4-151 | 2 d | 253.1 ± 56.0 | 15.4 ± 5.1 | 32.5 ± 10.0 |
| Testes4-150,4-151 | 2 wk | 178.3 ± 83.2 | 46.7 ± 22.2 | 34.6 ± 19.4 |
| Tissue . | Age of mice . | Wild type . | Csf1op/Csf1op . | Csf1r−/Csf1r− . |
|---|---|---|---|---|
| Bone marrow4-150,4-151 | 2 wk | 697.7 ± 11.7 | 210.5 ± 14.8 | 173.7 ± 41.0 |
| Liver4-150,4-151 | 2 d | 678.1 ± 108.4 | 413.4 ± 13.0 | 463.7 ± 45.3 |
| Liver4-150,4-151 | 4 mo | 372.9 ± 44.2 | 124.3 ± 7.2 | 150.0 ± 68.2 |
| Kidney4-150,4-151,‡ | 2 wk | 184.3 ± 83.0 | 34.5 ± 1.6 | 12.3 ± 0.4 |
| Thymus4-150,4-151 | 2 wk | 376.4 ± 40.1 | 110.6 ± 18.1 | 168.3 ± 15.2 |
| Langerhans cells4-150,4-151 | 2 d | 15.8 ± 1.34-153 | 3.0 ± 1.44-153 | 2.0 ± 1.44-153 |
| Dermis4-150,4-151 | 2 d | 253.1 ± 56.0 | 15.4 ± 5.1 | 32.5 ± 10.0 |
| Testes4-150,4-151 | 2 wk | 178.3 ± 83.2 | 46.7 ± 22.2 | 34.6 ± 19.4 |
Densities in cells/mm2 average of multiple counts (±SD, n = 4) of tissue sections from at least 2 mice per genotype.
Significant differences between wild type andCsf1op/Csf1op (P ≤ .05).
Significant differences between wild type andCsf1r−/Csf1r− (P≤ .05).
Significant differences betweenCsf1op/Csf1op andCsf1r−/Csf1r− (P≤ .05).
Densities in cells/mm average of multiple counts (±SD, n = 4) of tissue sections from at least 2 mice per genotype.
Hematopoietic progenitor cells inCsf1r−/Csf1r−mice
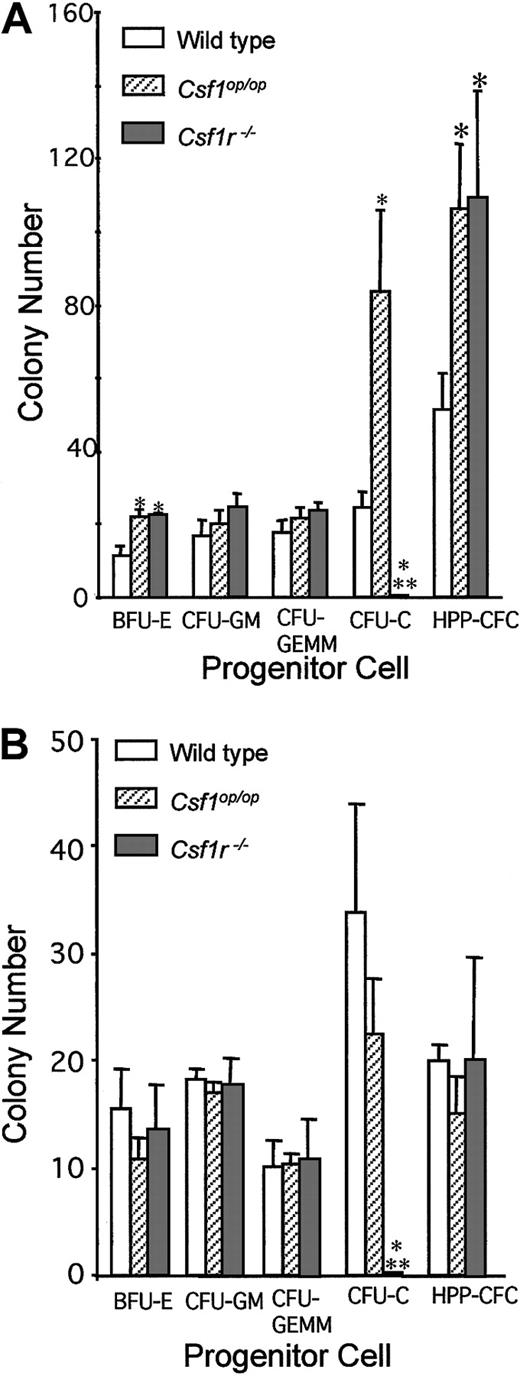
Previous experiments have shown that, consistent with their reduced space for marrow hematopoiesis (Table 3, Figures 2 and 3), 6-week-old Csf1op/Csf1opmice have a compensatory splenic hematopoiesis with elevated frequencies of CFU-Cs and HPP-CFCs.31 As in the case of the Csf1op/Csf1op mice (3.6 ± 0.2), the splenic weights (milligram per gram of body weight) of 6-week-old Csf1r−/Csf1r− mice (5.1 ± 0.5) were significantly elevated compared with those of littermate control wild-type mice (3.0 ± 0.3 [± SD], n = 3). The hematopoietic status of the wild-type,Csf1op/Csf1op, andCsf1r−/Csf1r− mice was examined by determining bone marrow and splenic frequency of hematopoietic progenitor cells at 6 weeks of age (Figure6). Splenic BFU-Es and HPP-CFCs were elevated to a similar extent in both types of mutant mouse. There was no significant effect of either mutation on CFU-GM or CFU-GEMM. As expected, in contrast to the elevated frequency of CSF-1–responsive splenic CFU-C in theCsf1op/Csf1op mice, there were no CSF-1–responsive progenitors in spleens of theCsf1r−/Csf1r− mice. In the bone marrow of bothCsf1op/Csf1op andCsf1r−/Csf1r− mice, there was no difference in the frequency of these progenitors compared with their frequency in wild-type mice, save for the absence of CFU-Cs in theCsf1r−/Csf1r− mice. These results reflect a negative regulatory role of CSF-1 on the in vivo frequencies of splenic BFU-Es and HPP-CFCs.
Hematopoietic progenitor cell concentrations in the spleen and bone marrow of 6-week-old mice.
(A) Splenic progenitor cell colony numbers per 105 cells (BFU-E, CFU-GM, CFU-GEMM, CFU-C) or per 104 cells (HPP-CFC). (B) Bone marrow progenitor cell colony numbers per 104 cells (BFU-E, CFU-GM, CFU-GEMM, CFU-C) or per 103 cells (HPP-CFC). Means ± SD (3 mice per genotype). *Significantly different from wild type; **Significantly different from wild type andCsf1op/Csf1op(P ≤ .01).
Hematopoietic progenitor cell concentrations in the spleen and bone marrow of 6-week-old mice.
(A) Splenic progenitor cell colony numbers per 105 cells (BFU-E, CFU-GM, CFU-GEMM, CFU-C) or per 104 cells (HPP-CFC). (B) Bone marrow progenitor cell colony numbers per 104 cells (BFU-E, CFU-GM, CFU-GEMM, CFU-C) or per 103 cells (HPP-CFC). Means ± SD (3 mice per genotype). *Significantly different from wild type; **Significantly different from wild type andCsf1op/Csf1op(P ≤ .01).
Reproductive phenotype ofCsf1r−/Csf1r−mice
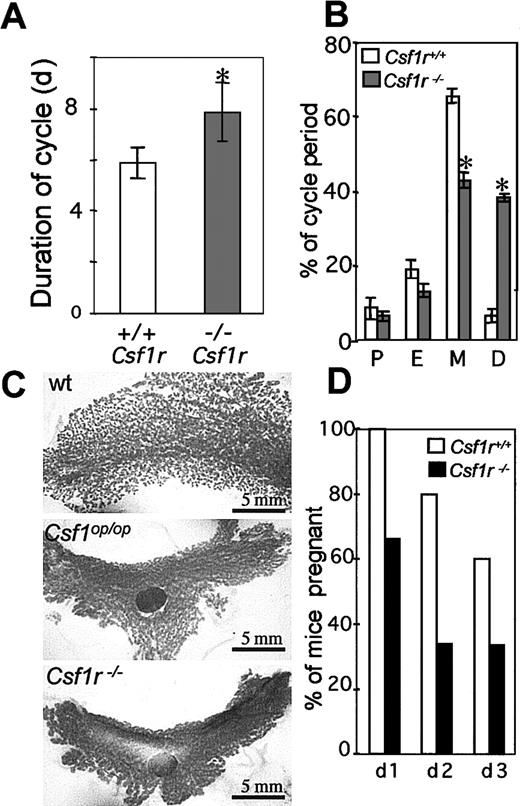
CSF-1 plays an important role in ovulation, preimplantation, placental function, regulation of the estrous cycle, and lactation.3,4,52-55 Estrous cycling times are altered in mature Csf1op/Csf1opmice, estrous occurring irregularly and more infrequently compared with the estrous times of normal mice (∼5 days).53 The duration of estrous in femaleCsf1r−/Csf1r− mice, determined by the appearance in the vagina of exfoliated anuclear cornified cells and the absence of macrophages, although lower (8.0 ± 2.0, n = 40; Figure 7A) than the previously reported cycling time forCsf1op/Csf1op mice (∼14.5 days), was significantly higher than the duration of estrus in wild-type mice (5.9 ± 0.5, n = 40; Figure 7 A). As previously reported for theCsf1op/Csf1op mice, this increase in cycling time was primarily due to an increase in the duration of the diestrus period (Figure 7B). In pregnantCsf1op/Csf1op mice, the lactating mammary gland fails to develop normally because of a failure of branching morphogenesis of the ductal epithelium.56 As shown in the whole-mount stained mammary glands in Figure 7C, a similar phenotype was observed for mammary glands from pregnantCsf1r−/Csf1r− mice.
Reproductive phenotype of
Csf1r−/Csf1r−mice. (A) Duration of estrus cycle in virgin female mice (5 mice per genotype, 8 cycles/mouse, ± SD). (B) Percentage time in proestrus (P), in estrus (E), in metestrus (M), and in diestrus (D) (± SD). (C) Whole-mount alum-carmine staining of the 4th inguinal mammary gland from 18-day pregnant mice. All panels are at the same magnification, approximately only one third of the wild-type gland is shown. LN indicates lymph node. (D) Percentage of successful pregnancies resulting from the consecutive daily mating of wild-type (open) andCsf1r−/Csf1r−(filled) male mice with superovulated virgin female mice. *Indicates significantly different from wild type; wt, wild type.
Reproductive phenotype of
Csf1r−/Csf1r−mice. (A) Duration of estrus cycle in virgin female mice (5 mice per genotype, 8 cycles/mouse, ± SD). (B) Percentage time in proestrus (P), in estrus (E), in metestrus (M), and in diestrus (D) (± SD). (C) Whole-mount alum-carmine staining of the 4th inguinal mammary gland from 18-day pregnant mice. All panels are at the same magnification, approximately only one third of the wild-type gland is shown. LN indicates lymph node. (D) Percentage of successful pregnancies resulting from the consecutive daily mating of wild-type (open) andCsf1r−/Csf1r−(filled) male mice with superovulated virgin female mice. *Indicates significantly different from wild type; wt, wild type.
Compared with normal males,Csf1op/Csf1op male mice had low testosterone levels, low libido, and reduced viable sperm numbers, as well as mating infrequently and displaying a long latency between mating when presented serially with female mice in estrous.42 A similar phenotype was observed forCsf1r−/Csf1r− mice, which mated less frequently with cycling females and produced fewer pregnant females on successive days following daily exposure to different superovulated females than wild-type males (Figure 7D).
Discussion
In this study, we have shown that BMM from mice in which bothCsf1r alleles have been targeted by the insertion of a PGKneo cassette containing stop codons into exon 3 fail to express detectable CSF-1R mRNA or protein. In contrast, BMM from wild- type mice express approximately 50 000 cell surface CSF-1R molecules per cell.7 Consistent with this observation, neither bone marrow nor splenic cells from theCsf1r−/Csf1r− mice are able to form CSF-1–dependent macrophage colonies in agar cultures. Furthermore, the circulating concentrations of CSF-1 in theCsf1r−/Csf1r− mice were 20-fold higher than the wild-type CSF-1 concentration, consistent with a failure of the previously established, CSF-1R–mediated clearance mechanism for circulating CSF-1.48 These results indicate that the Csf1r−/Csf1r− mice are truly CSF-1R nullizygous.
The phenotype of theCsf1r−/Csf1r− mice was very similar to the phenotype of theCsf1op/Csf1op mice lacking CSF-1. The growth rates and postnatal lethalities ofCsf1r−/Csf1r−,Csf1op/Csf1op, and the double-mutantCsf1r−/Csf1r−;Csf1op/Csf1opmice are indistinguishable. Of interest was the demonstration that postnatal lethality of both mutants was not detectable at day 1 but became apparent by 3 weeks of age (Table 1). BothCsf1r−/Csf1r− andCsf1op/Csf1op mice were toothless and severely osteopetrotic with the same osteopetrosis-associated skeletal abnormalities and with a reduced bone marrow cellularity that returns to normal levels by 8 months of age. There was a similar depletion of circulating monocytes, pleural and peritoneal cavity cells, peritoneal cavity Mac1+ cells, and tissue macrophages in both mutant mice. With the exception of CSF-1–dependent CFU-Cs, which cannot be measured in theCsf1r−/Csf1r− mice, there was no difference in their levels of bone marrow hematopoietic progenitor cells, and both mutants had splenic BFU-E and HPP-CFC levels that were significantly higher than in wild-type control mice. Furthermore, both mutants shared previously reported defects in reproductive function, including longer estrus cycles, failure of normal lactating mammary gland development, and a reduced male mating performance. As no phenotype in theCsf1op/Csf1op mice was more severe than in theCsf1r−/Csf1r− mice, these results indicate that all of the effects of CSF-1 are mediated via the CSF-1R that is encoded by the c-fms gene.
Despite the overall similarity of theCsf1r−/Csf1r− andCsf1op/Csf1op mouse phenotypes, aspects of theCsf1r−/Csf1r− phenotype were more severe than those of theCsf1op/Csf1op mouse. These aspects include a slightly more severe osteopetrosis in the femurs (Figures 2 and 3) and a more severe depletion of F4/80+ cells in the kidney (Figure 5, Table 4). In addition, there was a difference in the postnatal survival of mice homozygous for one allele and heterozygous for the other.Csf1op/Csf1op;Csf1r+/Csf1r−mouse survival was as expected for 2 independently segregating alleles with no associated lethality, whereas the survival ofCsf1r−/Csf1r−;Csf1+/Csf1opmice was one third that rate (Table 2). Because bothCsf1r+/Csf1r+;Csf1op/Csf1opandCsf1r−/Csf1r−;Csf1+/Csf1+mice also survived at one third of the expected rate, this finding suggests that a single copy of the CSF-1R gene may confer an advantage in the absence of CSF-1. This observation, with significant numbers of progeny, supports the concept of a CSF-1–independent protective effect of the CSF-1R, although it is not clear how 2 copies of the CSF-1R gene fail to protect. It is possible that some of these effects were due to partial rescue of theCsf1op/Csf1op mice by transplacental passage of CSF-1 from the heterozygotic mothers,57 which is not possible in the case of theCsf1r−/Csf1r− mice. However, because no difference was observed between the liver F4/80+cell densities of 2-day-oldCsf1op/Csf1op andCsf1r−/Csf1r− mice and nearly 80% of the fetal hepatic blood flow is derived directly from the umbilical vein,58 this seems unlikely. Furthermore, the increased severity of Csf1r−/Csf1r−phenotype has recently been confirmed by using the progeny of mice of a single strain on which both alleles have been backcrossed for more than 5 generations. On this background, theCsf1r−/Csf1r− phenotype is even more severe and the Csf1r−/Csf1r−mice all die by 3 weeks of age, whereas theCsf1op/Csf1op survival approximates their survival on the C57BL/6J × C3Heb/FeJ-a/a background. These observations suggest that the CSF-1R can respond to an additional ligand or can function in a ligand-independent fashion in some situations.
It has previously been suggested, in part because of the residual tissue macrophage production seen in certain tissues inCsf1op/Csf1op mice and in part because of the nature of the mutation, that this mouse is not completely devoid of a mutated yet active form of CSF-1.59However, as indicated above, theCsf1op/Csf1op andCsf1r−/Csf1r− phenotypes were very similar. In particular, with one exception, everyCsf1r−/Csf1r− tissue possessed macrophage concentrations that did not significantly differ from those of the correspondingCsf1op/Csf1op tissue. These observations strongly suggest that the residual tissue macrophage populations inCsf1op/Csf1op mice are not due to the action of a mutated less-efficient CSF-1. Thus, the present study indicates that other growth factors are involved in the regulation of macrophage production observed inCsf1op/Csf1op andCsf1r−/Csf1r− mice. Among the known growth factors most likely to be responsible for macrophage production in these mutant mice are GM-CSF and IL-3 and vascular endothelial growth factor. GM-CSF and IL-3 together are capable of correcting not only the osteopetrotic condition ofCsf1op/Csf1op mice but also macrophage deficiencies in several tissues.60Vascular endothelial growth factor is also able to correct the osteopetrotic condition ofCsf1op/Csf1op mice and appears to be responsible for the age-related increase in osteoclasts in these mice.33
It has previously been reported that the percentage of blood monocytes and lymphocytes is reduced34,37,49 and the percentage of blood granulocytes increased37,49 inCsf1op/Csf1op mice, without significant changes in the circulating leukocyte concentration. These changes in monocytes and granulocytes are not unexpected, given the specificity of CSF-1 for the mononuclear phagocytic lineage13,38 and the existence of progenitors capable of giving rise to both granulocytes and macrophages.61However, the decrease in the percentage of blood lymphocytes, which we also observed in bothCsf1op/Csf1op andCsf1r−/Csf1r− mice in the present study, is of interest. Recently, it has also been shown that the frequencies of stromal cell– and IL-7–dependent B-cell precursors and CFU–IL-7 in bone marrow were significantly diminished inCsf1op/Csf1op mice, as well as other osteopetrotic mice (Fos-nullizygous andmicrophthalmia (Mitfmi/Mitfmi). However, these precursors could be generated fromCsf1op/Csf1op bone marrow precursor cells on in vitro incubation with stromal cells in the presence of IL-7.62 Also, CSF-1 has been shown to reduce apoptosis in precursor B cells in whole mouse bone marrow cultures but not in cultures of B220+ B- lineage cells, implying that CSF-1 can indirectly regulate B lymphopoiesis.63 These results suggest that the decreased B lymphopoiesis inCsf1op/Csf1op mice results from alterations of the microenvironment rather than from a B-cell autonomous defect. The CSF-1R–nullizygous mice will be useful in analyzing direct and indirect effects of CSF-1 in B lymphopoiesis.
The development of the CSF-1R–nullizygous mouse permits new approaches to be taken to the analysis of CSF-1 action. In studies of CSF-1R signal transduction, a powerful approach to our understanding of CSF-1R structure-function has been to introduce mutated forms of the receptor into fibroblast or myeloid cell lines that do not express the CSF-1R.64-68 However, the functions affected by particular CSF-1R mutations have been found to differ substantially, indicating the importance of cellular context for experiments of this kind (reviewed in Hamilton69 70). By using BMM and progenitor cells from Csf1r−/Csf1r− mice and cell lines derived from them, as well as “knock-in” mutation approaches based on the “knock-out” construct used here, it is now possible to carry out these experiments in the correct cellular context.
Circulating CSF-1 is composed of both proteoglycan and glycoprotein forms.71 The analysis of these forms and their regulation has been limited by the amounts of mouse serum needed and the low specific activity of serum CSF-1 per unit serum protein. The 20-fold elevation of the concentration (and specific activity) of serum CSF-1 in Csf1r−/Csf1r− mice will greatly facilitate these investigations.
Perhaps the area of most immediate import is the assessment of the role of CSF-1 in the regulation of primitive hematopoietic cells. Previous studies have indicated that, although CFU-Cs are the most primitive cells that can be stimulated to proliferate and differentiate by CSF-1 alone in vitro, CSF-1 can cause the proliferation and differentiation of precursors of the CFU-Cs, by synergizing with other growth factors that alone have limited or no effect on the proliferation of the primitive cells.10,11,27,72 Indeed, the observation in the present study that splenic BFU-E and HPP-CFC levels inCsf1op/Csf1op andCsf1r−/Csf1r− mice are significantly elevated already suggests that CSF-1, via the CSF-1R, negatively regulates the maintenance of these precursor cells. Consistent with the observed elevation of BFU-Es and HPP-CFCs, CSF-1 has been shown to inhibit erythroid and erythroid/myeloid colony formation in vitro.73 CSF-1 has also been shown to inhibit the differentiation of ES cells to blood cells other than macrophages.74 In addition, it has recently been shown that ectopic expression of the CSF-1R in the multipotent hematopoietic EML cell line decreases their erythroid potential,75raising the possibility that CSF-1R regulates erythroid differentiation and/or commitment. Because of the cell autonomous nature of theCsf1r− mutation, it is now possible to study the role of the receptor in the generation of primitive cells of different lineages by using bone marrow transplantation. Such an approach permits testing of CSF-1/CSF-1R regulation of cell proliferation and differentiation in the absence of an endogenous osteopetrotic condition.
We thank members of the AECOM analytical imaging, FACS, histopathology, hybridoma, and transgenic and gene targeting facilities for assistance in different aspects of the work; Dr Paula E. Cohen for advice on the fertility analysis; Dr Sandy C. Marks Jr for advice on TRAP staining; David Gebhard of the AECOM FACS facility for assistance with the FACS analyses; Dr Thomas Graf for reviewing the manuscript; and Xiao-Hua Zong for technical assistance.
Supported by grant CA32551 from the National Institutes of Health (E.R.S.); grant 5P30-CA13330 from the Albert Einstein College of Medicine Cancer Center; an American Cancer Society Fellowship (A.J.H.); a Yamagiwa-Yoshida Memorial UICC International Cancer Study Grant (A.J.H.); and an American Society of Hematology fellowship (G.R.R.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
E. Richard Stanley, Dept of Developmental and Molecular Biology, Albert Einstein College of Medicine, 1300 Morris Park Ave, Bronx, NY 10461; e-mail: rstanley@aecom.yu.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal