Cytogenetic, genetic, and functional studies have demonstrated a direct link between deregulated Hoxa9 expression and acute myeloid leukemia (AML). Hoxa9 overexpression in mouse bone marrow cells invariably leads to AML within 3 to 10 months, suggesting the requirement for additional genetic events prior to AML. To gain further insight into how Hoxa9 affects hematopoietic development at the preleukemic stage, we have engineered its overexpression (1) in hematopoietic stem cells using retrovirus-mediated gene transfer and generated bone marrow transplantation chimeras and (2) in lymphoid cells using transgenic mice. Compared with controls, recipients ofHoxa9-transduced cells had an about 15-fold increase in transplantable lymphomyeloid long-term repopulating cells, indicating the capacity for this oncogene to confer a growth advantage to hematopoietic stem cells. In addition, overexpression ofHoxa9 in more mature cells enhanced granulopoiesis and partially blocked B lymphopoiesis at the pre–B-cell stage but had no detectable effect on T lymphoid development. Interestingly, despite specifically directing high expression of Hoxa9 in T and B lymphoid lineages, none of the Hoxa9 transgenic mice developed lymphoid malignancies for the observation period of more than 18 months.
Introduction
A growing body of evidence supports a role for members of the mammalian Hox homeobox gene family of transcription factors in the regulation of hematopoiesis.1In a subgroup of human myeloid leukemias, the HOXA9 gene is involved in a recurrent translocation between chromosomes 7 (NUP98) and 11 (HOXA9), resulting in a fusion transcript that encodes the NUP98-HOXA9 oncoprotein.2,3Recently, the expression of HOXA9 was also shown to be the single most highly correlating factor (of 6817 genes tested) for poor prognosis in human acute myeloid leukemia (AML),4 possibly indicating a broader role for this gene in human leukemia beyond that caused by its chromosomal translocations.
The leukemogenic potential of Hoxa9 was directly demonstrated by the development of AML in mouse bone marrow transplantation chimeras that received a graft of primitive hematopoietic cells engineered by retroviral gene transfer to overexpress Hoxa9.5,6 Although the latency for the development of AML in these chimeras is much shorter than reported for other similar Hox gene chimeras (ie, Hoxa10,Hoxb3, Hoxb8), overexpression of Hoxa9alone is not sufficient to induce leukemia, and additional somatic mutation(s) are needed.6-9 In leukemic transformation, genetic interactions have been demonstrated between Hoxa9and molecules such as Meis1 andE2A-PBX1,5,6 both of which can bind DNA cooperatively with Hoxa9.10 11 This suggests the presence of a leukemogenic complex, which includes Hoxa9 and potentially some of its DNA-binding cofactors.
In normal human CD34+ bone marrow cells, HOXA9is preferentially expressed in subfractions that are highly enriched for primitive long-term culture-initiating cells or myeloid progenitor cells, and its expression is significantly reduced in the erythroid lineage.12,13 This pattern of Hoxa9 expression is conserved in murine hematopoietic cells, with high levels detected in the Sca-1+ lineage-negative (Lin−) bone marrow subpopulation that is enriched for primitive hematopoietic cells, whereas much lower levels are present in the progenitor-depleted fraction (eg, Sca-1−Lin+).14,15Hoxa9 is also expressed in purified subpopulations of B and T lymphoid cells, although at levels much lower than those found in primitive bone marrow cells.14
Analysis of the Hoxa9 null mutant mice showed that lack of Hoxa9 caused a significant decrease in bone marrow myeloid and pre–B-cell progenitors, and Hoxa9−/− myeloid progenitors have a blunted response to granulocyte colony-stimulating factor, a growth factor known to enhance the production and release of granulocytes from the bone marrow.14 Although adult thymocyte development was only mildly abnormal inHoxa9−/− mice, the fetal thymus cellularity was reduced 5- to 10-fold, due in part to a delay in thymocyte progression from immature single-positive (T-cell receptor–negative) to double-positive (ie, CD4+CD8+) T cells.14,16 In contrast to lymphoid and myeloid progenitor cells in the Hoxa9−/− mice, no significant decrease was detected in more primitive cells measured as day 12 colony-forming unit-spleen (CFU-S) and long-term culture-initiating cells.14 However, recent studies using in vivo competitive assay have demonstrated profound anomalies in the ability of Hoxa9−/− hematopoietic stem cells (HSCs) to efficiently compete with wild-type cells in bone marrow transplantation experiments.17 These mice also have a prolonged suppression of hematopoiesis following sublethal irradiation, and quantitative studies have indicated a 4- to 12-fold reduction of HSC numbers in their bone marrow.17 These data, together with preferential expression of Hoxa9 in primitive cells, suggest a role for Hoxa9 in the regulation of primitive hematopoietic cells.
In this paper, we use 2 different mouse models (ie, transplantation chimeras and transgenics) to study the effects of Hoxa9overexpression on hematopoietic cells prior to the occurrence of leukemic transformation. We demonstrate, contrary to the phenotype of the Hoxa9−/− mice,17 that both the HSC and myeloid progenitor cell pools are significantly increased in the Hoxa9 transplantation chimeras. In both models, we also show that pre–B-cell progenitors are severely reduced by the overexpression of Hoxa9. In addition, evidence presented here suggests that in contrast to myeloid cells, lymphoid cells may be refractory to transformation induced by Hoxa9.
Materials and methods
Mice
Recipients of bone marrow cells were 7- to 12-week-old (C57BL/6J × C3H/HeJ)F1 ((B6C3)F1) mice, and donors were (C57BL/6Ly-Pep3b × C3H/HeJ)F1 ((PepC3)F1) mice. All mice were bred from parental strain breeders originally obtained from the Jackson Laboratories (Bar Harbor, ME), maintained in microisolator cages, and provided with sterilized food and acidified water at the animal facility of the Institut de recherches cliniques de Montréal. (B6C3)F1 and (PepC3)F1 mice are phenotypically distinguishable by their cell surface expression of different allelic forms of the Ly5 locus: (B6C3)F1 mice are homozygous for the Ly5.2 allotype, and (PepC3)F1 mice are heterozygous for the Ly5.1/Ly5.2 allotypes. For bone marrow transplantation procedures, lethally irradiated (900 cGy, 176 cGy/min, 137Cs γ rays; MARK1 irradiator from J. L. Shepherd and Associates, San Fernando, CA) (B6C3)F1(Ly5.2) mice were injected intravenously with 2 × 105 bone marrow cells from (PepC3)F1(Ly5.1) immediately after their cocultivation withHoxa9 or neo/EGFP viral producer cells as described previously.18
Generation of transgenic mice
The backbone pLIT3 vector has been previously described.19 Briefly, a 1.4 kilobase (kb) fragment from the Hoxa9 complementary DNA (cDNA) encompassing the complete reading frame was placed under the transcriptional control of the T-cell receptor (TCR) Vβ promoter and immunoglobulin (Ig) enchancer. Sequences upstream of the lckgene proximal promoter were also included. The human growth hormone (hGH) gene with a frameshift mutation in the coding region was inserted 3′ of the Hoxa9 cDNA to provide introns that appear to enhance the transgene expression. The transgene was injected into pronuclei of (C57Bl/6J × C3H)F2 hybrid zygotes. Pups resulting from transplantation of injected zygotes into pseudopregnant females were analyzed for the presence of the transgene by Southern blot analysis of tail DNA hybridized with the full-lengthHoxa9 cDNA. All experiments were performed on transgenic mice backcrossed into C57Bl/6J background for at least 5 generations.
Retroviral generation and infection of primary bone marrow cells
The generation of the MSCV-Hoxa9-pgk-neo retroviral vector has been described before,5 and theMSCV-Hoxa9-pgk-EGFP vector was generated by subcloning the full-length Hoxa9 cDNA at the EcoRI site upstream of a pgk-EGFP cassette into the murine stem cell virus (MSCV) retroviral vector. High-titer helper-free recombinantMSCV-Hoxa9-pgk-neo, MSCV-Hoxa9-pgk-EGFP,MSCV-pgk-neo, and MSCV-pgk-EGFP retroviruses were generated and tested as previously described.5 Bone marrow cells obtained from (PepC3)F1 mice injected 4 days earlier with 150 mg/kg of body weight of 5-fluorouracil were prestimulated in Dulbecco modified Eagle medium containing 15% fetal calf serum (FCS), 6 ng murine interleukin (IL)-3 per milliliter, 100 ng murine Steel factor per milliliter, and 10 ng human IL-6 per milliliter for 48 hours and then cocultivated on irradiated viral producer cells in the same medium with the addition of 6 μg of polybrene per milliliter. Loosely adherent and nonadherent cells were recovered and injected intravenously (2 × 105 cells per mouse) into (B6C3)F1 mice or directly cultivated in vitro as described below. Donor-derived repopulation in recipients was assessed by the proportion of leukocytes in peripheral blood, bone marrow, thymus, and spleen that expressed the Ly5.1 allelic form of the Ly5 locus, using the fluorescein isothiocyanate–conjugated anti-Ly5.1 antibody and flow cytometry.
Methylcellulose cultures and CRU assays
For myeloid clonogenic progenitor assays, cells were plated on 35 mm Petri dishes (Corning Costar, Cambridge, MA) in a 1.1 mL culture mixture containing 0.8% methylcellulose in α-medium supplemented with 10% FCS (Life Technologies, ON, Canada), 5.7% bovine serum albumin, 10−5 M β-mercaptoethanol (β-ME), 1 U human urinary erythropoietin per milliliter, 10% WEHI-conditioned medium (containing 50 ng IL-3 per milliliter) or 2% spleen cell–conditioned medium, 2 mM glutamine, 200 mg transferrin per milliliter, in the presence or the absence of 1.3 mg of G418 per milliliter. Bone marrow and spleen cells from the transplantation chimeras or the transgenic mice were plated at a concentration of 3 × 104 cells per milliliter (dish) and 2 × 106 cells per milliliter (dish), respectively. Colonies were scored on day 12 to 14 of incubation as derived from macrophage colony-forming units, granulocyte colony-forming units, granulocyte macrophage colony-forming units (CFU-GMs), or granulocyte, erythroid, macrophage, and megakaryocyte colony-forming units (CFU-GEMMs) according to standard criteria.20 For pre-B clonogenic progenitor assays, cells were plated at a concentration of 2 × 105/mL in 0.8% methylcellulose in α-medium supplemented with 30% FCS, 10−4 M β-ME, and 0.2 ng IL-7 per milliliter with or without 1.3 mg G418 per milliliter. Pre-B colonies were scored on day 7 of incubation. Evaluation of frequency of cells with long-term repopulating potential was done using the competitive repopulation unit (CRU) assay.21 Technical modifications of the procedure that were applied in this study have been reported previously.22 Briefly, bone marrow cells from primary Hoxa9-EGFP or EGFP(Ly5.1+) mice, killed at 4 weeks posttransplantation, were injected into lethally irradiated B6C3 (Ly5.1−) mice at varying dilutions (3 × 103 to 5 × 106bone marrow cells) along with 1 × 105 helper bone marrow cells from normal B6C3 (Ly5.1−) mice. The level of lymphoid and myeloid repopulation with Ly5.1+ donor-derived cells in these secondary recipients was evaluated at 16 weeks after transplantation by fluorescence-activated cell sorter (FACS) analysis of their peripheral blood. Recipients with more than 2% donor-derived peripheral blood lymphoid and myeloid leukocytes were considered to be repopulated by at least one CRU.
In vitro long-term culture assays for B cells
The frequency of primitive lymphoid B cells was determined with a limiting dilution analysis in the Whitlock-Witte in vitro culture system as described.23 Briefly, total bone marrow cells from control or Hoxa9 transgenic mice were cultured for 3 weeks on S17 stromal cells in RPMI medium containing 5% preselected FCS, 5 × 10−5 M β-ME, and 50 μg gentamicin per milliliter, and at concentrations ranging from 500 to 20 000 bone marrow cells per well. After 3 weeks each well was scored for the presence of nonadherent cells of B-cell origin. To confirm the B-cell origin of each culture, cells from multiple wells were stained with B220 and Mac1 and analyzed by FACS. The frequency of the Whitlock-Witte–initiating cells (WW-ICs) was then calculated by applying Poisson statistics to the proportion of negative wells at different dilutions.
Southern and Northern blot analyses
High molecular weight DNA isolated from bone marrow, spleen, and thymus of the transplantation chimeras was digested withKpnI, which cuts in the long terminal repeats (LTRs) to release the proviral genome or with EcoRI or NcoI to cut the provirus once to release DNA fragments specific to the proviral integration site(s). The genotyping of the Hoxa9transgenic mice was done by digesting genomic tail DNA withBglII. Total cellular RNA was isolated with the TRIzol reagent (Life Technologies) and Northern blot analysis performed as described.5 The probes used were aXhoI/SalI fragment of pMC1neo, the enhanced green fluorescent protein (EGFP) cDNA, or the full-length 1.4 kbHoxa9 cDNA, which had been labeled with 32P by random primer extension. To assess the relative amounts of total RNA loaded, membranes were probed for 18S RNA using end-labeled oligonucleotide 5′-ACGGTATCTGATCGTCCTCGAACC-3′.
Protein analysis
Preparation of cellular extracts of spleen and thymus derived from transgenics, Hoxa9, or control mice and Western analyses were performed as described.6 Briefly, 80 μg of proteins were separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred to Immobilon P membranes (Millipore, Bedford, MA), and then probed with polyclonal rabbit N-terminal Hoxa9 antibody (128-162 a.a.), kindly provided by Dr Takuro Nakamura, or with the monoclonal antibody PTP1D (Transduction Laboratories, Mississauga, ON, Canada). Bound antibodies were detected with horseradish peroxidase–conjugated antirabbit or antimouse antibodies (Sigma, St Louis, MO) followed by enhanced chemiluminescence (ECL; Amersham, Buckinghamshire, United Kingdom).
Results
Generation of bone marrow transplantation chimeras engineered to overexpress Hoxa9 in HSCs
In 2 separate experiments, bone marrow transplantation chimeras were generated with donor cells derived from congenic mice (Ly5.1+) and engineered by retroviral gene transfer to overexpress Hoxa9 (or a control “empty” vector) in primitive bone marrow cells. In both experiments, MSCV vectors were preferred over other Moloney-based retroviral vectors because the MSCV LTRs are modified to ensure high and persistent expression levels of the transgene in stem cells as well as in myeloid and lymphoid cells.24 25 The MSCV vectors used in these experiments contained 2 different selectable markers: the neor or the EGFP (Figure1A). The 2 experiments also differed in the dose of cells transplanted, with mice in the second experiment receiving a 10-fold higher number of Hoxa9-tranduced cells than those in experiment no. 1 (Table 1).
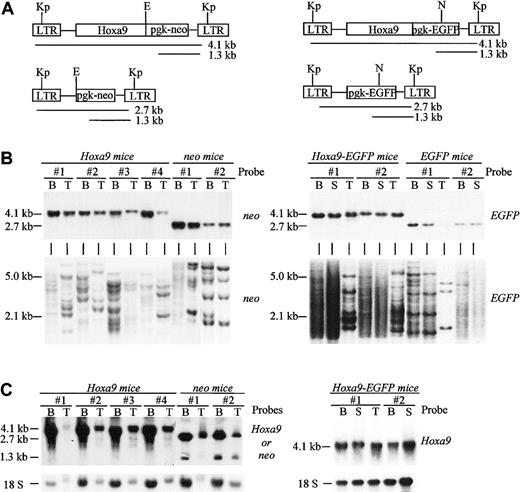
Demonstration of high-level regeneration of transplantation chimeras by retrovirally transduced cells.
(A) Diagrammatic representation of the integrated Hoxa9 and control proviruses used in this study. The expected sizes of the full-length LTR- and pgk-driven viral transcripts are shown. Restriction sites indicated are KpnI (Kp), EcoRI (E), and NcoI (N). (B) Southern blot analyses of genomic DNA isolated from the bone marrow, spleen, and thymus of Hoxa9and control chimeras. DNA was digested with KpnI to release the integrated Hoxa9-neo (4.1 kb), Hoxa9-EGFP(4.1 kb), neo (2.7 kb), and EGFP (2.7 kb) control viruses (upper panel). The DNA was also digested with eitherEcoRI or NcoI that cuts the integratedHoxa9 and neo or the Hoxa9-EGFP andEGFP proviruses once, respectively, thus generating a unique fragment for each proviral integration site (lower panel). The membranes were hybridized with a neo probe to detect theHoxa9-neo and neo proviruses and anEGFP probe to detect Hoxa9-EGFP andEGFP proviruses. (C) Northern blot analysis of total RNA (10 μg) isolated from bone marrow, thymus, and/or spleen of some of the mice described in panel B. The Hoxa9 probe detects the expected viral LTR-driven Hoxa9 messages and theneo probe both the LTR- and pgk-driven viral messages, as depicted in panel A. B indicates bone marrow; S, spleen; T, thymus.
Demonstration of high-level regeneration of transplantation chimeras by retrovirally transduced cells.
(A) Diagrammatic representation of the integrated Hoxa9 and control proviruses used in this study. The expected sizes of the full-length LTR- and pgk-driven viral transcripts are shown. Restriction sites indicated are KpnI (Kp), EcoRI (E), and NcoI (N). (B) Southern blot analyses of genomic DNA isolated from the bone marrow, spleen, and thymus of Hoxa9and control chimeras. DNA was digested with KpnI to release the integrated Hoxa9-neo (4.1 kb), Hoxa9-EGFP(4.1 kb), neo (2.7 kb), and EGFP (2.7 kb) control viruses (upper panel). The DNA was also digested with eitherEcoRI or NcoI that cuts the integratedHoxa9 and neo or the Hoxa9-EGFP andEGFP proviruses once, respectively, thus generating a unique fragment for each proviral integration site (lower panel). The membranes were hybridized with a neo probe to detect theHoxa9-neo and neo proviruses and anEGFP probe to detect Hoxa9-EGFP andEGFP proviruses. (C) Northern blot analysis of total RNA (10 μg) isolated from bone marrow, thymus, and/or spleen of some of the mice described in panel B. The Hoxa9 probe detects the expected viral LTR-driven Hoxa9 messages and theneo probe both the LTR- and pgk-driven viral messages, as depicted in panel A. B indicates bone marrow; S, spleen; T, thymus.
Absolute numbers of untransduced and transduced myeloid CFCs* transplanted per mouse
| Mouse group (n) . | Number of myeloid CFCs injected per mouse (×102)† . | |
|---|---|---|
| Untransduced . | G418r or EGFP+ . | |
| Experiment no. 1 | ||
| Neo (4) | 80 | 50 |
| Hoxa9 (4) | 65 | 45 |
| Experiment no. 2 | ||
| EGFP (4) | 180 | 360 |
| Hoxa9-EGFP (4) | 200 | 470 |
| Mouse group (n) . | Number of myeloid CFCs injected per mouse (×102)† . | |
|---|---|---|
| Untransduced . | G418r or EGFP+ . | |
| Experiment no. 1 | ||
| Neo (4) | 80 | 50 |
| Hoxa9 (4) | 65 | 45 |
| Experiment no. 2 | ||
| EGFP (4) | 180 | 360 |
| Hoxa9-EGFP (4) | 200 | 470 |
The number of transduced HSCs injected per mouse can be estimated based on our previous results, which determined the frequency of long-term repopulating cells at 1 per 100 CFCs,18 and estimation of gene transfer to long-term repopulating cells equal to that of CFCs.22
The number of transduced CFCs injected per mouse was determined as follows: (number of bone marrow cells injected per mouse) × (CFC frequency in the injected bone marrow inoculum) × (% of CFCs resistant to G418 or EGFP+).
As previously reported, overexpression of Hoxa9 eventually leads to AML in the transplantation chimeras,5 6 and recent studies have indicated that the time to leukemia onset is directly related to the dose of Hoxa9-transduced cells in the initial transplantation inoculum (U.T. and G.S., manuscript in preparation). To ensure that the effects of Hoxa9overexpression on hematopoietic regeneration would be assessed prior to the occurrence of leukemic transformation, bone marrow transplantation chimeras from experiments no. 1 and 2 were analyzed at 8 and 4 weeks posttransplantation, respectively. High levels of donor-derived repopulation (> 85% Ly5.1+ cells) were detected in the peripheral blood, bone marrow, thymus, and spleen in both groups of chimeras, and Southern blot analysis of DNA isolated from these same tissues demonstrated significant contribution to this regeneration by transduced cells (Figure 1B, upper panel). Clonal analysis performed with the same DNA but digested with a restriction enzyme that cuts once in the integrated proviral DNA indicated that numerous transduced clones contributed to both lymphoid (thymus and/or spleen) and myeloid (bone marrow) reconstitution in the Hoxa9 and control mice (Figure 1B, left side of lower left panel). Furthermore, the level of polyclonality was clearly related to the dose of transduced cells transplanted in each group of chimeras, with the Hoxa9-EGFPmice receiving the higher dose and being repopulated by a higher number of clones (Figure 1B, left side of lower right panel). Northern blot analysis of RNA extracted from these same organs demonstrated the high expression levels of the LTR-derived messenger RNA (Figure 1C).
Enhanced myelopoiesis and suppression of B lymphopoiesis by overexpression of Hoxa9
For all of the Hoxa9 mice analyzed at 4 or 8 weeks posttransplantation (Table 1), their bone marrow and thymic cellularity were within the normal range, whereas their spleens were mildly enlarged (0.15 ± 0.03 g for Hoxa9 vs 0.09 ± 0.01 g for controls). FACS analysis indicated that the relative and absolute numbers of mature myeloid cells (Mac1+) were slightly increased in the peripheral blood, bone marrow, and spleen of allHoxa9 chimeras (Figure 2A and data not shown). Morphologic evaluation of cell preparations from these organs indicated an increase in cells of the granulocyte lineage (data not shown). This effect of Hoxa9 on the myeloid lineage is further evidenced by detection of much higher proportion of EGFP-expressing Mac1+ cells (mostly granulocytes by morphology) in the Hoxa9-EGFP chimeras than in the controlEGFP mice (Figure 2A). Importantly, this “enhancing” effect of Hoxa9 appeared specific to the granulocytic lineage. This was evident by similar (or lower) proportions of EGFP-expressing cells in lymphoid lineages of Hoxa9 chimeras when compared with those detected in the control mice (Figure 2A for B cells [B220+] and data not shown for T cells [both CD4+ and CD8+]).
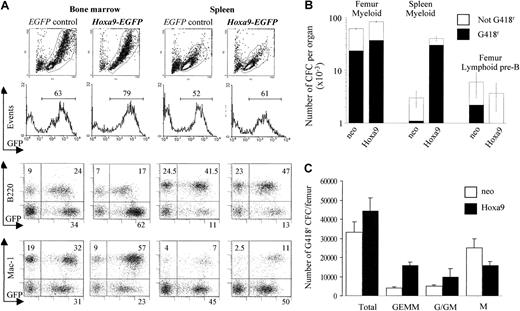
Overexpression of
Hoxa9 in the transplantation chimeras enhances myelopoiesis and suppresses B lymphopoiesis. (A) Flow cytometric analysis of hematopoietic cells from bone marrow and spleen of representative EGFP control and Hoxa9-EGFP mice 4 weeks after transplantation. For all dot-plots analyzed, EGFP expression is shown on the x-axis and that of Mac1 or B220 on the y-axis. Hoxa9 overexpression led to a significant increase in mature GFP+/Mac1+ myeloid cells in the bone marrow (Mac1+ myeloid cells ranged from 53%-56% as compared with 32%-38% for control GFP mice, n = 3 per group, P = .02) and a decrease in GFP+/B220+ (range 13%-25%) forHoxa9 mice compared to GFP controls (range 24%-38%,P = .05). (B) Results shown are the means ± SD of the numbers of transduced (G418r) and untransduced in vitro myeloid CFCs in bone marrow and spleen and of bone marrow pre-B CFCs in 4 neo and 4 Hoxa9 chimeras. (C) Analysis of the types of transduced (G418r) myeloid CFCs present in the bone marrow of Hoxa9 and neo chimeras. Well-isolated day 12 G418r colonies were randomly picked and examined after Wright-Giemsa staining. Results are expressed as the means ± SD of the colony types generated from bone marrow of 4Hoxa9 and 4 neo chimeras (20 colonies analyzed for each mouse). The granulocyte-erythrocyte-macrophage-megakaryocyte (GEMM) and granulocyte and granulocyte-macrophage (G/GM) colonies were significantly increased and the macrophage (M) significantly decreased in the Hoxa9 mice compared with the neocontrol mice.
Overexpression of
Hoxa9 in the transplantation chimeras enhances myelopoiesis and suppresses B lymphopoiesis. (A) Flow cytometric analysis of hematopoietic cells from bone marrow and spleen of representative EGFP control and Hoxa9-EGFP mice 4 weeks after transplantation. For all dot-plots analyzed, EGFP expression is shown on the x-axis and that of Mac1 or B220 on the y-axis. Hoxa9 overexpression led to a significant increase in mature GFP+/Mac1+ myeloid cells in the bone marrow (Mac1+ myeloid cells ranged from 53%-56% as compared with 32%-38% for control GFP mice, n = 3 per group, P = .02) and a decrease in GFP+/B220+ (range 13%-25%) forHoxa9 mice compared to GFP controls (range 24%-38%,P = .05). (B) Results shown are the means ± SD of the numbers of transduced (G418r) and untransduced in vitro myeloid CFCs in bone marrow and spleen and of bone marrow pre-B CFCs in 4 neo and 4 Hoxa9 chimeras. (C) Analysis of the types of transduced (G418r) myeloid CFCs present in the bone marrow of Hoxa9 and neo chimeras. Well-isolated day 12 G418r colonies were randomly picked and examined after Wright-Giemsa staining. Results are expressed as the means ± SD of the colony types generated from bone marrow of 4Hoxa9 and 4 neo chimeras (20 colonies analyzed for each mouse). The granulocyte-erythrocyte-macrophage-megakaryocyte (GEMM) and granulocyte and granulocyte-macrophage (G/GM) colonies were significantly increased and the macrophage (M) significantly decreased in the Hoxa9 mice compared with the neocontrol mice.
Consistent with the increase in mature myeloid cells, total myeloid progenitor numbers were increased both in the bone marrow and even more in the spleen of the Hoxa9 chimeras (Figure 2B). Most importantly, this elevation was largely caused by an increase in the absolute numbers of CFU-GMs and CFU-GEMMs, most of which were either resistant to G418 (Figure 2B,C) or expressed EGFP (data not shown). Together, these results suggest that the elevation in the number of mature granulocytes in the Hoxa9 chimeras is due to a significant increase in their progenitor cell numbers.
IL-7–responsive pre–B-cell progenitors (lymphoid pre-B) were also evaluated in the bone marrow of Hoxa9 mice. In contrast to the increase in Hoxa9-transduced myeloid progenitors, the total number of pre-B lymphoid progenitors was slightly reduced inHoxa9 mice, and virtually none were derived fromHoxa9-transduced cells (G418 resistant or EGFP+) (Figure 2B and data not shown). In addition, the fewHoxa9-transduced pre-B colonies detected were also smaller in size than either untransduced or neo-transduced pre-B colonies (data not shown). This suggests that while enhancing myelopoiesis Hoxa9 may selectively inhibit proliferation of pre-B progenitors. Because transplantation chimeras contain bothHoxa9-transduced and untransduced hematopoietic cells that can contribute to B lymphopoiesis, further analysis of thisHoxa9-induced B-cell defect was carried out in transgenic mice engineered to specifically express Hoxa9 in B and T lymphoid cells.
Regulated expression of Hoxa9 is required for normal B lymphopoiesis
Two different lines of Hoxa9 transgenic mice(Hoxa9-12 and Hoxa9-15) were produced using the pLIT3 vector, which was engineered to contain a number of lymphoid-specific regulatory elements19,26 (Figure3A). Previous expression studies have demonstrated that this vector directs high expression of the transgene in early and late B- (pro-B to mature peripheral B cells) and T-cell populations (double-negative [CD8−CD4−] to single-positive [CD8+ or CD4+]).19 26 In agreement with these studies, RNA levels of the Hoxa9 transgene were detected in the spleen (highest levels), thymus, and bone marrow of the 2Hoxa9 transgenic lines (Figure 3B). Hoxa9 protein levels were higher in the spleen of these transgenics than in controls (Figure 3C).
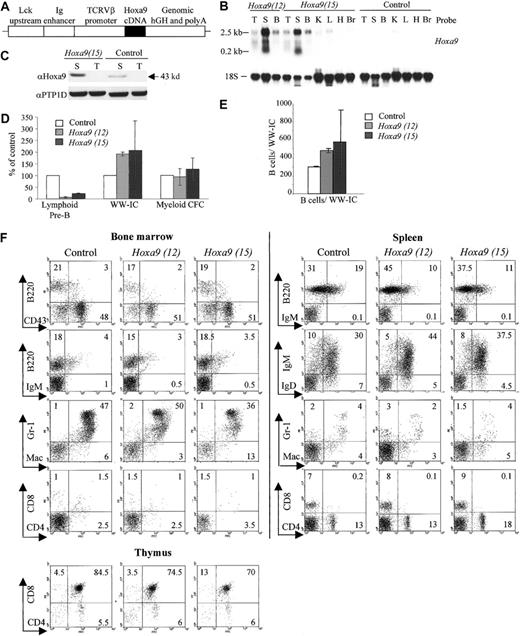
The numbers of lymphoid pre-B CFCs are severely reduced in
Hoxa9 transgenic mice. (A) Diagrammatic representation of the integrated Hoxa9 containing pLIT3 vector. This vector contains the full-length Hoxa9 cDNA, whose transcription is driven by the TCRVβgene, coupled to the μIgH chain enhancer and a fragment of the lck gene proximal promoter.19Sequences from the hGH gene, including splice sites and polyadenylation signal, are downstream of the Hoxa9 cDNA and are included in the primary transcript. (B) Total cellular RNA (10 μg) from the indicated tissues of homozygous mice from the 2Hoxa9 transgenic lines (12 and 15) and control littermates were analyzed by northern blot for the expression of theHoxa9 transgene using the full-length Hoxa9 cDNA as a probe. The expected 2.5 kb transgene transcript is detected mainly in bone marrow (B), spleen (S), and thymus (T), with low expression in lung (L), but is nondetectable in the kidney (K), heart (H), and brain (Br) tissues. A smaller transcript of 0.2 kb was detected in the spleen of both transgenic Hoxa9 lines. (C) Western blot analyses of total cell extracts from spleen (S) and thymus (T) obtained from transgenic Hoxa9 and control mice. Hoxa9 (upper panel) and PTP1D (loading control, bottom panel) levels are shown. (D) Evaluation of the numbers of lymphoid pre-B CFCs, WW-ICs, and myeloid CFCs in bone marrow of homozygous Hoxa9 transgenic mice. Results (mean ± SD) indicate percentages compared with controls (set at 100%) for 3 Hoxa9-12 and 3 Hoxa9-15 transgenic mice. (E) WW-IC–derived B220+ cells (at limiting dilution) were counted at day 21 of culture and are not statistically different (P < .13, 1-tailed Student t test with unequal variance, n = 4 mice in each group) between transgenics and controls. (F) Flow cytometric analysis of bone marrow, spleen, and thymic cells from the homozygous Hoxa9 transgenic mice.
The numbers of lymphoid pre-B CFCs are severely reduced in
Hoxa9 transgenic mice. (A) Diagrammatic representation of the integrated Hoxa9 containing pLIT3 vector. This vector contains the full-length Hoxa9 cDNA, whose transcription is driven by the TCRVβgene, coupled to the μIgH chain enhancer and a fragment of the lck gene proximal promoter.19Sequences from the hGH gene, including splice sites and polyadenylation signal, are downstream of the Hoxa9 cDNA and are included in the primary transcript. (B) Total cellular RNA (10 μg) from the indicated tissues of homozygous mice from the 2Hoxa9 transgenic lines (12 and 15) and control littermates were analyzed by northern blot for the expression of theHoxa9 transgene using the full-length Hoxa9 cDNA as a probe. The expected 2.5 kb transgene transcript is detected mainly in bone marrow (B), spleen (S), and thymus (T), with low expression in lung (L), but is nondetectable in the kidney (K), heart (H), and brain (Br) tissues. A smaller transcript of 0.2 kb was detected in the spleen of both transgenic Hoxa9 lines. (C) Western blot analyses of total cell extracts from spleen (S) and thymus (T) obtained from transgenic Hoxa9 and control mice. Hoxa9 (upper panel) and PTP1D (loading control, bottom panel) levels are shown. (D) Evaluation of the numbers of lymphoid pre-B CFCs, WW-ICs, and myeloid CFCs in bone marrow of homozygous Hoxa9 transgenic mice. Results (mean ± SD) indicate percentages compared with controls (set at 100%) for 3 Hoxa9-12 and 3 Hoxa9-15 transgenic mice. (E) WW-IC–derived B220+ cells (at limiting dilution) were counted at day 21 of culture and are not statistically different (P < .13, 1-tailed Student t test with unequal variance, n = 4 mice in each group) between transgenics and controls. (F) Flow cytometric analysis of bone marrow, spleen, and thymic cells from the homozygous Hoxa9 transgenic mice.
The effect of Hoxa9 on B lymphopoiesis was analyzed in 3- to 4-month-old transgenic mice and their littermate controls. In accordance with the Hoxa9 transplantation chimeras, the numbers of IL-7–responsive bone marrow pre-B progenitor cells were severely affected in both Hoxa9 transgenic lines (Figure3D). Furthermore, these transgenic pre-B progenitor cells also gave rise to much smaller colonies in vitro, demonstrating their reduced proliferative potential (data not shown). To determine whether more primitive cells of the B lineage were also affected in these mice, limiting dilution analysis was used to evaluate the number of bone marrow cells capable of initiating long-term B-cell cultures (WW-ICs23). The results failed to identify a decline in the number of WW-ICs in the bone marrow of both Hoxa9 lines (Figure 3D). Because the experimental conditions necessary to purify WW-ICs are still undefined, it cannot be excluded that absent or low transgene expression in WW-ICs is in part responsible for these results. The mature B-cell output per single WW-IC was also evaluated both quantitatively and qualitatively by FACS analysis. Each WW-IC from the Hoxa9 transgenics generated similar numbers of cells, most of which were B220+IgM−, demonstrating that the in vitro proliferation and differentiation potential of their WW-ICs was not different from that of their control littermates (Figure3E). These results indicate that high levels of Hoxa9 are selectively nonpermissive to a subset of B-cell progenitor(s) that are detected by in vitro culture conditions containing high levels of IL-7. This incomplete block in B-cell development at the stage of pre-B progenitors did not translate into reduced levels of mature B cells in the bone marrow or spleen of Hoxa9transgenics (Figure 3F).
In contrast to the Hoxa9 transplantation chimeras, there was no increase in bone marrow or spleen myeloid CFU-GMs or CFU-GEMMs (Figure 3D) or in the numbers of mature granulocytes in theseHoxa9 transgenic mice (Figure 3F). This is consistent with the selective expression of the transgene in the lymphoid compartment. Importantly, these results also excluded enhanced granulopoiesis as a contributing factor for the development of the above-described B-cell phenotype (either in Hoxa9 transgenic mice or transplantation chimeras).
Both lines of Hoxa9 transgenic mice have now been monitored for more than 18 months (n = 9 mice), and none of these mice have spontaneously developed any hematologic malignancies.
Together, these results and those presented above for the transplantation chimeras clearly indicate that inappropriately high levels of Hoxa9 expression perturb normal regulation of myeloid and pre-B lymphoid progenitor cells.
Enhanced CRU pool size in recipients ofHoxa9-transduced cells
Based on the preferential expression of Hoxa9 in primitive hematopoietic cells and the finding that HSC numbers inHoxa9 homozygous mutant mice are severely reduced,17 we sought to test the hypothesis that overexpression of Hoxa9 in primitive bone marrow cells might lead to expansion of their pool size.
The HSC numbers in recipients of control andHoxa9-transduced bone marrow cells were determined using the CRU assay previously validated to detect HSCs based on their functional definition, ie, being capable of both lymphoid and myeloid differentiation as well as long-term repopulation in transplanted mice (here secondary mice; see “Materials and methods”). At 4 weeks posttransplantation, the CRU frequency in bone marrow ofHoxa9 chimeras was increased about 15-fold over that measured for control chimeras killed at the same time and generated with equivalent numbers of EGFP-transduced CRUs (Figure4A). Hoxa9-induced expansion of CRUs was confirmed at numerous time points following bone marrow transplantation in several independent experiments (U.T. and L.J., unpublished observations, 2000).
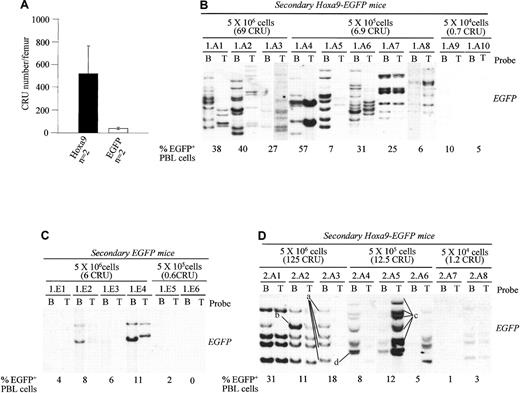
Overexpression of
Hoxa9 enhances self-renewal divisions of HSCs.(A) Evaluation of HSC numbers in bone marrow of Hoxa9-EGFPand EGFP chimeras using the CRU assay. Results are expressed as the mean ± SD of the CRU numbers in one femur ofHoxa9-EGFP (n = 2) and EGFP chimeras (n = 2) described in Table 1. (B-D) Southern blot analyses of proviral integration patterns in DNA isolated from bone marrow and thymus of the secondary recipients of (B) primary Hoxa9-EGFP (mouse Hoxa9-EGFP no. 1, Figure 1B); (C) primary EGFP mouse (mouse EGFP no. 1, Figure 1B) and (D) from a primary Hoxa9-EGFPrecipient receiving a lower dose on transduced cells (clonal analysis of this primary is not available). Secondary recipients were used to quantitate the CRU numbers and were all killed at 16 weeks posttransplantation. The DNA was digested with NcoI, which cuts once in the integrated provirus, generating a DNA fragment specific for each proviral integration site. The membranes were hybridized with an EGFP-specific probe to detect proviral fragments. Each secondary mouse is identified with a number. The number of bone marrow cells, as well as the estimated number of CRU cells received by each secondary recipient, is indicated above the blots. The percentages of EGFP expressing peripheral white blood cells in each of the secondary mice at 16 weeks after transplantation is indicated below each blot. The level of donor-derived (ie, Ly5.1+) peripheral blood leukocyte repopulation in these secondary mice shown were as follows: (B) 1.A1 (72%), 1.A2 (66%), 1.A3 (42%), 1.A4 (57%), 1.A5 (20%), 1.A6 (44%), 1.A7 (30%), 1.A8 (11%), 1.A9 (12%), 1.A10 (8%); (C) 1.E1 (11%), 1.E2 (14%), 1.E3 (10%), 1.E4 (15%), 1.E5 (5%), 1.E6 (0.5%); and (D) 2.A1 (55%), 2.A2 (25%), 2.A3 (47%), 2.A4 (12%), 2.A5 (15%), 2.A6 (11%), 2.A7 (5%), and 2.A8 (8%). The criteria used to define each stem cell clone are as follows: (i) same pattern of proviral integration in thymus (T cells) and bone marrow (myeloid and B cells); (ii) for clones containing more than one integrated provirus (eg, clone “a”), each band must be of the same intensity and always be transmitted together (eg, clone “a” in mouse 2.A3, 2.A1, 2.A8, and 2.A2 has 4 integrated proviruses; clone “b” also present in bone marrow of mouse 2.A2 is different because it does not always transmit with clone “a”; see thymus in mouse 2.A1 or mouse 2.A8 for examples). B indicates bone marrow; T, thymus; PBL, peripheral blood leukocytes.
Overexpression of
Hoxa9 enhances self-renewal divisions of HSCs.(A) Evaluation of HSC numbers in bone marrow of Hoxa9-EGFPand EGFP chimeras using the CRU assay. Results are expressed as the mean ± SD of the CRU numbers in one femur ofHoxa9-EGFP (n = 2) and EGFP chimeras (n = 2) described in Table 1. (B-D) Southern blot analyses of proviral integration patterns in DNA isolated from bone marrow and thymus of the secondary recipients of (B) primary Hoxa9-EGFP (mouse Hoxa9-EGFP no. 1, Figure 1B); (C) primary EGFP mouse (mouse EGFP no. 1, Figure 1B) and (D) from a primary Hoxa9-EGFPrecipient receiving a lower dose on transduced cells (clonal analysis of this primary is not available). Secondary recipients were used to quantitate the CRU numbers and were all killed at 16 weeks posttransplantation. The DNA was digested with NcoI, which cuts once in the integrated provirus, generating a DNA fragment specific for each proviral integration site. The membranes were hybridized with an EGFP-specific probe to detect proviral fragments. Each secondary mouse is identified with a number. The number of bone marrow cells, as well as the estimated number of CRU cells received by each secondary recipient, is indicated above the blots. The percentages of EGFP expressing peripheral white blood cells in each of the secondary mice at 16 weeks after transplantation is indicated below each blot. The level of donor-derived (ie, Ly5.1+) peripheral blood leukocyte repopulation in these secondary mice shown were as follows: (B) 1.A1 (72%), 1.A2 (66%), 1.A3 (42%), 1.A4 (57%), 1.A5 (20%), 1.A6 (44%), 1.A7 (30%), 1.A8 (11%), 1.A9 (12%), 1.A10 (8%); (C) 1.E1 (11%), 1.E2 (14%), 1.E3 (10%), 1.E4 (15%), 1.E5 (5%), 1.E6 (0.5%); and (D) 2.A1 (55%), 2.A2 (25%), 2.A3 (47%), 2.A4 (12%), 2.A5 (15%), 2.A6 (11%), 2.A7 (5%), and 2.A8 (8%). The criteria used to define each stem cell clone are as follows: (i) same pattern of proviral integration in thymus (T cells) and bone marrow (myeloid and B cells); (ii) for clones containing more than one integrated provirus (eg, clone “a”), each band must be of the same intensity and always be transmitted together (eg, clone “a” in mouse 2.A3, 2.A1, 2.A8, and 2.A2 has 4 integrated proviruses; clone “b” also present in bone marrow of mouse 2.A2 is different because it does not always transmit with clone “a”; see thymus in mouse 2.A1 or mouse 2.A8 for examples). B indicates bone marrow; T, thymus; PBL, peripheral blood leukocytes.
The expression of EGFP that was detected in hematopoietic cells of secondary Hoxa9-EGFP chimeras, positive for long-term donor-derived repopulation (Ly5.1+) in the CRU assay (Figure 4B, percentages shown below blots and data not shown), demonstrates that the enhanced CRU regeneration in theHoxa9-EGFP mice was indeed caused by transduced cells. A more definitive proof of this was obtained by Southern blot analysis of proviral integration sites in DNA isolated from hematopoietic tissues of secondary recipients from a representative primaryHoxa9-EGFP (Figure 4B). This analysis detected numerousHoxa9-transduced clones, some of which had both lymphoid (thymus) and myeloid (bone marrow) repopulating potential (ie, totipotent, Figure 4B, mice 1.A3, 1.A4, 1.A7, 1.A8). In contrast to theHoxa9-EGFP mice, similar analysis of the secondary recipients of the control EGFP mice (ie, EGFP expression and clonality by proviral integration) only detectedEGFP-transduced clones in the secondary recipients receiving the highest number (5 × 106 bone marrow cells) of donor cells (Figure 4C and data not shown). Furthermore, as might be expected from previous studies,27 only few totipotentEGFP-transduced clones could be detected in the secondary recipients of one of the control mice (Figure 4C). Thus,Hoxa9 overexpression greatly enhances the regenerative potential of CRUs (HSCs) following bone marrow transplantation.
Self-renewal divisions of Hoxa9-overexpressing HSCs (same totipotent clone detected in 2 or more secondary mice) could not be demonstrated in these mice, likely because the transplantation dose was too high. However, self-renewal could be shown using another cohort of primary mice in which each mouse was initially injected with only about 6 Hoxa9-transduced long-term repopulating cells (Figure 4D). Analysis of proviral integration sites by Southern blot analysis of DNA isolated from secondary recipients of one of these primary Hoxa9-EGFP mice detected 4 totipotentHoxa9-transduced clones (Figure 4D, clones “a”-“d”). Each of these 4 clones could be detected in 2 or more secondary recipients, thus demonstrating their self-renewal in the primaryHoxa9-EGFP mouse (Figure 4D). For example, the totipotent clone “a” is detected in 6 secondary mice (ie, mice 2.A1, 2.A2, 2.A3, 2.A4, 2.A6, 2.A8), clone “b” in 3 (2.A1, 2.A2, 2.A3) and clone “c” and “d” in 2 (2.A4, 2.A5).
At limiting dilution (eg, where only one transduced CRU is actively contributing to repopulation in a secondary recipient), it is possible to estimate the proliferative potential of individual CRUs, whether or not transduced by Hoxa9. This is done by measuring the level of donor-derived repopulation for lymphoid and myeloid cells in these selected mice. Donor-derived repopulation (ie, %Ly5.1+cells) was similar between secondary recipients of oneHoxa9- or EGFP-tranduced CRUs (2%-5%). This indicates that overexpression of Hoxa9 enhances CRU regeneration in vivo but does not enhance the mature end cell output of individual CRUs.
Discussion
In the studies presented here, we demonstrate that overexpression of Hoxa9 greatly enhances HSC regeneration in transplantation chimeras. Increased Hoxa9 levels also lead to the expansion of myeloid colony-forming cells (CFCs), resulting in a net increase of mature granulocytes present in these chimeras. In contrast to myeloid cells, overexpression of Hoxa9 was also shown to induce a partial block in B lymphopoiesis at the pre-B progenitor stage. These data, together with preferential expression ofHoxa9 in primitive hematopoietic cells12 and the reduction in stem cell numbers in Hoxa9 homozygous mutant mice,17 suggest that this gene might qualify as a regulator of primitive hematopoietic cells.
A variety of transcription factors such as SCL and AML-1 initially identified by their involvement in human leukemia-associated chromosomal rearrangements28,29 were later demonstrated by gene targeting to be key regulators of HSC proliferation and differentiation.30 The involvement of such transcription factors in leukemic transformation highlights the link between HSCs (their properties and specific genes they express) and leukemic transformation. One could thus hypothesize that factors that regulate HSC function might have targets in common with those involved in leukemic transformation. The evidence that Hoxa9 is involved in chromosomal translocations in human leukemias and that its levels regulate stem cell numbers suggests that Hoxa9 may perform critical roles in primitive bone marrow cells.
Intriguingly, overexpression of Hoxb4 in a similar transplantation model has also been observed to lead to marked enhancement in the regenerative potential of HSCs.18However, in sharp contrast, overexpression of Hoxb4 did not perturb differentiation of myeloid or lymphoid progenitors in vivo and did not change the proportion of mature cells in the various hemopoietic organs.18,22,34 Like Hoxb4 and 2 Hedgehog proteins recently reported,31 Hoxa9 can now be considered as one of the rare molecules with an intrinsic ability to expand stem cells. The mechanisms involved in this expansion could include enhancement in self-renewal of the stem cell population or enhanced survival of the stem cell pool.
Another significant outcome of Hoxa9 overexpression described here is its negative effect on early B-cell development, as evidenced by a decrease in the numbers of pre-B clonogenic progenitor cells both in the Hoxa9 transplantation chimeras and transgenic mice with targeted expression of Hoxa9 to the lymphoid compartment. This reduction of pre-B CFCs seems to be cell-autonomous because untransduced pre-B progenitors were normal in numbers and in their proliferative potential in these transplantation chimeras (Figure 2B and data not shown). With respect to the significant reduction of pre–B-cell progenitors in both of our mouse models, it is intriguing to observe that mature B cells are either slightly reduced (chimeras) or normal (transgenics). Although the nature of the discrepancy between progenitor and mature cell numbers is currently unknown, it may in part be explained by compensatory mechanisms known to regulate mature peripheral B-cell numbers.32 In contrast to the reduction of pre-B CFCs, the numbers of the earliest committed B lymphoid progenitors, detectable as WW-ICs, were within the normal range in the transgenic mice. This indicated that Hoxa9 overexpression does not alter B lymphocyte commitment (from HSCs to WW-ICs). In theHoxa9−/− mice, pre-B progenitors are also reduced in numbers and have reduced proliferative potential,14 suggesting that Hoxa9 levels need to be tightly regulated for normal B-cell development.
Collectively, our results show that Hoxa9 has the potential to expand HSCs when overexpressed in a bone marrow transplantation model. Its involvement in chromosomal translocation in human leukemia together with the evidence that mouse HSCs lacking Hoxa9compete poorly in a bone marrow transplantation model may suggest that this gene is a bona fide regulator of stem cell function. Our studies demonstrating Hoxa9-induced expansion of myeloid precursors may provide some initial clues on the apparently restricted ability of this oncogene to transform cells with myeloid differentiation capacities. The lack of lymphoid leukemia induced by Hoxa9 even in the context where it provides a marked selective advantage to lymphomyeloid long-term repopulating cells is most intriguing. This contrasts with other genes involved in human leukemias.33
The authors acknowledge Dr Trang Hoang for critical reading of this manuscript. Nadine Mayotte and Simon Girard are acknowledged for expert technical assistance and Marie-Eve Leroux and Stephane Matte for their expertise and help regarding the maintenance and manipulation of the animals kept at the SPF facility. Dr Robert G. Hawley is acknowledged for his MSCV vectors; and Dr Takuro Nakamura for providing us with the Hoxa9 antibody. The support of Nathalie Tessier and Eric Massicotte for FACS analyses is also acknowledged. We also thank Christian Charbonneau for his help in preparing the figures and Dr Qinzhang Zhu for the generation of transgenic Hoxa9 transgenic mice.
Supported by a grant from the National Cancer Institute of Canada to G.S. and National Institutes of Health grant RO1-DK48642 to H.J.L.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Guy Sauvageau, Clinical Research Institute of Montréal, 110 Pine Ave West, Montréal, Québec, Canada, H2W 1R7; e-mail: sauvagg@ircm.qc.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal