Abstract
CC chemokine receptor (CCR) 9, the receptor for the CC-chemokine CCL25/thymus-expressed chemokine (TECK), is mainly expressed by thymocytes and by intraepithelial (IEL) and lamina propria lymphocytes of the small intestine. To study the biologic role of CCR9, a mouse strain was generated in which the CCR9 gene was deleted. In spite of the high level of CCR9 found in double- and single-positive thymocytes and of the expression of its corresponding ligand on thymic stromal cells, CCR9 deletion had no major effect on intrathymic T-cell development. It was noted that there was only a one-day lag in the appearance of double-positive cells during fetal ontogeny in CCR9−/− thymi. When tested in chemotaxis assay, thymocytes isolated from CCR9−/− mice failed to respond to TECK/CCL25. Taken together, these results suggest that in thymocytes, CCR9 is the only physiologic receptor for TECK/CCL25, and that it is dispensable for proper T-cell development. Bone marrow pre-pro–B cells migrate in response to TECK/CCL25, but more mature B cells do not. Consistent with this observation, it was shown that there are fewer pre-pro–B cells in CCR9−/−mice than in wild-type mice. However, this diminution does not appear to have a detectable effect on the generation of a normal complement of mature B cells. Finally, it was shown that in the small intestine of CCR9-deficient mice, the intraepithelial T-cell–to–epithelial cell ratio is decreased, an observation that can be accounted for by a marked diminution of the T-cell receptor γδ+ compartment.
Introduction
During their development in the thymus, T cells migrate from the outer capsule to the inner medulla, a process that allows their sequential interaction with different types of stromal cells. Following their exportation to the periphery, a sophisticated network of chemokines control their appropriate navigation to and from secondary lymphoid organs.1-3 Likewise, the migration of developing T and B cells within their respective primary lymphoid organs is likely under the control of chemokines.4,5Consistent with the implication of G protein–coupled chemokine receptors in the intrathymic trafficking of T cells, the latter is inhibited by pertussis toxin.6 Moreover, developing thymocytes express several chemokine receptors and their corresponding ligands produced by thymic stromal cells.7
Recent studies, including our own, have described a novel thymus-expressed chemokine (TECK)/CCL258,9 and its receptor, CC chemokine receptor 9 (CCR9).9-12TECK/CCL25 is predominantly expressed by most thymic epithelial cortical cells, by a few thymic epithelial medullary cells, and by CD11b− thymic dendritic cells.9,13 The level of CCR9 transcripts undergoes a 10-fold increase during the double-negative (DN) to double-positive (DP) transition. Among thymocytes, DP thymocytes have the highest CCR9 expression and this correlates with their ability to migrate in response to TECK/CCL25. CCR9 transcripts subsequently decrease in single-positive (SP) thymocytes. Therefore, TECK/CCL25 may direct the centripetal intrathymic progression followed by developing T cells, and/or constitute a thymic retention factor since CD4+ SP cells have been shown to lose their ability to respond to TECK/CCL25 at the latest identifiable stage of thymocyte maturation. Chemokines are also postulated to play a role in the importation of lymphoid precursor cells to the thymus. Along this line, a number of chemokines (TECK/CCL25, stromal cell–derived factor 1 [SDF-1/CXCL12], and secondary lymphoid tissue chromoattractant [SLC/CCL21]) are expressed in the alymphoid thymic anlage present at day 12.5 of gestation.14 Moreover, TECK is readily detectable in fetal thymus at day 15 of gestation.9 14
TECK/CCL25 is also abundantly expressed in the epithelial cells lining the small intestine,9 and in humans most of the intestinal intraepithelial (IEL) and lamina propria T cells express CCR9.15-17 Furthermore, a small CCR9+subset of human peripheral T cells exists and is probably endowed with gut-homing properties.15-17 Therefore, in addition to its putative role during intrathymic T-cell development, TECK/CCL25 may play an additional role in the selective extravasation of memory intestinal T lymphocytes and/or in the migration of CCR9+lymphocytes once they have crossed the vascular endothelium and entered the intestinal tissue. To examine the spectrum of in vivo activities that are mediated by CCR9, we generated CCR9-deficient mice and characterized the effect of this mutation on the development of the B- and T-cell lineages.
Materials and methods
Generation of CCR9-deficient mice
Two mouse CCR9 genomic clones were isolated from a 129/Ola genomic phage library and used to determine the exon-intron organization of the CCR9 gene. In the targeting construct depicted in Figure 1A, a 2.4-kb fragment that includes exon 3 of the CCR9 gene was replaced by a 1.4-kb loxP-flanked neomycine resistance cassette. In the resulting construct, the neomycine cassette is flanked by homologous arms of 2.9 kb and 3.2 kb, and abutted to a thymidine kinase (TK) expression cassette. TheScaI-linearized construct was electroporated into CK35 129 mouse embryonic stem (ES) cells.18 Colonies resistant both to G418 (300 μg/mL) and to gancyclovir (2 μM) were screened by southern blot for homologous recombination using 5′ and 3′ single-copy probes. Using HindIII-digested DNA, the 5′ probe detects an 8.5-kb recombinant fragment and a 10.5-kb wild-type fragment, whereas on EcoRI-digested DNA, the 3′ probe hybridizes to a 9.5-kb recombinant fragment and to a 13-kb wild-type fragment. One recombinant ES clone (VC9) was found capable of germline transmission. The neomycine resistance gene present within the targeted chromosome was removed by crossing the resulting chimeric males to Deleter mice.19 Germline transmission of the targeted allele was detected by southern blot and polymerase chain reaction (PCR) using the following primers: 5′-CATCCACACTGTGAGTGTTCA-3′ (sense), 5′-GGCATTCAACCTCAGAATGTT-3′ (antisense). The CCR9-deficient mice were kept under specific pathogen-free conditions and backcrossed on a C57BL/6 background.
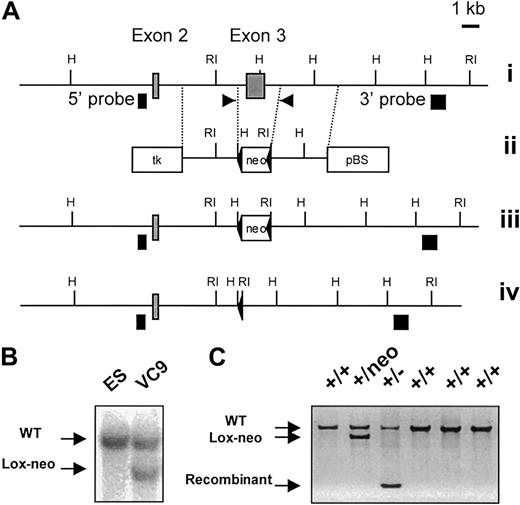
Generation and identification of CCR9-deficient mice.
(A) Partial restriction map of the wild-type CCR9 gene (i). ░, exons 2 and 3. The restriction sites are RI, EcoRI, and H,HindIII. Targeting vector used for the deletion of exon 3 (ii). Two diagnostic HindIII and EcoRI restriction sites were introduced at each end of the loxP-flanked neomycin resistance gene (neo). Open boxes correspond to the thymidine kinase expression cassette (tk), to the loxP-flanked neomycin gene, and to the pBluescript IIKS+ vector (pBS). Structure of the targeted allele following homologous recombination (iii). Final structure of the targeted allele after removal of the neomycin resistance gene via cre-mediated recombination (iv). The 5′ and 3′ single-copy probes used to verify the targeting events are indicated as ▪, and the position of the primers used to monitor the germline transmission of the intented mutation is indicated by arrows. (B) Southern blot analysis of the recombinant ES cell clone VC9 that gave germline transmission prior to deletion of the neomycin resistance gene. DNA was digested withEcoRI and hybridized with the 3′ single-copy probe. (C) DNA-PCR analysis of wild-type (+/+), neomycin-deleted heterozygous (+/−), and neomycin-nondeleted heterozygous (+/neo) littermates using the pair of primers shown in (A). After deletion of the neomycin resistance gene, the targeted CCR9 allele gave an amplified PCR product of 330 bp. Amplified products were run on an agarose gel and stained with ethidium bromide.
Generation and identification of CCR9-deficient mice.
(A) Partial restriction map of the wild-type CCR9 gene (i). ░, exons 2 and 3. The restriction sites are RI, EcoRI, and H,HindIII. Targeting vector used for the deletion of exon 3 (ii). Two diagnostic HindIII and EcoRI restriction sites were introduced at each end of the loxP-flanked neomycin resistance gene (neo). Open boxes correspond to the thymidine kinase expression cassette (tk), to the loxP-flanked neomycin gene, and to the pBluescript IIKS+ vector (pBS). Structure of the targeted allele following homologous recombination (iii). Final structure of the targeted allele after removal of the neomycin resistance gene via cre-mediated recombination (iv). The 5′ and 3′ single-copy probes used to verify the targeting events are indicated as ▪, and the position of the primers used to monitor the germline transmission of the intented mutation is indicated by arrows. (B) Southern blot analysis of the recombinant ES cell clone VC9 that gave germline transmission prior to deletion of the neomycin resistance gene. DNA was digested withEcoRI and hybridized with the 3′ single-copy probe. (C) DNA-PCR analysis of wild-type (+/+), neomycin-deleted heterozygous (+/−), and neomycin-nondeleted heterozygous (+/neo) littermates using the pair of primers shown in (A). After deletion of the neomycin resistance gene, the targeted CCR9 allele gave an amplified PCR product of 330 bp. Amplified products were run on an agarose gel and stained with ethidium bromide.
RNase protection assay
For multiplex chemokine receptor transcript analysis, total cellular RNA was isolated from thymi and spleens using TRIzol (Gibco-BRL Life Technologies, Paisley, United Kingdom), and analyzed by ribonuclease protection assay using an mCR-5 RiboQuant custom mouse template set (BD Pharmingen, Heidelberg, Germany), in which CCR9, CCR7, and CCR6 templates were included. Briefly,32P-labeled riboprobes were mixed with 20 μg of RNA, incubated at 56°C for 12 to 16 hours, and then treated with a mixture of RNases A and T1 and proteinase K. RNase-protected32P-labeled RNA fragments were separated on denaturing polyacrylamide gels and the intensity of the bands evaluated with a Fuji imaging plate system.
Flow cytometric analysis
Flow cytometric analyses were performed on mice between 4 weeks and 8 weeks of age as described.20 All the antibodies used in this study were purchased from BD Pharmingen. Prior to their analysis, bone marrow B cells from wild-type, heterozygous, or CCR9-deficient mice were enriched by positive selection using MACS mouse B220 microbeads (Miltenyi Biotech, Auburn, CA). The purified cells were analyzed using 2- and 3-color flow cytometry for the expression of B220, CD4, HSA, CD43, and CD25.
Mice
Timed matings of CCR9-deficient mice were set up as described.21 Briefly, CCR9-deficient males were left overnight with 3 CCR9-deficient females. The next morning was termed day 0.5. Fetal thymi were analyzed between day 14.5 and day 18.5 of gestation (referred to as E14.5, E18.5, etc.).
Chemotaxis assay
Migration assays were performed in 24-well Transwell plates (Corning Costar, Cambridge, MA) with 5-μm pore polycarbonate filters. Thymocytes from wild-type and CCR9-deficient mice were resuspended at a density of 107 cells/mL in RPMI containing 0.5% bovine serum albumin. After incubation at 37°C for 1 hour, 100 μL of each cell suspension was placed in the upper chamber, and 600 μL of medium or of a given dilution of recombinant mouse TECK/CCL25 (R&D Systems, Minneapolis, MN) was placed in the lower chamber. After incubation for 4 hours at 37°C, the upper chamber was removed, and the cells in the lower chamber were resuspended and transferred to tubes. After centrifugation, cells were resuspended in 100 μL of medium containing phycoerythrin (PE)–conjugated anti-CD4 and allophycocyanin (APC)–conjugated anti-CD8 monoclonal antibodies (mAbs). After staining, 30 μL of the suspension was used for manual counting, and the remainder was counted and analyzed on a FACSCalibur (Becton Dickinson Biosciences, Heidelberg, Germany).
Proliferation assay
Spleen cells were placed in 96-well flat-bottomed microtiter plates at 0.5 × 105 splenic T lymphocytes per well in 200 μL of culture medium. For anti-CD3 stimulation, serial dilutions of the 2C11 mAb were precoated overnight on microtiter plates prior to the addition of spleen cells. Stimulations with staphyloccocal enterotoxins and mitogens were performed using the following final concentrations: staphyloccocal enterotoxin A (SEA; SERVA, Feinbiochemica, Heidelberg, Germany), 100 ng/mL; staphyloccocal enterotoxin B (SEB; Toxin Technology, Sarasota, FL), 1 μg/mL; and Conamycin (Sigma, St Louis, MO), 2.5 μg/mL. In each experiment, cells were also assayed for their ability to respond to a combination of PMA (5 ng/mL, Sigma) and ionomycin (250 ng/mL, Sigma). After 40 hours at 37°C, proliferation was assayed by incorporation of [3H] thymidine (0.037 MBq [1 μCi]/well). After incubation for 8 hours at 37°C, cells were transferred onto glass fiber filters (Packard, Meriden, CT) by an automated cell harvester and incorporation of [3H] thymidine was measured with a scintillation counter.
IEL isolation and histologic studies
IELs from the small intestine were isolated as previously described.22 In brief, Peyer patches were removed and, after flushing with phosphate-buffered saline (PBS), the gut was opened on a wet linen square. The mucosa was scraped with a scalpel, then dissociated by stirring in 50 mL of Medium 199 (Gibco-BRL Life Technologies) containing 10% newborn calf serum and dithioerythritol (1 mM) for 15 minutes at room temperature. After centrifugation, the pellet was resuspended in PBS containing 10% newborn calf serum, vortexed for 3 minutes, and rapidly passed through a glass wool column (1.6 g packed in a 20-mL syringe; Fisher Bioblock Scientific, Illkirch, France). IELs were further purified on a Ficoll/Isopaque gradient (Nyco-Prep 1.077A; Nycomed Amersham, Buckinghamshire, United Kingdom) and stained with mAbs for flow cytometric analysis. For histologic studies, a 1-cm piece of small intestine, taken 3 cm below the pylorus, was properly oriented on filter paper and fixed in Carnoy fluid for 24 hours. Paraffin-embedded sections were prepared and stained with periodic acid Schiff (PAS) and hematoxylin.
Results
Generation of CCR9-deficient mice
Exon 3 of the CCR9 gene, encoding 362 of 369 amino acids, was deleted through homologous recombination using the strategy depicted in Figure 1A. Targeted ES cells were used to generate mice that transmitted the intended mutation through the germline (Figure 1C). Considering that many CC chemokine receptor genes are clustered, and that the introduction of a neomycin resistance cassette within a given gene cluster may inadvertently affect the expression of contiguous genes, the neomycin cassette used to select the recombinant ES cells was flanked with loxP sites and subsequently deleted from the chromosome by crossing chimeric males onto females of the Deleter strain (Figure 1C). Heterozygous mutants with a deleted neomycin cassette were interbred to generate homozygous mutant mice. The CCR9-deficient mice were viable, fertile, and showed no gross morphologic or developmental abnormalities.
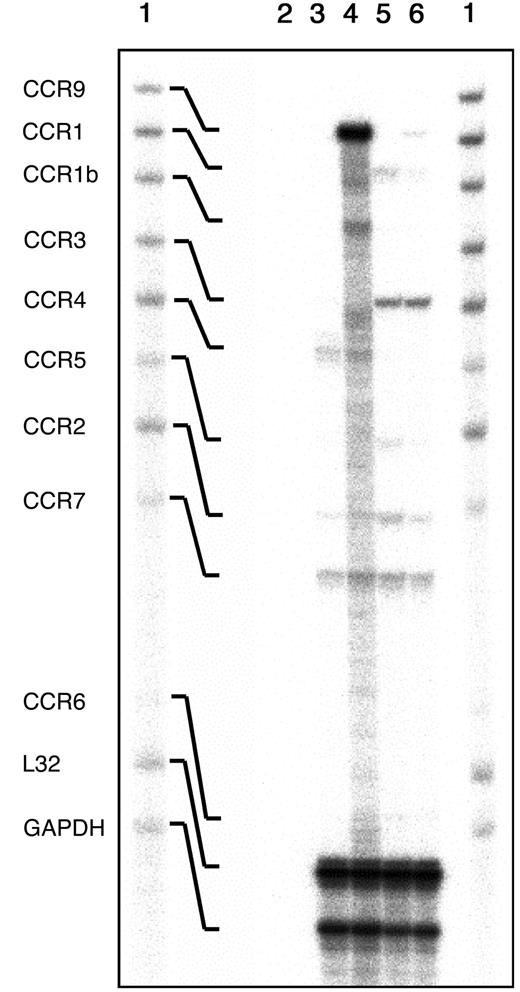
Semiquantitative reverse transcriptase (RT)–PCR indicated that the mRNA for CCR9 was absent from the thymus of CCR9-deficient mice (data not shown). To document whether the lack of CCR9 was compensated by up-regulation of other chemokine receptors, a multiprobe RNase protection assay was used to analyze the levels of transcripts of 9 chemokine receptors including CCR9 (Figure2). CCR9 transcripts were abundant in wild-type thymi, reduced in CCR9-heterozygous thymocytes (data not shown), and not detectable in CCR9-deficient thymocytes. Whereas most of the analyzed CC chemokine receptor transcripts, except the ones corresponding to CCR9, appeared expressed at similar levels in CCR9-deficient and wild-type thymi, transcripts corresponding to CCR1b and CCR5 were consistently down-regulated in CCR9-deficient thymocytes. However, this result, for which we do not have any explanation, does not extend to CCR9-deficient splenic T cells (Figure 2), and does not appear to detectably affect the response of CCR9-deficient thymocytes to CCR1b and CCR5 ligands (data not shown).
Analysis of the chemokine receptor transcripts expressed in freshly isolated thymocytes.
Total RNA extracted from thymi and spleen from CCR9-deficient and wild-type mice were analyzed by multiprobe ribonuclease protection assay using a mCR-5 RiboQuant mouse template set (BD Pharmingen). The autoradiogram shows the mCR-5–probe set not treated with RNases (lane 1), and the RNase-protected bands following hybridization with control yeast tRNA (line 2) with RNA isolated from CCR9-deficient (lane 3), or wild-type thymi (lane 4). Also shown are the RNase-protected bands obtained after hybridization with RNA isolated from CCR9-deficient spleens (lane 5), or wild-type spleens (lane 6). The identity of the various protected bands is indicated on the left.
Analysis of the chemokine receptor transcripts expressed in freshly isolated thymocytes.
Total RNA extracted from thymi and spleen from CCR9-deficient and wild-type mice were analyzed by multiprobe ribonuclease protection assay using a mCR-5 RiboQuant mouse template set (BD Pharmingen). The autoradiogram shows the mCR-5–probe set not treated with RNases (lane 1), and the RNase-protected bands following hybridization with control yeast tRNA (line 2) with RNA isolated from CCR9-deficient (lane 3), or wild-type thymi (lane 4). Also shown are the RNase-protected bands obtained after hybridization with RNA isolated from CCR9-deficient spleens (lane 5), or wild-type spleens (lane 6). The identity of the various protected bands is indicated on the left.
T-cell development and fetal thymic ontogeny
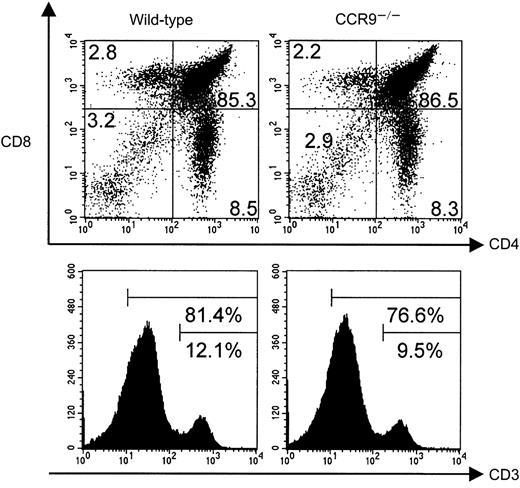
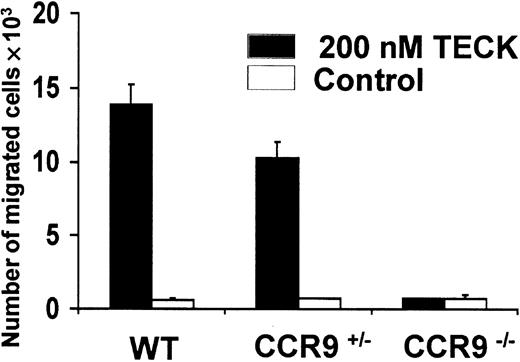
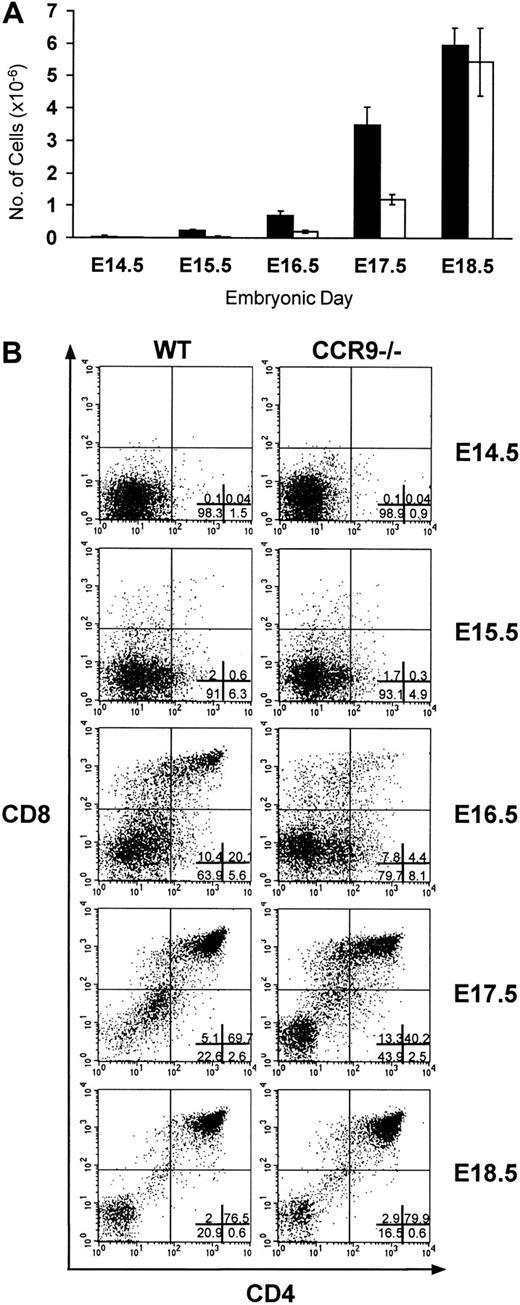
Wild-type and CCR9-deficient thymocytes were stained with mAbs specific for CD4, CD8, and CD3, and analyzed by 3-color flow cytometry. Even though CCR9 is highly expressed at the DP stage of T-cell development, lack of CCR9 affects neither the percentage nor the cellularity of the various subpopulations defined on the basis of CD4/CD8 expression (Figure 3). Moreover, there is no detectable change in the level of T-cell receptor (TCR)/CD3 complex expressed at the cell surface of CCR9-deficient thymocytes when compared to wild-type thymocytes. There was also no detectable change in γδ T cells, and in the expression of the CD25, CD44, CD5, CD62L, and CD69 cell surface markers (data not shown). Considering that T-cell maturation was not detectably altered in CCR9-deficient mice, a chemotaxis assay was performed to determine whether TECK/CCL25 was still able to induce the migration of CCR9-deficient thymocytes via another chemokine receptor. As shown in Figure 4, CCR9-deficient thymocytes were unable to migrate to TECK/CCL25, unlike to wild-type thymocytes. Note that the response to TECK/CCL25 is slightly impeded in CCR9-heterozygous thymocytes and may reflect some gene dosage effect. These results suggest that, in thymocytes, CCR9 is the only physiologic receptor for TECK/CCL25 and that it is dispensable for proper T-cell development. Given the absence of detectable abnormal phenotypes when the thymi found in young and adult CCR9-deficient mice were analyzed under steady-state conditions, we analyzed next the kinetics of appearance of the first DP cells during thymic fetal ontogeny. As shown in Figure 5A, from embryonic day (E) 14.5 to E17.5, CCR9-deficient thymi contained 3-fold fewer lymphoid cells than wild-type thymi. This difference in cellularity is lost at E18.5 and in young and adult mice (Figure 5A and data not shown). The differences in cellularity noted at E16.5 between mutant and wild-type thymi correlate with a marked reduction of the percentage of DP cells among CCR9-deficient thymocytes (Figure 5B), resulting on day 16.5 in 15-to 20-fold fewer DP cells in the mutant thymus compared to the wild type. Therefore, when analyzed under the dynamic conditions that occur during thymic fetal ontogeny, the absence of CCR9 resulted in a 1-day lag in the appearance of DP cells.
T-cell development in CCR9-deficient mice.
Thymocytes from wild-type and CCR9-deficient (CCR9−/−) mice were analyzed by 3-color flow cytometry for the expression of CD4, CD8, and CD3. The percentage of cells found in each quadrant is indicated, and the percentage of CD3high, and of CD3high plus CD3intermediate thymocytes indicated for the single-color histograms is shown in the lower panel.
T-cell development in CCR9-deficient mice.
Thymocytes from wild-type and CCR9-deficient (CCR9−/−) mice were analyzed by 3-color flow cytometry for the expression of CD4, CD8, and CD3. The percentage of cells found in each quadrant is indicated, and the percentage of CD3high, and of CD3high plus CD3intermediate thymocytes indicated for the single-color histograms is shown in the lower panel.
Chemotactic responses to TECK/CCL25.
Freshly isolated thymocytes from CCR9-deficient (CCR9−/−), CCR9-heterozygous (CCR9+/−) and wild-type mice were placed in the upper chamber of a 24-well Transwell plate. Recombinant TECK/CCL25 (200 nM) or culture medium, as a negative control, were placed in the lower chamber. After 4 hours of incubation, cells in the lower chamber were counted both manually and by flow cytometry.
Chemotactic responses to TECK/CCL25.
Freshly isolated thymocytes from CCR9-deficient (CCR9−/−), CCR9-heterozygous (CCR9+/−) and wild-type mice were placed in the upper chamber of a 24-well Transwell plate. Recombinant TECK/CCL25 (200 nM) or culture medium, as a negative control, were placed in the lower chamber. After 4 hours of incubation, cells in the lower chamber were counted both manually and by flow cytometry.
Fetal thymic ontogeny in CCR9-deficient mice.
(A) The number of lymphoid cells found in CCR9-deficient (□) and wild-type (■) thymi are represented at different ages of embryonic life. (B) CD4/CD8 staining profiles on total thymocytes from wild-type and from CCR9-deficient mice at different ages of embryonic life. The percentage of cells within each quadrant is indicated in the lower-right panel.
Fetal thymic ontogeny in CCR9-deficient mice.
(A) The number of lymphoid cells found in CCR9-deficient (□) and wild-type (■) thymi are represented at different ages of embryonic life. (B) CD4/CD8 staining profiles on total thymocytes from wild-type and from CCR9-deficient mice at different ages of embryonic life. The percentage of cells within each quadrant is indicated in the lower-right panel.
Peripheral T lymphocytes in CCR9-deficient mice
The size and cellularity of the spleen and lymph nodes found in CCR9-deficient mice were comparable to those found in wild-type mice. When TCRαβ+ and TCRγδ+ lymphocytes were analyzed, we noted consistent 2- to 3-fold higher percentages and absolute numbers of TCRγδ+ cells (Figure6) in CCR9-deficient mice compared with wild-type mice. To determine whether the αβ T lymphocytes that populate the secondary lymphoid organs were functional, splenic T lymphocytes were stimulated using anti-CD3 mAb, the staphyloccocal enterotoxin superantigens SEA and SEB, ConA, and PMA/ionomycin as a positive control. No detectable differences were observed between CCR9-deficient and wild-type cell populations (data not shown). These results suggest that CCR9-deficient peripheral αβ T lymphocytes are fully capable of responding via TCR triggering. Moreover, the ability of CCR9-deficient cells to respond to SEA and SEB suggests that there is probably no major bias in the TCRαβ repertoire that is exported to the periphery of CCR9-deficient mice.
Peripheral TCRαβ and TCRγδ cells in CCR9-deficient mice.
Lymphocytes isolated from lymph nodes and spleens of CCR9-deficient and wild-type mice were analyzed by 4-color flow cytometry. Two-color plots represent the TCRαβ versus TCRγδ staining observed on CD3-positive cells. The percentages of TCRαβ and TCRγδ cells found in each organ are indicated.
Peripheral TCRαβ and TCRγδ cells in CCR9-deficient mice.
Lymphocytes isolated from lymph nodes and spleens of CCR9-deficient and wild-type mice were analyzed by 4-color flow cytometry. Two-color plots represent the TCRαβ versus TCRγδ staining observed on CD3-positive cells. The percentages of TCRαβ and TCRγδ cells found in each organ are indicated.
Early B-cell differentiation in CCR9-deficient mice
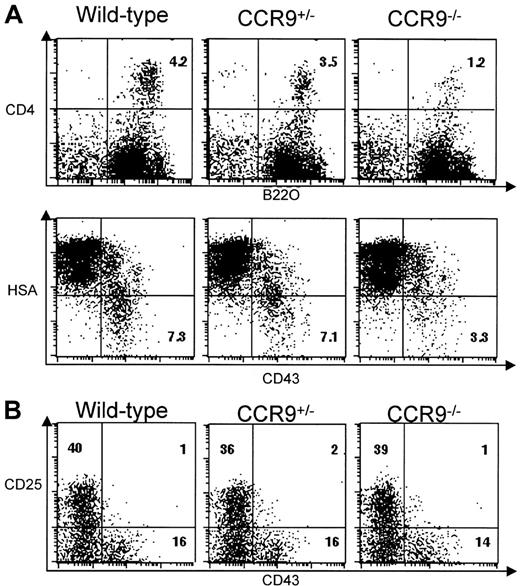
In humans, CCR9 is expressed on a subset of CD19+ peripheral B lymphocytes,15 whereas in mice, bone marrow cells displaying a pre-pro-B-cell phenotype and corresponding to both the A1 (CD4+B220+) and A2 (CD4−B220+) fractions (according to Hardy classification23,24) migrate in response to TECK/CCL25, a capacity that is lost when they progress to later stages of development.5 Despite the presence of normal numbers of mature B cells in the spleen and lymph nodes of CCR9-deficient mice, we wondered whether TECK/CCL25-responsive pre-pro-B cells were affected by the absence of CCR9. To focus on this minor B-cell compartment, bone marrow B cells were first enriched using B220-coupled microbeads, and then subjected to 2-color flow cytometric analysis using the combination of cell surface markers specified in Figure7A. The percentage of CD4+B220+ B cells corresponding to fraction A1 is less in CCR9-deficient mice (1.2%) than in wild-type mice (4.2%). This percentage is also slightly lower in heterozygous animals (3.5%) than in wild-type mice. Moreover, B220+CD43+HSAlow/− pre-pro-B cells corresponding to fractions A1 and A2 represent only 3.3% in CCR9-deficient mice compared with 7.3% in wild-type mice. Considering that mutant and wild-type bone marrow samples yielded almost the same number of B220+ cells, the differences in percentage we observed are likely to reflect differences in absolute cell number. These results indicate that lack of CCR9 specifically affects the CD4+B220+ B cells corresponding to fraction A1. As shown in Figure 7B, the percentage of more mature bone marrow pro-B B220+CD43+CD25− cells or of bone marrow pre-B B220+CD43−CD25+cells are unaffected in mutant mice. Thus, the lack of CCR9 results in 3-fold fewer pre-pro-B cells than in wild type, but, as in fetal thymus, some homeostatic adjustment appears to occur subsequent to this developmental step.
B-cell development in CCR9-deficient mice.
(A) Two-color plots show the CD4/B220 and HSA/CD43 profiles found on B220-enriched bone marrow B cells from CCR9-deficient, CCR9-heterozygous, and wild-type mice. Also shown is the percentage of CD4+B220+ and CD43+HAS− B cells. (B) Two-color plots show the CD43/CD25 profile of B220-enriched bone marrow B cells from CCR9-deficient, CCR9-heterozygous, and wild-type mice. Numbers indicate the percentage of cells within this quadrant.
B-cell development in CCR9-deficient mice.
(A) Two-color plots show the CD4/B220 and HSA/CD43 profiles found on B220-enriched bone marrow B cells from CCR9-deficient, CCR9-heterozygous, and wild-type mice. Also shown is the percentage of CD4+B220+ and CD43+HAS− B cells. (B) Two-color plots show the CD43/CD25 profile of B220-enriched bone marrow B cells from CCR9-deficient, CCR9-heterozygous, and wild-type mice. Numbers indicate the percentage of cells within this quadrant.
IEL populations in CCR9-deficient mice
In normal mice, the CD3+ IELs found in the small intestine can be subdivided into TCRαβ+ cells that express either CD4 or CD8αβ molecules, and into TCRαβ+ or TCRγδ+ cells that coexpress CD8αα homodimers. Both latter subsets of IELs appear capable of differentiating in part via an extrathymic pathway and to follow rules of TCR repertoire selection that differ from those of the thymo-dependent TCRαβ+CD8αβ+ (or CD4+) gut IEL subsets. Considering that, at least in humans, all IELs express CCR9,15 16 we expected that a large fraction of IELs would be affected by the lack of CCR9. Wild-type and CCR9-deficient small intestines were first subjected to comparative histologic studies and the number of IELs counted and normalized per epithelial cells (EC). The total IEL to EC ratio was diminished 2-fold in CCR9-deficient mice compared with wild-type mice (Figure8A). Flow cytometry analysis of isolated IELs showed that such a decrease in IEL cellularity was mainly due to the presence of low numbers of TCRγδ+ IELs (Figure 8B and C). Based on these analyses, it can be calculated that the absolute number of TCRγδ+ IEL is 5-fold lower in CCR9-deficient than in wild-type mice.
Gut IELs in CCR9-deficient mice.
(A) Analysis of tissue sections allowed us to determine the number of total IELs and to normalize the number to 100 epithelial cells.29 (B) Comparison of the various CD3+IEL subsets found in CCR9-deficient and in wild-type mice. IELs were labeled with the specified combination of antibodies and analyzed by flow cytometry. (C) IELs from CCR9-deficient or wild-type mice were analyzed by 2-color flow cytometry for the expression of TCRδ versus CD3. The percentage of TCRγδ+ cells is indicated.
Gut IELs in CCR9-deficient mice.
(A) Analysis of tissue sections allowed us to determine the number of total IELs and to normalize the number to 100 epithelial cells.29 (B) Comparison of the various CD3+IEL subsets found in CCR9-deficient and in wild-type mice. IELs were labeled with the specified combination of antibodies and analyzed by flow cytometry. (C) IELs from CCR9-deficient or wild-type mice were analyzed by 2-color flow cytometry for the expression of TCRδ versus CD3. The percentage of TCRγδ+ cells is indicated.
Discussion
Considering that CCR9 is highly expressed at the surface of DP thymocytes and down-regulated on CD4+ SP cells prior to their exit from the thymus, it might have been anticipated that the disruption of the CCR9 gene would have markedly affected intrathymic T-cell development. Moreover, we showed that in adult thymocytes, CCR9 is the only physiologic receptor for TECK/CCL25. Despite this one-to-one relationship, CCR9 appears dispensable for proper T-cell development. During fetal thymic ontogeny, we observed that in CCR9-deficient mice, the emergence of DP cells and of TCR αβ+ cells was delayed by approximately a day when compared with wild-type littermates. At E15.5, fetal thymi are only made of DN cells. In this context, it remains to be established whether the diminished cellularity also noted at this early time point in the absence of CCR9 reflects the contribution of TECK/CCL25 to the colonization of the thymus anlage. In any instance, our data show that the lack of CCR9 has only limited effects on T-cell development. Similarly, B-cell maturation is only modestly altered in CCR9-deficient mice, and the 3-fold decrease noted in the number of CCR9-deficient pre-pro-B lymphocytes is congruent with their unique ability to respond to TECK/CCL25 in in vitro chemotactic assays.5
Carramolino and colleagues25 recently raised a polyclonal antisera specific for CCR9 and showed that the pattern of expression of CCR9 protein is largely consistent with previous analyses of the tissue distribution of CCR9 transcripts. They further showed that the expression of CCR9 persists at low levels on the CD8+ SP found in the thymus and in the spleen, whereas it is absent on CD4+ SP thymocytes. Consistent with this observation, we noted that the CD8+ SP found in the thymus and the periphery of CCR9-deficient mice lose their ability to respond to TECK/CCL25 when analyzed in in vitro chemotactic assays (data not shown). Therefore, our results suggest that, at least on thymocytes and CD8+ SP cells, CCR9 is the only physiologic ligand for TECK/CCL25.
In the periphery, the absence of CCR9 resulted in a 2- to 3-fold increase in the number of TCRγδ+ cells present in lymph nodes and spleen, and in a 3- to 5-fold decrease in the number of small intestine TCRγδ+ IELs. To date, TCRγδ+cells have not been formally shown to express CCR9. However, the fact that all human small intestine IELs are intensely stained with an anti-CCR9 monoclonal antibody15 strongly suggests that TCRγδ+ cells are indeed CCR9+. In this context, it is tempting to speculate that the increase in TCRγδ+ cells found in the spleen and lymph nodes of CCR9-deficient mice may correlate with their impaired ability to migrate to the intestinal mucosal epithelium. However, the moderately abnormal phenotypes noted both in secondary lymphoid organs and the small intestine of CCR9-deficient mice suggest that some substantial functional redundancy may exist with other chemokine receptors. Along this line, it is important to note that the chemokine receptor CCR11, primarily expressed in heart, lung and, more relevant for the present study, small intestine, has recently been found capable of binding to several chemokines including TECK/CCL25.26-28Finally, because our CCR9-deficient mice have been kept in specific pathogen-free conditions, it is not known whether these mice exhibit an increased susceptibility to enteric pathogens. Furthermore, it remains to be established whether the absence of CCR9 might influence inflammatory bowel disease conditions.
We thank Philippe Naquet, Pierre Golstein, and Rodolphe Guinamard for discussion, Anne Gillet for advice during blastocyst injection, Chantal Kress and Charles Babinet for the CK35 129 mouse ES cells, Ian Clark-Lewis for kindly providing us with several chemokine samples, and Noëlle Guglietta for editing the manuscript. M.-A.W. was supported by doctoral fellowships from Ligue Nationale Contre le Cancer and from Ministère de l'Education Nationale de la Recherche et de la Technologie.
Supported by institutional grants from Centre National de la Recherche Scientifique (CNRS) and Institut National de la Santé et de la Recherche Scientifique (INSERM), and by specific grants from Association pour la Recherche contre le Cancer, Villejuif, France; Association Francaise sur les Myopathies/Genethon, Evry, France; and the European Communities, Brussels, Belgium (project QLG1-CT1999-00202).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Bernard Malissen, Centre d'Immunologie de Marseille-Luminy, INSERM-CNRS–Universite de la Mediterranee, Parc Scientifique de Luminy, Case 906, 13288 Marseille Cedex 9, France; e-mail: bernardm@ciml.univ-mrs.fr.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal