Conventional karyotypes performed before any treatment in 208 patients with multiple myeloma were reviewed by the Groupe Français de Cytogénétique Hématologique. A total of 138 patients displayed complex chromosomal abnormalities (CCAs). According to the chromosome number pattern, a first group of 75 patients had a hyperdiploid karyotype. A second group of 63 patients referred to as the hypodiploid group had either pseudodiploid, hypodiploid, or near-tetraploid karyotypes. Of 159 treated patients available for survival analysis, 116 had an abnormal karyotype. The comparison of overall survival (OS) between hyperdiploid and hypodiploid patients showed a highly significant difference (median OS 33.8 vs 12.6 months, respectively, P < .001). The presence of 14q32 rearrangements (36 of 116 patients) worsened the prognosis (median OS 17.6 vs 29.9 months, P < .02). The presence of chromosome 13q abnormalities (13qA, 63 patients) did not modify OS in CCA patients (median OS 20.6 vs 27.8 months,P < .59). However, taking into account the whole series including normal karyotypes, 13qA (63 of 159 patients) had a significant impact on OS (median 20.6 vs 37.1 months,P < .04). In the same way, the presence of a hypodiploid karyotype (52 of 159 patients) had a strong prognostic value (OS 12.8 vs 44.5 months, P < .000 01). A multivariate analysis including stage, β2-microglobulin, bone marrow plasmocytosis, treatment type, 13qA, and hyperdiploidy and hypodiploidy showed that a hypodiploid karyotype was the first independent factor for OS (P < .001), followed by treatment approach. These results confirm that the chromosome number pattern of malignant plasma cells is a very powerful prognostic factor in newly diagnosed multiple myeloma patients.
Introduction
Multiple myeloma (MM) is a clonal B-cell neoplasia characterized by the accumulation in bone marrow of malignant plasma cells producing a monoclonal immunoglobulin.1Survival ranges from a few months to more than 10 years,1,2 in keeping with the great heterogeneity observed in this disease.3 A median survival of about 2 years was obtained with conventional chemotherapy, but new trials using high-dose therapy followed by autologous stem cell transplantation have improved the remission rate, event-free survival, and overall survival (OS) in younger patients.4 Therefore, it appears essential, at diagnosis, to recognize clinical or biological parameters predicting the outcome and identifying patients for whom an aggressive therapy would be indicated.2,5 Several prognostic factors have been reported, either related to the proliferative rate of plasma cells as the labeling index6 or to tumor burden: β2-microglobulin level, percentage of bone marrow plasma cells, and lytic bone lesions. Among the factors related to clinical aggressiveness, there is increasing evidence for the importance of cytogenetic defects. There is discordance in published results: the adverse prognostic impact of the loss or partial deletion of chromosome 13 (13qA) is reported by several groups using fluorescence in situ hybridization techniques7-9 or conventional cytogenetics10-13 but is not confirmed by others.14-18 We report the results of a multicenter study, using conventional cytogenetics, undertaken by the Groupe Français de Cytogénétique Hématologique (GFCH) to assess the impact of cytogenetic abnormalities on OS in an independent series of 208 patients.
Patients, materials, and methods
Patients
From October 1994 to October 1999, 16 laboratories of the GFCH (listed in the ) performed conventional cytogenetics in 208 MM patients before any chemotherapy.
This prospective collaborative study enrolled 100 female and 108 male patients. The median age was 64 years (range, 25-96). According to Durie and Salmon staging,19 37 (18%) and 24 (11%) patients were stage I and II, respectively, while 147 (71%) were stage III MM. A total of 196 patients were karyotyped at the time of diagnosis and 12 during evolution of untreated stage I or smoldering MM.20 The monoclonal component was immunoglobulin G (IgG) in 126 patients, IgA in 53, IgD in 1, IgM in 1, 25 expressed only light chains, and 2 were nonsecretory MM. Twenty-nine patients had renal failure (22 stage III, 3 stage II, 4 stage I).
Twenty-four patients (11%) had a previous history of monoclonal gammopathy of undetermined significance.21 Thirteen had another malignancy, either known before MM or diagnosed in association with MM. The main clinical, hematologic, and biochemical characteristics of these patients are listed in Table1.
Main clinical and biological data in total series and in normal/abnormal karyotypes
| . | Total series n = 208 (%) . | Normal karyotypes n = 70 (%) . | Abnormal karyotypes n = 138 (%) . | Hyperdiploid n = 75 (%) . | Hypodiploid/pseudodiploid/∼4n n = 63 (%) . |
|---|---|---|---|---|---|
| Median age, y | 64 | 67 | 63 | 65 | 60 |
| Sex | |||||
| F | 100 (48) | 35 (50) | 65 (47) | 38 (51) | 27 (43) |
| M | 108 (52) | 35 (50) | 73 (53) | 37 (49) | 36 (57) |
| Stage | |||||
| I | 37 (18) | 23 (33) | 14 (10) | 10 (13) | 4 (6) |
| II | 24 (11) | 11 (16) | 13 (9) | 7 (9) | 6 (9.5) |
| III | 147 (71) | 36 (51) | 111 (81) | 58 (77) | 53 (84) |
| Paraprotein | |||||
| IgG | 126 (60.5) | 45 (64) | 81 (59) | 52 (69) | 29 (46) |
| IgA | 53 (25) | 20 (28.5) | 33 (24) | 18 (24) | 15 (24) |
| Only light chain | 25 (12) | 5 (7) | 20 (14) | 4 (5) | 16 (25) |
| Light chains | |||||
| Kappa | 126 (60.5) | 47 (67) | 79 (57) | 47 (63) | 32 (51) |
| Lambda | 80 (38.4) | 23 (33) | 57 (41) | 28 (37) | 29 (46) |
| Hemoglobin, g/dL | 10.57 | 11.09 | 10.30 | 10.63 | 9.91 |
| Calcium, μM | 2.51 | 2.39 | 2.58 | 2.50 | 2.67 |
| Creatinine, μM/L | 148 | 123.7 | 160.2 | 119.6 | 208.6 |
| β2-microglobulin (mg/L) | 6.21 | 4.24 | 7.25 | 5.83 | 9.12 |
| C-reactive protein | |||||
| Norm. | 106 (51) | 36 (51) | 70 (51) | 42 (56) | 28 (44) |
| ➚ | 68 (33) | 19 (27) | 49 (35.5) | 24 (32) | 25 (40) |
| %VM plasmocytosis | 40.6 | 27.7 | 47.4 | 40.8 | 55.4 |
| Lytic lesions | 124 (60) | 34 (48.5) | 90 (65) | 48 (64) | 42 (67) |
| Treatment | |||||
| Abstention | 33 (16) | 18* (26) | 15 (11) | 6† (8) | 9‡ (14) |
| Chemotherapy | 100 (48) | 32 (46) | 68 (49) | 39 (52) | 29 (46) |
| HDC SCT | 71 (34) | 20 (28) | 51 (37) | 28 (37) | 23 (37) |
| Others | 4 (2) | 0 (0) | 4 (3) | 2 (3) | 2 (3) |
| . | Total series n = 208 (%) . | Normal karyotypes n = 70 (%) . | Abnormal karyotypes n = 138 (%) . | Hyperdiploid n = 75 (%) . | Hypodiploid/pseudodiploid/∼4n n = 63 (%) . |
|---|---|---|---|---|---|
| Median age, y | 64 | 67 | 63 | 65 | 60 |
| Sex | |||||
| F | 100 (48) | 35 (50) | 65 (47) | 38 (51) | 27 (43) |
| M | 108 (52) | 35 (50) | 73 (53) | 37 (49) | 36 (57) |
| Stage | |||||
| I | 37 (18) | 23 (33) | 14 (10) | 10 (13) | 4 (6) |
| II | 24 (11) | 11 (16) | 13 (9) | 7 (9) | 6 (9.5) |
| III | 147 (71) | 36 (51) | 111 (81) | 58 (77) | 53 (84) |
| Paraprotein | |||||
| IgG | 126 (60.5) | 45 (64) | 81 (59) | 52 (69) | 29 (46) |
| IgA | 53 (25) | 20 (28.5) | 33 (24) | 18 (24) | 15 (24) |
| Only light chain | 25 (12) | 5 (7) | 20 (14) | 4 (5) | 16 (25) |
| Light chains | |||||
| Kappa | 126 (60.5) | 47 (67) | 79 (57) | 47 (63) | 32 (51) |
| Lambda | 80 (38.4) | 23 (33) | 57 (41) | 28 (37) | 29 (46) |
| Hemoglobin, g/dL | 10.57 | 11.09 | 10.30 | 10.63 | 9.91 |
| Calcium, μM | 2.51 | 2.39 | 2.58 | 2.50 | 2.67 |
| Creatinine, μM/L | 148 | 123.7 | 160.2 | 119.6 | 208.6 |
| β2-microglobulin (mg/L) | 6.21 | 4.24 | 7.25 | 5.83 | 9.12 |
| C-reactive protein | |||||
| Norm. | 106 (51) | 36 (51) | 70 (51) | 42 (56) | 28 (44) |
| ➚ | 68 (33) | 19 (27) | 49 (35.5) | 24 (32) | 25 (40) |
| %VM plasmocytosis | 40.6 | 27.7 | 47.4 | 40.8 | 55.4 |
| Lytic lesions | 124 (60) | 34 (48.5) | 90 (65) | 48 (64) | 42 (67) |
| Treatment | |||||
| Abstention | 33 (16) | 18* (26) | 15 (11) | 6† (8) | 9‡ (14) |
| Chemotherapy | 100 (48) | 32 (46) | 68 (49) | 39 (52) | 29 (46) |
| HDC SCT | 71 (34) | 20 (28) | 51 (37) | 28 (37) | 23 (37) |
| Others | 4 (2) | 0 (0) | 4 (3) | 2 (3) | 2 (3) |
Age, hemoglobin, calcium, creatinine, β2-microglobulin, and bone marrow (BM) plasmocytosis are given with the average value. Norm indicates normal value; ➚, increased value; chemotherapy, monochemotherapy or polychemotherapy.
Including 6 early deaths.
Including 2 early deaths.
Including 7 early deaths.
Treatment regimens
Patients were treated according to several different clinical trials. Approval was obtained from the institutional review board for these studies. Informed consent was provided according to the Declaration of Helsinki. A total of 100 patients received conventional-dose chemotherapy: either monochemotherapy, mainly with melphalan prednisone or cyclophosphamide prednisone (63 patients), or polychemotherapy consisting of vincristine, doxorubicin, and dexamethasone or vincristine, cyclophosphamide, melphalan, and prednisone (37 patients). Seventy-one patients were enrolled in high-dose chemotherapy regimens followed by 1 or 2 autologous bone marrow or peripheral blood stem cell transplantations (HDC SCT). One patient received an allogeneic bone marrow transplant. Thirty-three did not receive any therapy because of either the presence of an asymptomatic disease (18 patients: 11 stage I, 3 stage II, 4 stage III) or the occurrence of an early death (15 patients). For 3 patients, information on the treatment was not available (Table 1).
Cytogenetic analysis
Conventional chromosome studies were undertaken before any treatment, mainly on bone marrow (202 cases), seldom on peripheral blood (4 cases), and in 2 cases on bone tumor.
Initially, the trial required 2 simultaneous culture protocols to precisely determine the best conditions to detect the MM malignant clone: a 3-day unstimulated culture and a 4- to 7-day culture with interleukin-6 and granulocyte-macrophage colony-stimulating factor.16,22,23 However, the analysis of the first 100 patients enrolled confirmed that the use of cytokines did not improve the malignant clone detection,24 and it was decided that the following inclusions would require only a 3-day unstimulated culture. RHG banding was used for all karyotypes. In 10 cases whole chromosome painting was performed to precisely determine the origin of deleted or derivative chromosomes. Chromosomal abnormalities were described according to ISCN.25 Standard criteria to define a clone were applied, but 12 karyotypes with only 1 MM characteristic abnormal metaphase were included: 9 were hyperdiploid (cases 3, 7, 22, 27, 31, 46, 50, 60, 71) and 3 were pseudodiploid or hypodiploid with an 11;14 translocation (cases 80, 98, 104). A minimum of 30 fully analyzed metaphases was required to include a normal karyotype. Each karyotype was critically reviewed by the participants of the GFCH at 2 successive workshops before it was recorded in the study.
Statistical methods
Because patients included in this multicenter protocol received different treatment regimens, no standard definition of response, partial remission, or complete remission was applicable.26 Thus, OS from time of diagnosis was chosen as the single end point, whatever the cause of death. A total of 159 patients were eligible for survival analysis. Exclusion criteria were unknown or no treatment, allogeneic bone marrow transplantation, or karyotype performed during evolution of an untreated myeloma. Curves were plotted according to Kaplan and Meier27 and were compared using the log-rank test. Multivariate analysis of different prognostic factors was carried out using the Cox regression model.28 Cut-off values for biological parameters were chosen according to previously reported series and were 3 mg/L for β2-microglobulin, 200 μM/L for creatinine, and 20% for bone marrow plasmocytosis. Analysis was performed using the JPSI statistical software (developed by P. Kwiatkowski, Centre J Perrin, Clermont Ferrand, France).
Results
Myeloma clone detection
A total of 138 (66%) of 208 MM patients displayed numerical and structural complex chromosomal abnormalities (CCAs) related to myeloma. The remaining 70 karyotypes (34%) were considered not representative of the MM cell population because 55 were normal karyotypes and 15 showed either an isolated numerical or structural abnormality, mainly loss or gain of sexual chromosomes (9 cases), loss or gain of one autosome (2 cases), or a single rearranged chromosome (4 cases). A mean of 24.5 mitoses was studied per patient (range, 6-94) with a mean of 10.4 abnormal mitoses (range, 1-38). All but 14 abnormal karyotypes showed a mixture of normal and abnormal cells.
CCA detection increased with disease stage: 14 (38%) of 37 for stage I patients, 13 (54%) of 24 for stage II, and 111 (75.5%) of 147 for stage III patients. Complete karyotypes are listed in Table2.
Karyotypes classified according to the chromosome number pattern
| Hyperdiploid karyotypes | |
| 1 | 48,XY,+5,der(7)t(7;8)(p22;q13),+der(7)t(7;8)(p22;q13),−8,−13,der(16)t(1;16)(q12;q11),+der(19)t(1;19)(q12;q11)×2,add(22)(p13)[3]/48,idem,−7,der(7)t(1;7) (q12;p22),t(8;14)(q13;q31),−der(16)t(1;16)(q12;q11)[11]/48,idem,del(6)(q15),−7,+der(7)t(1;7)(q12;p22),t(8;14)(q13;q31),−der(16)(1;16)(q12;q11)[8]/96, idemx2,del(6)(q15),der(7)t(1;7)(q12;p22),t(8;14)(q13;q31),−der(16)t(1;16)(q12;q11)[1]/46,XY[5] |
| 2 | 49,XY,+5,+9,der(12)t(3;12)(p21;p13),del(14)(q24q32),+19,del(20)(q11)[8]/46,XY[15] |
| 3 | 51,XY,add(4)(qter),+5,del(5)(q21q34),del(6)(q13q27),+6,+9,del(11)(p11),del(13)(q13q22),+15,add(17)(q?),del(17)(q?),+19,+20,−22[1]/46,XY[101] |
| 4 | 53,X,−X,t(2;6)(q33;p23),+3,+5,+6,+7,+9,+15,+18,+19[2]/54,idem,+21[4]/56,idem,+X,+8,+21[12]/46,XX[30] |
| 5 | 52,XY,+9,+add(11)(p11),+15,+18,+19,+21,der(22)t(11;22)(p12;p12)[16]/49,XY,add(8)(p11),+add(11)(p11),−13,+15,der(16)t(1;16)(q12;q11),+18,+21, der(22)t(11;22)(p12;p12)[5]/98,idemx2[1]/98,idemx2,del(6)(q11),del(11)(q13)[1]/46,XY[8] |
| 6 | 54,X,−Y,+3,+5,+9,+9,+11,+11,+15,+15,+19[3]/55,idem,+r1[3]/55,idem,+r2[2]/55,XY,+3,+5,+9,+9,+11,+15,+19,+r2[9]/55,idem,−r2,+r1[9] |
| 7 | 47,XX,+9,−13,−14,+19,+mar[1]/46,XX[40] |
| 8 | 54,XX,+3,i(4)(p10)×2,del(6)(q?21),+7,+del(9)(p22),+11,del(12)(p13),−13,+15,+15,−16,−16,add(17)(p11),+del(20)(q?23)×2,+mar1,+mar2[cp4]/54,idem, +mar3,+mar4[cp2]/54,idem,tas(15;15)(p13;p13)[cp3]/46,XX[21] |
| 9 | 52,XY,+5,+9,+11,−13,+15,−17,+19,+21,+mar1,+mar2[2]/46,XY[32] |
| 10 | 54,X,−X,+3,+5,der(8)t(7;8)(q1?1;p1?1).ish(wcp8+),+9,+11,+15,+15,+19,+der(21)t(1;21)(q11,q22).ish(wcp1+)[16]/54,idem,add(14)(p11)[1]/46,XX[28] |
| 11 | 55,XX,+3,+4,+5,+6,+7,+9,+11,+15,+21[5]/46,XX[38] |
| 12 | 49,XY,t(1;6)(q1?1;q2?5),+der(6)t(1;6)(q1?1;q2?5),+9,+18,−21,add(22)(p1?1),+mar[cp4]/98,idemx2[1]/46,XY[89] |
| 13 | 54,XX,+3,+5,+7,+9,+9,+15,+19,+21[4]/46,XX[25] |
| 14 | 54,XY,del(1)(p12p31),+add(1)(p11),+3,+5,+7,add(8)(p22),+9,+15,+19,+21[5]/46,XY[8] |
| 15 | 54,XY,add(4)(p1?6),+?del(3)(p11p23),+5,+9,+9,+11,+15,+15,+19,+19[1]/54,XY,t(1;3;?)(q23;p21;?),+?del(3)(p11p23),+5,add(8)(p?22),del(9)(q22q34), +9,+9,+11,+15,+15,+19[5]/54,idem,−7,+mar,inc[2]/46,XY[43] |
| 16 | 52,XY,+3,+5,dic(8;18)(p11;p11),+9,+add(11)(p15),+15,+15,+19[8]/46,XY[47] |
| 17 | 74∼80〈3n〉,XXX,add(9)(q?)×2,+mar,inc[7]/46,XX[18] |
| 18 | 54,XY,del(1)(p21p32),+3,+5,+6,+7,+9,−13,+15,+der(19)t(1;19)(q12;q13),+21,+21[5]/46,XY[17] |
| 19 | 50,X,−Y,del(1)(p22p31),+3,+der(5)t(1;5)(q12;q35),+7,+9,del(11)(p11p12),add(12)(p13),−13,−14,+15,+19,−22,+mar1,+mar2[7]/49,idem,−5[6]/46,XY[8] |
| 20 | 51,XY,add(1)(?p11),+5,+9,+11,+add(15)(p11),−16,+r,+mar[4]/53,idem,+9,+r[3]/46,XY[32] |
| 21 | 51,XY,+5,+add(7)(q36),+9,−13,+18,+19,del(20)(q12),+mar[2]/51,idem,t(5;10)(q35;q24)[2]/46,XY[36] |
| 22 | 47,X,−X,del(1)(p12p34),−4,del(5)(q13q31),+7,+11,+der(1;?)(q10;?)[1]/46,XX[19] |
| 23 | 56∼58,XX,add(1)(q11),add(1)(q21),+3,+5,der(6)t(1;6)(q12;q12)×2,+7,+9,add(13)(p11),−14,−15,add(15)(p11),+18,+19,+19,−21,+22,+5mar[cp13]/46,XX[16] |
| 24 | 47,X,add(X)(p22),del(4)(q24),+add(5)(q11),der(6)t(6;8)(q11;q11),+7,−8,+dup(9)(p11p24),+add(11)(q23),−12,−13,add(13)(q14),−16,−17,+19,+19[16] |
| 25 | 52,X,−Y,+3,+5,+9,add(14)(q32),+15,+15,+19,+21[2]/46,XY[6] |
| 26 | 54,XY,t(1;22)(p34;q11),add(2)(p25),+del(3)(p12),+5,der(6)t(?1;6)(q21;q23),+6,der(7)t(7;11)(p15;q13),del(9)(q?12q34),+9,+15,+der(19)t(15;19)(q1?4;q12), +21,+21[26] |
| 27 | 53,XY,+3,+5,+9,+11,del(13)(q?),add(14)(q32),+19,+mar1,+mar2[1]/46,XY[17] |
| 28 | 50,X,−X,+3,ins(5;17)(p13;q22q25).ish(wcp17+),+7,+9,+15,+del(17)(q22).ish(wcp17+)[18]/49,idem,der(8;22)(q10;p10).ish(wcp22+)[11]/49,idem,−13, der(?8;22)(p10;q10)[3]/49,idem,t(1;13)(p12-13;q32),−7,der(1;22)(q10;q10)[cp4]/46,XX[29] |
| 29 | 47,XX,add(8)(p23),add(18)(q22),+19,der(22)t(1;22)(q12;q13)[3]/46,idem,−X,add(9)(q?),+11,−17[1]/46,XX[4] |
| 30 | 57,XY,del(1)(p12),add(2)(q32)×2,+3,+add(5)(q35),+7,add(8)(p21),add(8)(p11)(q24),+inv(9),+11,+11,+15,+17,+add(19)(p12),+21[12]/46,XY,inv(9)[23] |
| 31 | 51∼53,XX,del(1)(q22),+3,+5,+6,+7,add(8)(p22),+9,−14,+mar1,+mar2,+mar3[inc3]/46,XX[20] |
| 32 | 53,XX,del(1)(p21p31),add(2)(q34),+3,+5,+add(8)(q24),+add(11)(p14),+15,−16,+19,+19,+del(22)(q1?2)[5]/46,XX[10] |
| 33 | 54∼57,XX,t(1;5)(q21;p15),+3,+der(5)t(1;5)[5],add(7)(p15)[2],+9,+9,+11,−13,t(13;15)(q10;q10),+15,+18,+18[3],+19,+21,+21[cp12]/54∼57,idem,−t(13;15) (q10;q10),−13,der(19)t(13;19)(q21;p13)[2]/46,XX[9] |
| 34 | 59∼75〈3n〉,XXX,−1,+7,+6,−12,+19[cp2]/46,XX[35] |
| 35 | 49,XX,−2,+3,add(5)(q35),del(6)(q21),+9,+12,−13,del(14)(q24q32),i(18)(q10),+3mar[1]/46,XX[49] |
| 36 | 46,XY,add(2)(p15),add(2)(q22)[6]/49∼52,XY,add(1)(p31)(q41),del(2)(p11p22),add(3)(p24)(q21)×2,−4,del(4)(p14?),+5,add(6)(q26),+del(7)(q31),−8,+9, +11,der(12)t(9;12)(q11;q24),+15,add(16)(q11),+18,+19,+1∼3mar[cp12]/46,XY(19] |
| 37 | 52,XX,del(6)(q2?1),+7,t(11;14)(q13;q32),+der(14)t(11;14)(q13;q32),+15,+18,+19,+21[5]/51,idem,−X,−del(6)(q2?1)[9]/51,idem,−X,−del(6)(q2?1), dmin[17]/46,XX[2] |
| 38 | 51,X,−X,+5,−8,+9,+11,+15,+15,+19,+der(3;8)(q10;q10)[25]/102,idemx2[2]/46,XX[5] |
| 39 | 50,XX,del(1)(p21p31),+5,+9,+15,+19[15]/46,XX[10] |
| 40 | 52,XY,+X,+3,+5,+11,+15,−16,+19,+21[3]/46,XY[3] |
| 41 | 49,X,−X,−1,del(5)(q21q34),+der(6)t(6;12)(q21;q13),+7,+9,−12,add(15)(q25).ish(wcp15+,wcp15−),add(15)(q25).ish(wcp15+,wcp15−),der(16)t(1;16) (p34;q24).ish(wcp1+),+18,+19,+19,−22,+der(1;15)(q10;q10).ish(wcp1+,wcp15+),0∼2dmin[8]/49,idem,inv(2)(p2?3q3?3)[3]/49,idem,add(X)(p2?2), del(2)(p1?4p24),del(12)(q11q23),−add(15)(q25),+mar1[4]/46,XX[13] |
| 42 | 59〈3n〉,XXX,−1,t(2;8)(p12;q24),−4,−8,+9,−10,−12,−13,−14,+15,−16,−17,−18,der(19)t(1;19)(q12;p12),−20,−21,del(22)(q?)[5]/68,idem,+t(2;8)(p12;q24), +5,+7,+9,+10,+15,+17,+der(19)t(1;19)(q12;p12),+20[15] |
| 43 | 55,XY,t(1;19)(q24;porqter),+5,+del(6)(q15q27),+7,+9,+add(11)(q23),+add(15)(q15),−16,add(17)(p11),+19,+19,+21,+mar[13]/56,idem,+7[4]/46,XY[17] |
| 44 | 53∼55,X,−X,del(1)(p12p31),+2,+3,+5,+7,+9,−10,add(10)(q26),+11,−12,add(12)(q24),+15,+15,+mar1,+mar2,+mar3[cp3]/46,XX[47] |
| 45 | 52,XY,+5,+9,+11,add(12)(q24),+15,add(16)(q2?3),+19,+21[8]/46,XY[11] |
| 46 | 53,XX,+5,+9,+9,+11,−13,+15,+15,+2mar[1]/46,XX[37] |
| 47 | 55,XX,+3,+5,+7,+9,+11,+15,+15,+19,+21[17]/46,XX[1] |
| 48 | 53∼55,X,−X,+3,+5,+6,+7,+8,+9qh+,+9,+11,−13,+15,+15,−16,+19,+mar1[cp3]/46,XX,9qh+[15] |
| 49 | 52,XX,+3,+5,+der(6)t(1;6)(q12;q11),+9,−13,+15,+19,+20[12]/103〈4n〉,idem,−11[1]/46,XX[15] |
| 50 | 55,XX,+X,+3,+5,+9,+12,add(14)(q32),+15,add(17)(p11),+19,+19,+21[1]/46,XX[28] |
| 51 | 53,XY,del(1)(p12p21),+del(3)(p12p26),+4,+9,+11,t(11;14)(q13;q32),+15,+19,+22[2]/52,X,−Y,idem[8]/46,XY[30] |
| 52 | 72〈3n〉,X,−X,−X,−1,t(2;?8;?17)(p12;q24;q12).ish(wcp2+),+5,del(6)(q22),+i(6)(p10),+7,der(8)t(8;17)(q24;q12),+9,−10,der(10)t(1;10)(q12;q26),+11, add(11)(p15),−13,+15,der(16)t(1;16)(q12;q11),+19,+21,−22[13]/46,XX[15] |
| 53 | 55,X,−X,+i(1)(q10),+3,+5,+7,+9,+9,+11,−13,+15,+18,+19,+19[8]/46,XX[8] |
| 54 | 48,X,−Y,−1,del(1)(q31),del(2)(p23),−4,+7,+9,add(16)(p12),add(17)(p11),+add(18)(p11),+19,+mar[cp6]/46,XY[21] |
| 55 | 51,XY,del(1)(p21p31),+5,+7,+9,−13,−14,+15,der(16)t(1;16)(q12;q11),+19,der(19)t(1;19)(p21;q13),+21,+mar[8]/46,XY[12] |
| 56 | 54,X,−Y,+der(1)t(1;8)(p13;q12),t(1;8)(?p22;q24),del(1)(p21p22),+3,t(4;16)(q13;p10),+5,+add(5)(p15),+7,−8,+9,+11,+15,der(8;16)(q11;q11),+19,+21[11]/ 46,XY[11] |
| 57 | 51,X,−X,+3,+5,+9,dup(11)(q12q25),+15,der(16)t(11;16)(q12;p13),+19,+21[17]/46,XX[1] |
| 58 | 47∼49,XY,der(1)t(1;4)(p22;q13),+del(1)(q12),+3,−4,del(4)(q13),+7,add(8)(p11),der(10)t(1;10)(q10;p13),−13,−16,−18,add(22)(q11),+2∼4mar,inc[cp3]/46,XY[17] |
| 59 | 53,XY,der(2)t(1;2)(q12;q37),+3,i(8)(q10),+9,+11,+15,−18,+19,+19,−21,+mar1,+mar2[7]/46,XY[4] |
| 60 | 49,XY,+2,+3,del(5)(q21q31),+del(5)(q21q31),−6,+7,−8,−11,−15,+16,−17,+21,−22,+3mar,inc[1]/46,XY[21] |
| 61 | 52,XX,add(8)(p11),+add(8)(p11)×2,+9,t(11;14)(q13;q32),+der(11)t(11;14)(q13;q32),+18,+21,del(22)(q11)[18] |
| 62 | 55∼56,XY,+2,+add(3)(p13),+5,+add(6)(q27),+del(7)(?p13),add(8)(p11),+9,+10,+11,−13,der(15)t(1;15)(?q12;p11),+15,+15,der(17)t(1;17)(?q12;p12),+18[4]/ 46,XY[37] |
| 63 | 55,XX,+del(1)(p11p34),+3,+5,+7,+7,+9,+9,+11,−13,+15,+19[7]/56,idem,+del(1)(p11),?del(9)(q1?2q3?1)[3]/46,XX[11] |
| 64 | 54,XY,+3,+5,del(6)(?q16?q27),+7,der(8;18)(q10;p10),+der(8;18)(q10;p10),+del(9)(p13),+11,der(12;18)(q10;q10),+15,add(17)(p1?2),+19[10]/54,idem,del(13) (q14q34)[1]/56,XY,idem,+Y,+21[1]/111∼113,idemx2[2]/46,XY[1] |
| 65 | 51,XY,del(1)(q21),add(1)(?p11),+add(1)(?p11),+3,+add(5)(q12),add(7)(p12),−8,+9,add(10)(q24),add(11)(q14),+del(11)(q13),add(12)(p11),−14,+15,−18,+19, +mar[2]/51∼52,XY,+3,+5,−8,+9,+add(11)(p11),add(11)(q14)×2,+del(11)(q13),−12,−14,+15,+19,+2∼3mar[4]/46,XY[10] |
| 66 | 51,X,−X,+5,del(6)(q21q26)×2,+6,+7,dup(8)(q12q21),+9,−13,+15,+15,−16,+19,+mar[30]/51,idem,del(1)(p22p32-33)[1]/46,XX[9] |
| 67 | 48,XX,del(1)(p21p34),−4,del(8)(q11q22),inv(9)(p12p24),+12,+15,+19,der(1;22)(q10;q10)[6]/49,idem,+20[3]/46,XX[32] |
| 68 | 58,XY,+3,+5,+7,add(8)(p11),+9,+9,+11,+11,+15,+15,+19,+21,+22[cp2]/46,XY[18] |
| 69 | 53,XY,del(1)(p13p31),+3,+5,del(6)(q16q27),+9,+11,add(11)(p15),+15,+15,+19[16]/46,XY[27] |
| 70 | 46∼56,XY,+2,+3,+5,+6,del(6)(q23)×2,+7,+9,der(9)t(1;9)(q12;qter)×2,+11,+15,+17,+19,+mar[cp5]/46,XY[15] |
| 71 | 55,X,−X,+1,+4,−5,+9,+10,+11,−12,+15,+15,+16,+18,+19,+20,+22,+22[1]/46,XX[24] |
| 72 | 55,XY,t(3;5)(q21;q31),+3,+5,+6,+7,+9,+9,+11,+15,+19[8]/45,X,−Y[10]/46,XY[40] |
| 73 | 48,X,−Y,der(1)t(1;4)(p13;p14),−4,+5,del(6)(q15),+add(11)(p15),add(16)(?p11)×2,add(17)(p11),+19,+22[7]/46,XY[12] |
| 74 | 58,X,−X,+3,+3,+5,+5,+9,+9,+11,+15,+15,+19,+21,+mar1,+mar2[2]/46,XX[5] |
| 75 | 55,XX,t(2;6)(q36;p12),+3,+4,+5,+7,+9,+9,+11,+15,+19[11]/46,XX[38] |
| Pseudodiploid karyotypes | |
| 76 | 46,XY,del(13)(q1?3q14),der(14)t(11;14)(q13;q32)[cp2]/46,XY[24] |
| 77 | 46,XX,del(11)(q22)[2]/46,idem,t(del(11)(q22);18)(p15;p113)[27]/45,XX,add(1)(q44),−2,−16,−20,+mar1,+mar2,1dmin[3]/46,XX[10] |
| 78 | 46,XY,del(6)(q12),t(11;14)(q13;q32)[11]/46,XY[9] |
| 79 | 46,XY,+9,der(14)t(11;14)(q13;q32),−20[2]/45,idem,−13,−18,+mar[2]/46,XY[13] |
| 80 | 46,XX,t(11;14)(q13;q32)[1]/46,XX[36] |
| 81 | 46,XX,del(2)(p23),−3,del(3)(p21),add(9)(q34),der(14)t(11;14)(q13;q32),+mar[4]/46,XX[26] |
| 82 | 46,XX,t(11;14)(q13;q32)[2]/46,XX[54] |
| 83 | 46,X,−Y,dup(1)(q12q41),add(1)(p11),+3,+add(7)(p13),t(8;14)(q24;q32),t(9;11)(p13;p13),add(12)(q24),−13,t(16;20)(p13;q13)[20] |
| 84 | 46,XY,t(8;14)(q24;q32),der(14)t(11;14)(q13;q32)[15]/46,XY[5] |
| Hypodiploid karyotypes with or without hypotetraploid duplicated mitoses | |
| 85 | 45,X,−Y,del(13)(q13q21),t(11;14)(q13;q32)[14]/45,idem,t(4;8)(q24;q22)[1]/46,XY[1] |
| 86 | 41∼46,X,−X[10],t(4;9)(p16;q12)[5],−5[18],del(8)(q2?2q2?4)[5],−8[14],−9[6],t(11;14)(q13;q32)[18],−16[10],−18[10],−20[9],−21[5],add(22)(p11)[12],−22[6], +mar1[18],+mar2[18],1dmin[12][cp18]/46,XX[12] |
| 87 | 42,−X,Y,der(1)dic(1;?)(?;?),del(2)(p24),add(2)(q37),add(3)(q26),add(6)(q24),add(7)(p13),der(10)add(10)(p11)add(10)(q24),−12,−13,del(14)(q23q32),dic(16;?) (q13;?),del(17)(p12),del(19)(q12),dic(21;?),−21,+r[11]/42,idem,−add(2)(q37),−del(19)(q12),+add(19)(q12),+mar1[3]/42,idem,add(2)(q13),−add(2)(q37), −18,+mar2[3] |
| 88 | 45,XY,add(3)(q26),t(6;12)(p24;q13),t(11;14)(q13;q32),−13,−14,add(15)(q26),−21,+mar1,+mar2[cp2]/46,XY[13] |
| 89 | 44,X,−X,del(1)(p22p31),del(4)(p13p1?5),der(4)t(1;4)(q11;p16),del(6)(q13q26),add(8)(p11),del(9)(p1?3),del(11)(q13q2?3),−13[cp9]/46,XX[22] |
| 90 | 45,XY,del(1)(p13p31),del(4)(q24q28),add(12)(q24),−13,−14,add(17)(p11),−19,+mar1,+mar2[cp9]/46,XY[22] |
| 91 | 43,X,−X,i(1)(q10),del(6)(q12q16),−13,−14,der(20)t(X;20)(p12;p11)[9]/46,XX[1] |
| 92 | 45,X,−X,t(2;18)(q3?2;q12),t(11;14)(q13;q32),del(13)(q12q21)[12]/45,idem,dup(1)(q?12q43)[2]/45,idem,?del(6)(q11q13)[3]/45,idem,der(20)t(1;20)(q12;p13)[1]/ 46,XX[2] |
| 93 | 40∼42,X,−Y,add(1)(p32),−2,add(2)(q31),add(3)(q26),del(4)(p12p15),add(5)(q15),−6,del(6)(q11q25),add(8)(p11)×2,+add(8)(p11),der(10)t(10;13)(p1?3;q1?4), −13,t(13;15)(q13;q21),−14,add(15)(q24),−16,der(17)t(1,17)(q12;p12),−20,add(20)(porq),−21,−21,−22,+del(22)(q11),+r(?),+mar1,+mar2[cp9]/46,XY[1] |
| 94 | 43,X,−X,del(1)(p12p31),del(9)(p13p21),t(11;14)(q13;q32),del(11)(q14q22),del(12)(p11p13),−13,del(14)(q21q32),−22[21]/46,XX[41] |
| 95 | 44∼45,X,−X,t(1;8)(p13;q24),der(2)t(2;7)(q24;q21),der(8)t(8;13)(p?22;q?13),−13,add(14)(q32),der(14)t(1;14)(q12;q32),+mar[cp10] |
| 96 | 45,XX,del(6)(q12),−8,t(11;14)(q13;q32),+der(11)t(11;14)(q13;q32),−13,−16,+22[3]/46,idem,+18[2]/46,XX[15] |
| 97 | 45,XY,del(11)(q14q2?2),−13,add(14)(q32),der(17)t(1;17)(q12;q2?2)[2]/44,XY,der(8;12)(q10;q10),del(11)(q14q2?2),−13,del(14)(q11q32),der(17)t(1;17)(q12;q2?2) [13]/46,XY(12] |
| 98 | 44,X,−X,t(11;14)(q13;q32),−13[1]/46,XX[43] |
| 99 | 40,XY,−1,del(1)(q11q41).ish(wcp1+),der(2)t(1;2)(?;q22).ish(wcp1+),del(4)(p15.2p16),add(4)(p16),add(5)(q12).ish(wcp5+),add(5)(q14).ish(wcp5+),add(6)(q26), del(7)(q10q21),−8,der(8)t(1;8)(p?;p10).ish(wcp1+)?dup(8)(q?),add(9)(q34),der(11)t(11;13)(p12;q13).ish(wcp11+),add(12)(p13),−12,del(13)(q?12?q32).ish (wcp13+),der(13)t(5;13)(q14;q12).ish(wcp5+,wcp13+),−14,dic(15;19)(p10;q13.3).ish(wcp15+,wcp19+),add(15)(p10).ish(wcp15+,wcp15−),der(16)t(1;16) (?;q24).ish(wcp1+),der(17)t(?12;17)(p13;p13),del(19)(p13).ish(wcp19+),−20,der(21)t(1;21)(?;p13).ish(wcp1+),−22,+mar[22] |
| 100 | 45,X,−Y,del(1)(p11p33),add(8)(p21),der(12)t(1;12)(q12;q21),del(15)(q15q25),add(16)(q12),der(19)t(1;19)(q11;porqter)[3]/44,X,−Y,add(8)(q24),der(12)t(1;12) (q12;q21),−13,der(19)t(1;19)(q11;qorpter)[cp2]/46,XY[7] |
| 101 | 44∼46,XY,del(1)(q32q43),add(2)(q23),der(11)t(8;11)(q14;q13),der(12)t(12;16)(p12;p11),−13,add(14)(p11),−16,−18,der(18)t(18;21)(q10;q10)×2,−21,+22[cp25]/ 46,XY[6] |
| 102 | 44,X,−X,der(7)t(1;?;7)(q11;?;q35),−13[1]/44,idem,add(8)(q24),−13,+mar[3]/46,XX[10] |
| 103 | 43,XX,−13,−14,−20,−21,+mar1[1]/43,XX,dup(5)(q31q34),−13,−14,−20,−21,+mar[10]/46,XX[9] |
| 104 | 44,X,−X,−8,−10,t(11;14)(q13;q32),−22,+2mar[1]/46,XX[42] |
| 105 | 45,XY,−5,add(11)(q2?5),t(11;14)(q13;q32),der(13)t(5;13)(q23q;q34),add(20)(q12)[20]/46,XY[2] |
| 106 | 44,XY,−1,t(3;12)(p21;q13),del(6)(p22),+7,−13,−14,der(18)t(1;18)(q12;q2?3),del(19)(p12),del(22)(q11q13)[cp10]/46,idem,t(5;16)(q11;q13)[cp4]/46,XY[6] |
| 107 | 43,X,−Y,der(8)t(Y;8)(q1?1;p1?1),−12,−13,der(14)(12qter→pter::14p11→qter),add(16)(p12),del(16)(q21)[23]/43,idem,der(Y;4)(q10;p10),del(5)(q14q23), −der(8)t(Y;8)(q1?1;p1?1),der(4;8)(q10;q10)[4]/46,XY[12] |
| 108 | 43∼44,XY,del(1)(p21p31)[8],−4[8],del(6)(q24)[6],add(7)(p11)[4],del(11)(p15)[5],−13[8],−14[8],der(16)t(1;16)(q12;q11)[8],−17[8],+19[6],+mar1[8],+mar2[3] [cp8]/46,XY[5] |
| 109 | 45,XY,del(1)(p21p31),−1,der(2)t(1;2)(q12;qter),t(2;7)(p12;q11),del(6)(q21),der(6)t(3;6)(q21;q15),trip(8)(q22q23),del(9)(p11),del(12)(q22q23),del(13)(q13q31), +19,der(20)t(1;20)(p12;q12),der(1;22)(?q10;?q10),+mar1,+mar2[cp14]/46,XY[2] |
| 110 | 44,XX,−1,der(8)t(1;8)(p11-p12;q24),t(11;14)(q13;q32),del(14)(q21q32),der(15)t(1;15)(q12;p11),add(16)(q10),der(18)(18pter→18p11::1?::14q32::11q13→11qter), −22,+mar[16] |
| 111 | 44,XY,−13,t(12;14)(p;q),−14,add(16)(q?1)[6]/46,XY[16] |
| 112 | 43,XY,−3,−5,add(6)(p25),+8,−13,−15,−18,+der(5;15)(p10;q10)[20] |
| 113 | 43,XX,add(1)(q44),add(5)(p15),−8,−10,−12,−13,del(14)(q22q23),−16,add(17)(p11),−20,+r,+der(?)t(?;8)(?;q1?2),+mar,1∼2dmin[cp9]/46,XX[15] |
| 114 | 43,XY,add(1)(p1?2),del(2)(q31q36).ish(wcp2+),der(6)t(6;13)(?q21;?q12).ish(wcp6+,wcp13+),del(12)(p11),−13,−13,del(14)(q22q32),−22[3]/43,idem,der(9) t(1;9)(q12;p22)[18]/46,XY[8] |
| 115 | 45,X,−X,t(11;14)(q13;q32)[5]/46,XX[21] |
| 116 | 45,XX,dup(9)(q12qter).ish(wcp9+),del(12)(q21q24),−13,−14,del(16)(q21q23),+der(16)t(1;16)(q12;q11).ish(wcp1+,wcp16+)[25] |
| 117 | 42,X,−X,i(1)(q10),add(2)(q32),t(3;16)(q21;p13),del(6)(q21q25),del(8)(q24),−13,−14,−22[cp4]/84,idemx2,−7,−12,add(19)(qter),add(20)(?),+mar[1]/46,XX[20] |
| 118 | 44,XX,del(1)(p21p31),add(4)(q34),add(7)(p13),add(8)(p12),−13,−14,−15,−18,+mar[cp2]/46,XX[20] |
| 119 | 45,X,der(X)t(X;12)(q25;q11),add(10)(q26),t(11;14)(q13;q32),der(12)t(X;12)(q25;q11),der(16)t(1;16)(q12;q12),add(17)(q2?2)[10]/46,XX[17] |
| 120 | 44,XY,del(2)(p13),add(5)(p?),−7,−10,t(11;14)(q13;q32),add(17)(q25)[3]/46,XY[33] |
| 121 | 43,XY,der(2)t(1;2)(q12;p25),del(6)(q16),−7,der(7;12)(q10;q10),add(8)(q23),−13,der(17)t(1;17)(q12;q25),−22[3]/46,XY[22] |
| 122 | 43,XX,1qh+(c),der(1)t(1;9)(q43;q11).ish(wcp9+),−9,−13,−14,add(15)(p11)[2]/44∼45,XX,1qh+(c),der(9)t(9;9)(p23;q11).ish(wcp9+),−13,−14,add(15)(p11) [cp9]/46,XX[79] |
| 123 | 44,X,−X,del(1)(p13),+i(1)(q10),der(5)del(5)(p11p15)t(5;8)(q33;q24),add(6)(q15),der(8)t(6;8)(q15;q24)ins(8;5)(q24;?q33?q35),del(9)(q13q33),−13,add(17) (p13),add(21)(q22),der(21;22)(q10;q10)[7]/44,idem,der(1)i(1)(q10)del(1)(q11q21)[17] |
| 124 | 43,X,−Y,del(1)(p12p31),del(6)(q13q27),add(6)(q21),der(12)t(1;12)(q12;q24),−14,−15[6]/44,idem,+19[7]/var[3]/46,XY[2] |
| 125 | 45,XY,del(1)(p21p31),del(9)t(1;9)(q11;q34),del(12)(q13q24),−13,add(14)(q32),del(16)(q2?1),+19[26]/44,idem,del(6)(q16),t(add(14)(q32);15)(q10;q10)[15]/ 90,idemx2[2]/46,XY[3] |
| 126 | 40∼47,X,−Y,del(1)(p12),del(3)(q27),add(4)(q12),−7,der(7)t(2;7)(q22;q31),−8,−8,add(11)(p11),der(11)t(1;11)(p21;q23),−12,add(14)(q32),der(16)t(?;16) t(?;q10),−17,−18,add(19)(?p13),−20,add(20)(p11),−21,del(22)(q11),+mar1,+mar2,+mar3[12]/37∼40,idem,t(13;13)(p10;p10)[2]/76∼80,idemx2[6]/85, idem,t(13;13)(p10;p10)×2[1] |
| 127 | 43,X,−Y,der(1)t(1;1)(p35;q12),del(1)(p13p32),−13,−16,del(22)(q11)[2]/86〈4n〉,idemx2[2]/46,XY[33] |
| 128 | 44∼45,X,−X,+del(1)(q21?),−5,−6,−12,−13,−15,−16,+mar1,+mar2,+mar3,+mar4,+mar5[cp8]/81,idemx2[cp3]/46,XX[6] |
| 129 | 30,X,+3,+7,+9,+11,−13,+18,+19,+mar1,+mar2[7]/57∼58,idemx2,add(11)(q?23),+mar3[3]/46,XY[3] |
| 130 | 40∼41,XY,−1,−2,add(2)(q11),−13,−14,add(16)(p13),der(17)t(2;17)(?q11q12;p12p13),−20,i(22)(q10)[6]/40,idem,derdic(1)t(1;?8)(q32;?p11),−8, derdic(13)t(8;13)(p11;p11)[6]/40,idem,del(7)(p12)[4]/88,idemx2[4]/46,XY[3] |
| 131 | 43,X,del(X)(q12q26),i(1)(q10),der(4)t(1;4)(q12;p13),−6,del(8)(q11q22),−10,−13,add(13)(q33),−14,der(16)t(6;16)(p11;q12),+mar[1]/85〈4n〉,idemx2,−8[1]/ 42,X,del(X)(12q26),i(1)(q10),der(4)t(1;4)(q12p13),−6,t(7;14)(q22;q32),del(8)(q11q22),−10,del(12)(q13q24),−13,add(13)(q33),−14,der(16)t(6,16)(p11;q12) [3]/84〈4n〉,idemx2[11]/46,XX[7] |
| 132 | 44,XY,del(2)(p2?2),add(3)(q27),del(3)(p21),dup(4)(q13q31),−8,t(11;14)(q13;q32),del(12)(q22),−13,+der(16)t(1;16)(q12;q12),add(18)(q21),−19,+mar[13]/ 88〈4n〉,idemx2[1]/46,XY[21] |
| 133 | 45,XX,del(1)(p21p31),del(1)(p32p34),der(2)t(2;13)(q34;q14),del(2)(q31q34),t(3;14)(p11;q32),del(5)(q13q22),der(7)(qter→cen::q21→qter),−13,der(13)t(7;13) (p10;q10),−20,add(22)(q12),+mar[13]/44,idem,−X[5]/90,idemx2[6] |
| Hypotetraploid karyotypes | |
| 134 | 84∼90〈4n〉,XXYY,del(1)(p12p31)×2,−2,del(6)(q11q26)×2,del(7)(q32)×2,−8,−10,add(12)(p13),−13,−14,−14,add(14)(q32),−15,−15,−16,−17,−18,−19, −20,−20,+2∼5mar[cp10]/46,XY[1] |
| 135 | 75∼80〈4n〉,X,del(X)(p11),Y,−Y,−1,del(1)(p21p31),−4,−8,−9,−9,−10,−11,add(12)(p11)×2,−13,−13,der(13)t(9;13)(q11;p11)×2,add(14)(q32)×2,der(16) t(1;16)(q12;q11)×2,−19,−19,−20,−21,−22,−22,+2mar[7]/46,XY[32] |
| 136 | 82〈4n〉,X,−X,−Y,−Y,i(1)(q10)×2,−4,−5,der(5)t(1;5)(q12;q11),−6,+der(9)t(9;17)(p11;q11),−13,−13,add(13)(p11)×2,−14,i(16)(p10),−16,−17[21]/46,XY[6] |
| 137 | 76∼86〈4n〉,XXXX,i(1)(q10),add(1)(q11)×2,add?(3)(q27),del(6)(q13q16),−7,−7,−8,−11,−11,t(11;14)(q13;q32),−12,−13,−13,der(14)(?::14p11→q11:: 14q13→14q32::11q13→11qter),der(14)add(14)(p11)del(14)(q11q13)×2,i(15)(q10)×2,−16,−16,add?(17)(p11),i(21)(q10),−22,−22,+mar[10]/46,XX[8] |
| 138 | 88∼90,XX,−Y,−Y,del(1)(?p21p31)×2,−4,−6,−6,−7,−8,+9,+9,del(11)(?q13),−13,−13,add(14)(?q32),der(?16)t(1;?16)(q12;q11),+19,+19,−20,−20, +5∼10mar[cp3]/46,XY[40] |
| Hyperdiploid karyotypes | |
| 1 | 48,XY,+5,der(7)t(7;8)(p22;q13),+der(7)t(7;8)(p22;q13),−8,−13,der(16)t(1;16)(q12;q11),+der(19)t(1;19)(q12;q11)×2,add(22)(p13)[3]/48,idem,−7,der(7)t(1;7) (q12;p22),t(8;14)(q13;q31),−der(16)t(1;16)(q12;q11)[11]/48,idem,del(6)(q15),−7,+der(7)t(1;7)(q12;p22),t(8;14)(q13;q31),−der(16)(1;16)(q12;q11)[8]/96, idemx2,del(6)(q15),der(7)t(1;7)(q12;p22),t(8;14)(q13;q31),−der(16)t(1;16)(q12;q11)[1]/46,XY[5] |
| 2 | 49,XY,+5,+9,der(12)t(3;12)(p21;p13),del(14)(q24q32),+19,del(20)(q11)[8]/46,XY[15] |
| 3 | 51,XY,add(4)(qter),+5,del(5)(q21q34),del(6)(q13q27),+6,+9,del(11)(p11),del(13)(q13q22),+15,add(17)(q?),del(17)(q?),+19,+20,−22[1]/46,XY[101] |
| 4 | 53,X,−X,t(2;6)(q33;p23),+3,+5,+6,+7,+9,+15,+18,+19[2]/54,idem,+21[4]/56,idem,+X,+8,+21[12]/46,XX[30] |
| 5 | 52,XY,+9,+add(11)(p11),+15,+18,+19,+21,der(22)t(11;22)(p12;p12)[16]/49,XY,add(8)(p11),+add(11)(p11),−13,+15,der(16)t(1;16)(q12;q11),+18,+21, der(22)t(11;22)(p12;p12)[5]/98,idemx2[1]/98,idemx2,del(6)(q11),del(11)(q13)[1]/46,XY[8] |
| 6 | 54,X,−Y,+3,+5,+9,+9,+11,+11,+15,+15,+19[3]/55,idem,+r1[3]/55,idem,+r2[2]/55,XY,+3,+5,+9,+9,+11,+15,+19,+r2[9]/55,idem,−r2,+r1[9] |
| 7 | 47,XX,+9,−13,−14,+19,+mar[1]/46,XX[40] |
| 8 | 54,XX,+3,i(4)(p10)×2,del(6)(q?21),+7,+del(9)(p22),+11,del(12)(p13),−13,+15,+15,−16,−16,add(17)(p11),+del(20)(q?23)×2,+mar1,+mar2[cp4]/54,idem, +mar3,+mar4[cp2]/54,idem,tas(15;15)(p13;p13)[cp3]/46,XX[21] |
| 9 | 52,XY,+5,+9,+11,−13,+15,−17,+19,+21,+mar1,+mar2[2]/46,XY[32] |
| 10 | 54,X,−X,+3,+5,der(8)t(7;8)(q1?1;p1?1).ish(wcp8+),+9,+11,+15,+15,+19,+der(21)t(1;21)(q11,q22).ish(wcp1+)[16]/54,idem,add(14)(p11)[1]/46,XX[28] |
| 11 | 55,XX,+3,+4,+5,+6,+7,+9,+11,+15,+21[5]/46,XX[38] |
| 12 | 49,XY,t(1;6)(q1?1;q2?5),+der(6)t(1;6)(q1?1;q2?5),+9,+18,−21,add(22)(p1?1),+mar[cp4]/98,idemx2[1]/46,XY[89] |
| 13 | 54,XX,+3,+5,+7,+9,+9,+15,+19,+21[4]/46,XX[25] |
| 14 | 54,XY,del(1)(p12p31),+add(1)(p11),+3,+5,+7,add(8)(p22),+9,+15,+19,+21[5]/46,XY[8] |
| 15 | 54,XY,add(4)(p1?6),+?del(3)(p11p23),+5,+9,+9,+11,+15,+15,+19,+19[1]/54,XY,t(1;3;?)(q23;p21;?),+?del(3)(p11p23),+5,add(8)(p?22),del(9)(q22q34), +9,+9,+11,+15,+15,+19[5]/54,idem,−7,+mar,inc[2]/46,XY[43] |
| 16 | 52,XY,+3,+5,dic(8;18)(p11;p11),+9,+add(11)(p15),+15,+15,+19[8]/46,XY[47] |
| 17 | 74∼80〈3n〉,XXX,add(9)(q?)×2,+mar,inc[7]/46,XX[18] |
| 18 | 54,XY,del(1)(p21p32),+3,+5,+6,+7,+9,−13,+15,+der(19)t(1;19)(q12;q13),+21,+21[5]/46,XY[17] |
| 19 | 50,X,−Y,del(1)(p22p31),+3,+der(5)t(1;5)(q12;q35),+7,+9,del(11)(p11p12),add(12)(p13),−13,−14,+15,+19,−22,+mar1,+mar2[7]/49,idem,−5[6]/46,XY[8] |
| 20 | 51,XY,add(1)(?p11),+5,+9,+11,+add(15)(p11),−16,+r,+mar[4]/53,idem,+9,+r[3]/46,XY[32] |
| 21 | 51,XY,+5,+add(7)(q36),+9,−13,+18,+19,del(20)(q12),+mar[2]/51,idem,t(5;10)(q35;q24)[2]/46,XY[36] |
| 22 | 47,X,−X,del(1)(p12p34),−4,del(5)(q13q31),+7,+11,+der(1;?)(q10;?)[1]/46,XX[19] |
| 23 | 56∼58,XX,add(1)(q11),add(1)(q21),+3,+5,der(6)t(1;6)(q12;q12)×2,+7,+9,add(13)(p11),−14,−15,add(15)(p11),+18,+19,+19,−21,+22,+5mar[cp13]/46,XX[16] |
| 24 | 47,X,add(X)(p22),del(4)(q24),+add(5)(q11),der(6)t(6;8)(q11;q11),+7,−8,+dup(9)(p11p24),+add(11)(q23),−12,−13,add(13)(q14),−16,−17,+19,+19[16] |
| 25 | 52,X,−Y,+3,+5,+9,add(14)(q32),+15,+15,+19,+21[2]/46,XY[6] |
| 26 | 54,XY,t(1;22)(p34;q11),add(2)(p25),+del(3)(p12),+5,der(6)t(?1;6)(q21;q23),+6,der(7)t(7;11)(p15;q13),del(9)(q?12q34),+9,+15,+der(19)t(15;19)(q1?4;q12), +21,+21[26] |
| 27 | 53,XY,+3,+5,+9,+11,del(13)(q?),add(14)(q32),+19,+mar1,+mar2[1]/46,XY[17] |
| 28 | 50,X,−X,+3,ins(5;17)(p13;q22q25).ish(wcp17+),+7,+9,+15,+del(17)(q22).ish(wcp17+)[18]/49,idem,der(8;22)(q10;p10).ish(wcp22+)[11]/49,idem,−13, der(?8;22)(p10;q10)[3]/49,idem,t(1;13)(p12-13;q32),−7,der(1;22)(q10;q10)[cp4]/46,XX[29] |
| 29 | 47,XX,add(8)(p23),add(18)(q22),+19,der(22)t(1;22)(q12;q13)[3]/46,idem,−X,add(9)(q?),+11,−17[1]/46,XX[4] |
| 30 | 57,XY,del(1)(p12),add(2)(q32)×2,+3,+add(5)(q35),+7,add(8)(p21),add(8)(p11)(q24),+inv(9),+11,+11,+15,+17,+add(19)(p12),+21[12]/46,XY,inv(9)[23] |
| 31 | 51∼53,XX,del(1)(q22),+3,+5,+6,+7,add(8)(p22),+9,−14,+mar1,+mar2,+mar3[inc3]/46,XX[20] |
| 32 | 53,XX,del(1)(p21p31),add(2)(q34),+3,+5,+add(8)(q24),+add(11)(p14),+15,−16,+19,+19,+del(22)(q1?2)[5]/46,XX[10] |
| 33 | 54∼57,XX,t(1;5)(q21;p15),+3,+der(5)t(1;5)[5],add(7)(p15)[2],+9,+9,+11,−13,t(13;15)(q10;q10),+15,+18,+18[3],+19,+21,+21[cp12]/54∼57,idem,−t(13;15) (q10;q10),−13,der(19)t(13;19)(q21;p13)[2]/46,XX[9] |
| 34 | 59∼75〈3n〉,XXX,−1,+7,+6,−12,+19[cp2]/46,XX[35] |
| 35 | 49,XX,−2,+3,add(5)(q35),del(6)(q21),+9,+12,−13,del(14)(q24q32),i(18)(q10),+3mar[1]/46,XX[49] |
| 36 | 46,XY,add(2)(p15),add(2)(q22)[6]/49∼52,XY,add(1)(p31)(q41),del(2)(p11p22),add(3)(p24)(q21)×2,−4,del(4)(p14?),+5,add(6)(q26),+del(7)(q31),−8,+9, +11,der(12)t(9;12)(q11;q24),+15,add(16)(q11),+18,+19,+1∼3mar[cp12]/46,XY(19] |
| 37 | 52,XX,del(6)(q2?1),+7,t(11;14)(q13;q32),+der(14)t(11;14)(q13;q32),+15,+18,+19,+21[5]/51,idem,−X,−del(6)(q2?1)[9]/51,idem,−X,−del(6)(q2?1), dmin[17]/46,XX[2] |
| 38 | 51,X,−X,+5,−8,+9,+11,+15,+15,+19,+der(3;8)(q10;q10)[25]/102,idemx2[2]/46,XX[5] |
| 39 | 50,XX,del(1)(p21p31),+5,+9,+15,+19[15]/46,XX[10] |
| 40 | 52,XY,+X,+3,+5,+11,+15,−16,+19,+21[3]/46,XY[3] |
| 41 | 49,X,−X,−1,del(5)(q21q34),+der(6)t(6;12)(q21;q13),+7,+9,−12,add(15)(q25).ish(wcp15+,wcp15−),add(15)(q25).ish(wcp15+,wcp15−),der(16)t(1;16) (p34;q24).ish(wcp1+),+18,+19,+19,−22,+der(1;15)(q10;q10).ish(wcp1+,wcp15+),0∼2dmin[8]/49,idem,inv(2)(p2?3q3?3)[3]/49,idem,add(X)(p2?2), del(2)(p1?4p24),del(12)(q11q23),−add(15)(q25),+mar1[4]/46,XX[13] |
| 42 | 59〈3n〉,XXX,−1,t(2;8)(p12;q24),−4,−8,+9,−10,−12,−13,−14,+15,−16,−17,−18,der(19)t(1;19)(q12;p12),−20,−21,del(22)(q?)[5]/68,idem,+t(2;8)(p12;q24), +5,+7,+9,+10,+15,+17,+der(19)t(1;19)(q12;p12),+20[15] |
| 43 | 55,XY,t(1;19)(q24;porqter),+5,+del(6)(q15q27),+7,+9,+add(11)(q23),+add(15)(q15),−16,add(17)(p11),+19,+19,+21,+mar[13]/56,idem,+7[4]/46,XY[17] |
| 44 | 53∼55,X,−X,del(1)(p12p31),+2,+3,+5,+7,+9,−10,add(10)(q26),+11,−12,add(12)(q24),+15,+15,+mar1,+mar2,+mar3[cp3]/46,XX[47] |
| 45 | 52,XY,+5,+9,+11,add(12)(q24),+15,add(16)(q2?3),+19,+21[8]/46,XY[11] |
| 46 | 53,XX,+5,+9,+9,+11,−13,+15,+15,+2mar[1]/46,XX[37] |
| 47 | 55,XX,+3,+5,+7,+9,+11,+15,+15,+19,+21[17]/46,XX[1] |
| 48 | 53∼55,X,−X,+3,+5,+6,+7,+8,+9qh+,+9,+11,−13,+15,+15,−16,+19,+mar1[cp3]/46,XX,9qh+[15] |
| 49 | 52,XX,+3,+5,+der(6)t(1;6)(q12;q11),+9,−13,+15,+19,+20[12]/103〈4n〉,idem,−11[1]/46,XX[15] |
| 50 | 55,XX,+X,+3,+5,+9,+12,add(14)(q32),+15,add(17)(p11),+19,+19,+21[1]/46,XX[28] |
| 51 | 53,XY,del(1)(p12p21),+del(3)(p12p26),+4,+9,+11,t(11;14)(q13;q32),+15,+19,+22[2]/52,X,−Y,idem[8]/46,XY[30] |
| 52 | 72〈3n〉,X,−X,−X,−1,t(2;?8;?17)(p12;q24;q12).ish(wcp2+),+5,del(6)(q22),+i(6)(p10),+7,der(8)t(8;17)(q24;q12),+9,−10,der(10)t(1;10)(q12;q26),+11, add(11)(p15),−13,+15,der(16)t(1;16)(q12;q11),+19,+21,−22[13]/46,XX[15] |
| 53 | 55,X,−X,+i(1)(q10),+3,+5,+7,+9,+9,+11,−13,+15,+18,+19,+19[8]/46,XX[8] |
| 54 | 48,X,−Y,−1,del(1)(q31),del(2)(p23),−4,+7,+9,add(16)(p12),add(17)(p11),+add(18)(p11),+19,+mar[cp6]/46,XY[21] |
| 55 | 51,XY,del(1)(p21p31),+5,+7,+9,−13,−14,+15,der(16)t(1;16)(q12;q11),+19,der(19)t(1;19)(p21;q13),+21,+mar[8]/46,XY[12] |
| 56 | 54,X,−Y,+der(1)t(1;8)(p13;q12),t(1;8)(?p22;q24),del(1)(p21p22),+3,t(4;16)(q13;p10),+5,+add(5)(p15),+7,−8,+9,+11,+15,der(8;16)(q11;q11),+19,+21[11]/ 46,XY[11] |
| 57 | 51,X,−X,+3,+5,+9,dup(11)(q12q25),+15,der(16)t(11;16)(q12;p13),+19,+21[17]/46,XX[1] |
| 58 | 47∼49,XY,der(1)t(1;4)(p22;q13),+del(1)(q12),+3,−4,del(4)(q13),+7,add(8)(p11),der(10)t(1;10)(q10;p13),−13,−16,−18,add(22)(q11),+2∼4mar,inc[cp3]/46,XY[17] |
| 59 | 53,XY,der(2)t(1;2)(q12;q37),+3,i(8)(q10),+9,+11,+15,−18,+19,+19,−21,+mar1,+mar2[7]/46,XY[4] |
| 60 | 49,XY,+2,+3,del(5)(q21q31),+del(5)(q21q31),−6,+7,−8,−11,−15,+16,−17,+21,−22,+3mar,inc[1]/46,XY[21] |
| 61 | 52,XX,add(8)(p11),+add(8)(p11)×2,+9,t(11;14)(q13;q32),+der(11)t(11;14)(q13;q32),+18,+21,del(22)(q11)[18] |
| 62 | 55∼56,XY,+2,+add(3)(p13),+5,+add(6)(q27),+del(7)(?p13),add(8)(p11),+9,+10,+11,−13,der(15)t(1;15)(?q12;p11),+15,+15,der(17)t(1;17)(?q12;p12),+18[4]/ 46,XY[37] |
| 63 | 55,XX,+del(1)(p11p34),+3,+5,+7,+7,+9,+9,+11,−13,+15,+19[7]/56,idem,+del(1)(p11),?del(9)(q1?2q3?1)[3]/46,XX[11] |
| 64 | 54,XY,+3,+5,del(6)(?q16?q27),+7,der(8;18)(q10;p10),+der(8;18)(q10;p10),+del(9)(p13),+11,der(12;18)(q10;q10),+15,add(17)(p1?2),+19[10]/54,idem,del(13) (q14q34)[1]/56,XY,idem,+Y,+21[1]/111∼113,idemx2[2]/46,XY[1] |
| 65 | 51,XY,del(1)(q21),add(1)(?p11),+add(1)(?p11),+3,+add(5)(q12),add(7)(p12),−8,+9,add(10)(q24),add(11)(q14),+del(11)(q13),add(12)(p11),−14,+15,−18,+19, +mar[2]/51∼52,XY,+3,+5,−8,+9,+add(11)(p11),add(11)(q14)×2,+del(11)(q13),−12,−14,+15,+19,+2∼3mar[4]/46,XY[10] |
| 66 | 51,X,−X,+5,del(6)(q21q26)×2,+6,+7,dup(8)(q12q21),+9,−13,+15,+15,−16,+19,+mar[30]/51,idem,del(1)(p22p32-33)[1]/46,XX[9] |
| 67 | 48,XX,del(1)(p21p34),−4,del(8)(q11q22),inv(9)(p12p24),+12,+15,+19,der(1;22)(q10;q10)[6]/49,idem,+20[3]/46,XX[32] |
| 68 | 58,XY,+3,+5,+7,add(8)(p11),+9,+9,+11,+11,+15,+15,+19,+21,+22[cp2]/46,XY[18] |
| 69 | 53,XY,del(1)(p13p31),+3,+5,del(6)(q16q27),+9,+11,add(11)(p15),+15,+15,+19[16]/46,XY[27] |
| 70 | 46∼56,XY,+2,+3,+5,+6,del(6)(q23)×2,+7,+9,der(9)t(1;9)(q12;qter)×2,+11,+15,+17,+19,+mar[cp5]/46,XY[15] |
| 71 | 55,X,−X,+1,+4,−5,+9,+10,+11,−12,+15,+15,+16,+18,+19,+20,+22,+22[1]/46,XX[24] |
| 72 | 55,XY,t(3;5)(q21;q31),+3,+5,+6,+7,+9,+9,+11,+15,+19[8]/45,X,−Y[10]/46,XY[40] |
| 73 | 48,X,−Y,der(1)t(1;4)(p13;p14),−4,+5,del(6)(q15),+add(11)(p15),add(16)(?p11)×2,add(17)(p11),+19,+22[7]/46,XY[12] |
| 74 | 58,X,−X,+3,+3,+5,+5,+9,+9,+11,+15,+15,+19,+21,+mar1,+mar2[2]/46,XX[5] |
| 75 | 55,XX,t(2;6)(q36;p12),+3,+4,+5,+7,+9,+9,+11,+15,+19[11]/46,XX[38] |
| Pseudodiploid karyotypes | |
| 76 | 46,XY,del(13)(q1?3q14),der(14)t(11;14)(q13;q32)[cp2]/46,XY[24] |
| 77 | 46,XX,del(11)(q22)[2]/46,idem,t(del(11)(q22);18)(p15;p113)[27]/45,XX,add(1)(q44),−2,−16,−20,+mar1,+mar2,1dmin[3]/46,XX[10] |
| 78 | 46,XY,del(6)(q12),t(11;14)(q13;q32)[11]/46,XY[9] |
| 79 | 46,XY,+9,der(14)t(11;14)(q13;q32),−20[2]/45,idem,−13,−18,+mar[2]/46,XY[13] |
| 80 | 46,XX,t(11;14)(q13;q32)[1]/46,XX[36] |
| 81 | 46,XX,del(2)(p23),−3,del(3)(p21),add(9)(q34),der(14)t(11;14)(q13;q32),+mar[4]/46,XX[26] |
| 82 | 46,XX,t(11;14)(q13;q32)[2]/46,XX[54] |
| 83 | 46,X,−Y,dup(1)(q12q41),add(1)(p11),+3,+add(7)(p13),t(8;14)(q24;q32),t(9;11)(p13;p13),add(12)(q24),−13,t(16;20)(p13;q13)[20] |
| 84 | 46,XY,t(8;14)(q24;q32),der(14)t(11;14)(q13;q32)[15]/46,XY[5] |
| Hypodiploid karyotypes with or without hypotetraploid duplicated mitoses | |
| 85 | 45,X,−Y,del(13)(q13q21),t(11;14)(q13;q32)[14]/45,idem,t(4;8)(q24;q22)[1]/46,XY[1] |
| 86 | 41∼46,X,−X[10],t(4;9)(p16;q12)[5],−5[18],del(8)(q2?2q2?4)[5],−8[14],−9[6],t(11;14)(q13;q32)[18],−16[10],−18[10],−20[9],−21[5],add(22)(p11)[12],−22[6], +mar1[18],+mar2[18],1dmin[12][cp18]/46,XX[12] |
| 87 | 42,−X,Y,der(1)dic(1;?)(?;?),del(2)(p24),add(2)(q37),add(3)(q26),add(6)(q24),add(7)(p13),der(10)add(10)(p11)add(10)(q24),−12,−13,del(14)(q23q32),dic(16;?) (q13;?),del(17)(p12),del(19)(q12),dic(21;?),−21,+r[11]/42,idem,−add(2)(q37),−del(19)(q12),+add(19)(q12),+mar1[3]/42,idem,add(2)(q13),−add(2)(q37), −18,+mar2[3] |
| 88 | 45,XY,add(3)(q26),t(6;12)(p24;q13),t(11;14)(q13;q32),−13,−14,add(15)(q26),−21,+mar1,+mar2[cp2]/46,XY[13] |
| 89 | 44,X,−X,del(1)(p22p31),del(4)(p13p1?5),der(4)t(1;4)(q11;p16),del(6)(q13q26),add(8)(p11),del(9)(p1?3),del(11)(q13q2?3),−13[cp9]/46,XX[22] |
| 90 | 45,XY,del(1)(p13p31),del(4)(q24q28),add(12)(q24),−13,−14,add(17)(p11),−19,+mar1,+mar2[cp9]/46,XY[22] |
| 91 | 43,X,−X,i(1)(q10),del(6)(q12q16),−13,−14,der(20)t(X;20)(p12;p11)[9]/46,XX[1] |
| 92 | 45,X,−X,t(2;18)(q3?2;q12),t(11;14)(q13;q32),del(13)(q12q21)[12]/45,idem,dup(1)(q?12q43)[2]/45,idem,?del(6)(q11q13)[3]/45,idem,der(20)t(1;20)(q12;p13)[1]/ 46,XX[2] |
| 93 | 40∼42,X,−Y,add(1)(p32),−2,add(2)(q31),add(3)(q26),del(4)(p12p15),add(5)(q15),−6,del(6)(q11q25),add(8)(p11)×2,+add(8)(p11),der(10)t(10;13)(p1?3;q1?4), −13,t(13;15)(q13;q21),−14,add(15)(q24),−16,der(17)t(1,17)(q12;p12),−20,add(20)(porq),−21,−21,−22,+del(22)(q11),+r(?),+mar1,+mar2[cp9]/46,XY[1] |
| 94 | 43,X,−X,del(1)(p12p31),del(9)(p13p21),t(11;14)(q13;q32),del(11)(q14q22),del(12)(p11p13),−13,del(14)(q21q32),−22[21]/46,XX[41] |
| 95 | 44∼45,X,−X,t(1;8)(p13;q24),der(2)t(2;7)(q24;q21),der(8)t(8;13)(p?22;q?13),−13,add(14)(q32),der(14)t(1;14)(q12;q32),+mar[cp10] |
| 96 | 45,XX,del(6)(q12),−8,t(11;14)(q13;q32),+der(11)t(11;14)(q13;q32),−13,−16,+22[3]/46,idem,+18[2]/46,XX[15] |
| 97 | 45,XY,del(11)(q14q2?2),−13,add(14)(q32),der(17)t(1;17)(q12;q2?2)[2]/44,XY,der(8;12)(q10;q10),del(11)(q14q2?2),−13,del(14)(q11q32),der(17)t(1;17)(q12;q2?2) [13]/46,XY(12] |
| 98 | 44,X,−X,t(11;14)(q13;q32),−13[1]/46,XX[43] |
| 99 | 40,XY,−1,del(1)(q11q41).ish(wcp1+),der(2)t(1;2)(?;q22).ish(wcp1+),del(4)(p15.2p16),add(4)(p16),add(5)(q12).ish(wcp5+),add(5)(q14).ish(wcp5+),add(6)(q26), del(7)(q10q21),−8,der(8)t(1;8)(p?;p10).ish(wcp1+)?dup(8)(q?),add(9)(q34),der(11)t(11;13)(p12;q13).ish(wcp11+),add(12)(p13),−12,del(13)(q?12?q32).ish (wcp13+),der(13)t(5;13)(q14;q12).ish(wcp5+,wcp13+),−14,dic(15;19)(p10;q13.3).ish(wcp15+,wcp19+),add(15)(p10).ish(wcp15+,wcp15−),der(16)t(1;16) (?;q24).ish(wcp1+),der(17)t(?12;17)(p13;p13),del(19)(p13).ish(wcp19+),−20,der(21)t(1;21)(?;p13).ish(wcp1+),−22,+mar[22] |
| 100 | 45,X,−Y,del(1)(p11p33),add(8)(p21),der(12)t(1;12)(q12;q21),del(15)(q15q25),add(16)(q12),der(19)t(1;19)(q11;porqter)[3]/44,X,−Y,add(8)(q24),der(12)t(1;12) (q12;q21),−13,der(19)t(1;19)(q11;qorpter)[cp2]/46,XY[7] |
| 101 | 44∼46,XY,del(1)(q32q43),add(2)(q23),der(11)t(8;11)(q14;q13),der(12)t(12;16)(p12;p11),−13,add(14)(p11),−16,−18,der(18)t(18;21)(q10;q10)×2,−21,+22[cp25]/ 46,XY[6] |
| 102 | 44,X,−X,der(7)t(1;?;7)(q11;?;q35),−13[1]/44,idem,add(8)(q24),−13,+mar[3]/46,XX[10] |
| 103 | 43,XX,−13,−14,−20,−21,+mar1[1]/43,XX,dup(5)(q31q34),−13,−14,−20,−21,+mar[10]/46,XX[9] |
| 104 | 44,X,−X,−8,−10,t(11;14)(q13;q32),−22,+2mar[1]/46,XX[42] |
| 105 | 45,XY,−5,add(11)(q2?5),t(11;14)(q13;q32),der(13)t(5;13)(q23q;q34),add(20)(q12)[20]/46,XY[2] |
| 106 | 44,XY,−1,t(3;12)(p21;q13),del(6)(p22),+7,−13,−14,der(18)t(1;18)(q12;q2?3),del(19)(p12),del(22)(q11q13)[cp10]/46,idem,t(5;16)(q11;q13)[cp4]/46,XY[6] |
| 107 | 43,X,−Y,der(8)t(Y;8)(q1?1;p1?1),−12,−13,der(14)(12qter→pter::14p11→qter),add(16)(p12),del(16)(q21)[23]/43,idem,der(Y;4)(q10;p10),del(5)(q14q23), −der(8)t(Y;8)(q1?1;p1?1),der(4;8)(q10;q10)[4]/46,XY[12] |
| 108 | 43∼44,XY,del(1)(p21p31)[8],−4[8],del(6)(q24)[6],add(7)(p11)[4],del(11)(p15)[5],−13[8],−14[8],der(16)t(1;16)(q12;q11)[8],−17[8],+19[6],+mar1[8],+mar2[3] [cp8]/46,XY[5] |
| 109 | 45,XY,del(1)(p21p31),−1,der(2)t(1;2)(q12;qter),t(2;7)(p12;q11),del(6)(q21),der(6)t(3;6)(q21;q15),trip(8)(q22q23),del(9)(p11),del(12)(q22q23),del(13)(q13q31), +19,der(20)t(1;20)(p12;q12),der(1;22)(?q10;?q10),+mar1,+mar2[cp14]/46,XY[2] |
| 110 | 44,XX,−1,der(8)t(1;8)(p11-p12;q24),t(11;14)(q13;q32),del(14)(q21q32),der(15)t(1;15)(q12;p11),add(16)(q10),der(18)(18pter→18p11::1?::14q32::11q13→11qter), −22,+mar[16] |
| 111 | 44,XY,−13,t(12;14)(p;q),−14,add(16)(q?1)[6]/46,XY[16] |
| 112 | 43,XY,−3,−5,add(6)(p25),+8,−13,−15,−18,+der(5;15)(p10;q10)[20] |
| 113 | 43,XX,add(1)(q44),add(5)(p15),−8,−10,−12,−13,del(14)(q22q23),−16,add(17)(p11),−20,+r,+der(?)t(?;8)(?;q1?2),+mar,1∼2dmin[cp9]/46,XX[15] |
| 114 | 43,XY,add(1)(p1?2),del(2)(q31q36).ish(wcp2+),der(6)t(6;13)(?q21;?q12).ish(wcp6+,wcp13+),del(12)(p11),−13,−13,del(14)(q22q32),−22[3]/43,idem,der(9) t(1;9)(q12;p22)[18]/46,XY[8] |
| 115 | 45,X,−X,t(11;14)(q13;q32)[5]/46,XX[21] |
| 116 | 45,XX,dup(9)(q12qter).ish(wcp9+),del(12)(q21q24),−13,−14,del(16)(q21q23),+der(16)t(1;16)(q12;q11).ish(wcp1+,wcp16+)[25] |
| 117 | 42,X,−X,i(1)(q10),add(2)(q32),t(3;16)(q21;p13),del(6)(q21q25),del(8)(q24),−13,−14,−22[cp4]/84,idemx2,−7,−12,add(19)(qter),add(20)(?),+mar[1]/46,XX[20] |
| 118 | 44,XX,del(1)(p21p31),add(4)(q34),add(7)(p13),add(8)(p12),−13,−14,−15,−18,+mar[cp2]/46,XX[20] |
| 119 | 45,X,der(X)t(X;12)(q25;q11),add(10)(q26),t(11;14)(q13;q32),der(12)t(X;12)(q25;q11),der(16)t(1;16)(q12;q12),add(17)(q2?2)[10]/46,XX[17] |
| 120 | 44,XY,del(2)(p13),add(5)(p?),−7,−10,t(11;14)(q13;q32),add(17)(q25)[3]/46,XY[33] |
| 121 | 43,XY,der(2)t(1;2)(q12;p25),del(6)(q16),−7,der(7;12)(q10;q10),add(8)(q23),−13,der(17)t(1;17)(q12;q25),−22[3]/46,XY[22] |
| 122 | 43,XX,1qh+(c),der(1)t(1;9)(q43;q11).ish(wcp9+),−9,−13,−14,add(15)(p11)[2]/44∼45,XX,1qh+(c),der(9)t(9;9)(p23;q11).ish(wcp9+),−13,−14,add(15)(p11) [cp9]/46,XX[79] |
| 123 | 44,X,−X,del(1)(p13),+i(1)(q10),der(5)del(5)(p11p15)t(5;8)(q33;q24),add(6)(q15),der(8)t(6;8)(q15;q24)ins(8;5)(q24;?q33?q35),del(9)(q13q33),−13,add(17) (p13),add(21)(q22),der(21;22)(q10;q10)[7]/44,idem,der(1)i(1)(q10)del(1)(q11q21)[17] |
| 124 | 43,X,−Y,del(1)(p12p31),del(6)(q13q27),add(6)(q21),der(12)t(1;12)(q12;q24),−14,−15[6]/44,idem,+19[7]/var[3]/46,XY[2] |
| 125 | 45,XY,del(1)(p21p31),del(9)t(1;9)(q11;q34),del(12)(q13q24),−13,add(14)(q32),del(16)(q2?1),+19[26]/44,idem,del(6)(q16),t(add(14)(q32);15)(q10;q10)[15]/ 90,idemx2[2]/46,XY[3] |
| 126 | 40∼47,X,−Y,del(1)(p12),del(3)(q27),add(4)(q12),−7,der(7)t(2;7)(q22;q31),−8,−8,add(11)(p11),der(11)t(1;11)(p21;q23),−12,add(14)(q32),der(16)t(?;16) t(?;q10),−17,−18,add(19)(?p13),−20,add(20)(p11),−21,del(22)(q11),+mar1,+mar2,+mar3[12]/37∼40,idem,t(13;13)(p10;p10)[2]/76∼80,idemx2[6]/85, idem,t(13;13)(p10;p10)×2[1] |
| 127 | 43,X,−Y,der(1)t(1;1)(p35;q12),del(1)(p13p32),−13,−16,del(22)(q11)[2]/86〈4n〉,idemx2[2]/46,XY[33] |
| 128 | 44∼45,X,−X,+del(1)(q21?),−5,−6,−12,−13,−15,−16,+mar1,+mar2,+mar3,+mar4,+mar5[cp8]/81,idemx2[cp3]/46,XX[6] |
| 129 | 30,X,+3,+7,+9,+11,−13,+18,+19,+mar1,+mar2[7]/57∼58,idemx2,add(11)(q?23),+mar3[3]/46,XY[3] |
| 130 | 40∼41,XY,−1,−2,add(2)(q11),−13,−14,add(16)(p13),der(17)t(2;17)(?q11q12;p12p13),−20,i(22)(q10)[6]/40,idem,derdic(1)t(1;?8)(q32;?p11),−8, derdic(13)t(8;13)(p11;p11)[6]/40,idem,del(7)(p12)[4]/88,idemx2[4]/46,XY[3] |
| 131 | 43,X,del(X)(q12q26),i(1)(q10),der(4)t(1;4)(q12;p13),−6,del(8)(q11q22),−10,−13,add(13)(q33),−14,der(16)t(6;16)(p11;q12),+mar[1]/85〈4n〉,idemx2,−8[1]/ 42,X,del(X)(12q26),i(1)(q10),der(4)t(1;4)(q12p13),−6,t(7;14)(q22;q32),del(8)(q11q22),−10,del(12)(q13q24),−13,add(13)(q33),−14,der(16)t(6,16)(p11;q12) [3]/84〈4n〉,idemx2[11]/46,XX[7] |
| 132 | 44,XY,del(2)(p2?2),add(3)(q27),del(3)(p21),dup(4)(q13q31),−8,t(11;14)(q13;q32),del(12)(q22),−13,+der(16)t(1;16)(q12;q12),add(18)(q21),−19,+mar[13]/ 88〈4n〉,idemx2[1]/46,XY[21] |
| 133 | 45,XX,del(1)(p21p31),del(1)(p32p34),der(2)t(2;13)(q34;q14),del(2)(q31q34),t(3;14)(p11;q32),del(5)(q13q22),der(7)(qter→cen::q21→qter),−13,der(13)t(7;13) (p10;q10),−20,add(22)(q12),+mar[13]/44,idem,−X[5]/90,idemx2[6] |
| Hypotetraploid karyotypes | |
| 134 | 84∼90〈4n〉,XXYY,del(1)(p12p31)×2,−2,del(6)(q11q26)×2,del(7)(q32)×2,−8,−10,add(12)(p13),−13,−14,−14,add(14)(q32),−15,−15,−16,−17,−18,−19, −20,−20,+2∼5mar[cp10]/46,XY[1] |
| 135 | 75∼80〈4n〉,X,del(X)(p11),Y,−Y,−1,del(1)(p21p31),−4,−8,−9,−9,−10,−11,add(12)(p11)×2,−13,−13,der(13)t(9;13)(q11;p11)×2,add(14)(q32)×2,der(16) t(1;16)(q12;q11)×2,−19,−19,−20,−21,−22,−22,+2mar[7]/46,XY[32] |
| 136 | 82〈4n〉,X,−X,−Y,−Y,i(1)(q10)×2,−4,−5,der(5)t(1;5)(q12;q11),−6,+der(9)t(9;17)(p11;q11),−13,−13,add(13)(p11)×2,−14,i(16)(p10),−16,−17[21]/46,XY[6] |
| 137 | 76∼86〈4n〉,XXXX,i(1)(q10),add(1)(q11)×2,add?(3)(q27),del(6)(q13q16),−7,−7,−8,−11,−11,t(11;14)(q13;q32),−12,−13,−13,der(14)(?::14p11→q11:: 14q13→14q32::11q13→11qter),der(14)add(14)(p11)del(14)(q11q13)×2,i(15)(q10)×2,−16,−16,add?(17)(p11),i(21)(q10),−22,−22,+mar[10]/46,XX[8] |
| 138 | 88∼90,XX,−Y,−Y,del(1)(?p21p31)×2,−4,−6,−6,−7,−8,+9,+9,del(11)(?q13),−13,−13,add(14)(?q32),der(?16)t(1;?16)(q12;q11),+19,+19,−20,−20, +5∼10mar[cp3]/46,XY[40] |
Numerical abnormalities
Patients with abnormal karyotypes belong to 1 of 2 clearly different groups according to the chromosome number pattern. A first group of 75 patients (54%) had hyperdiploid karyotypes (mean 52.5 chromosomes, range, 47-75) characterized by recurrent gains of chromosomes 3, 5, 7, 9, 11, 15, and 19. The most common resulting trisomies were, in decreasing order, trisomies 9, 19, 15, 5, 11, 3, and 7 (Table 3). A total of 45 (60%) of 75 patients had at least 5 of the 7 recurrent chromosome gains. A second group of 63 patients (hypodiploid group, 46%) had either pseudodiploid karyotypes (9 cases, 7%), hypodiploid karyotypes (49 cases, 35%) with duplicated hypotetraploid mitoses in 9 cases, or only hypotetraploid karyotypes (5 cases, 4%). The mean chromosome number was 43.4 (range, 30-45) and 83 (range, 75-90) for hypodiploid and hypotetraploid karyotypes, respectively.
Trisomies distribution according to the chromosome number pattern
| . | Chromosome No. . | ||||||
|---|---|---|---|---|---|---|---|
| 3 . | 5 . | 7 . | 9 . | 11 . | 15 . | 19 . | |
| Hyperdiploid (n = 75) | 42 (53%) | 54 (74%) | 38 (49%) | 63 (86%) | 41 (58%) | 55 (75%) | 59 (80%) |
| Hypodiploid/pseudodiploid/∼4n, (n = 63) | 1 | 0 | 1 | 4 | 1 | 0 | 3 |
| . | Chromosome No. . | ||||||
|---|---|---|---|---|---|---|---|
| 3 . | 5 . | 7 . | 9 . | 11 . | 15 . | 19 . | |
| Hyperdiploid (n = 75) | 42 (53%) | 54 (74%) | 38 (49%) | 63 (86%) | 41 (58%) | 55 (75%) | 59 (80%) |
| Hypodiploid/pseudodiploid/∼4n, (n = 63) | 1 | 0 | 1 | 4 | 1 | 0 | 3 |
Structural abnormalities
Structural abnormalities were present in all but 4 hyperdiploid cases. The number of structural abnormalities per cell was higher in the hypodiploid group (average 9.1) than in the hyperdiploid group (average 5.1). The same abnormalities were present in the 2 groups, with identical or different frequencies (Table4).
Type and distribution of structural abnormalities in the total series and according to the chromosome number pattern
| Abnormalities . | Total series (n = 138) . | Hyperdiploid group (n = 75) . | Hypodiploid group (n = 63) . | P4-150 . |
|---|---|---|---|---|
| Ig regions | ||||
| 14q32 | 43 (31%) | 8 (11%) | 35 (56%) | <.001 |
| 22q11 | 10 (7%) | 5 (7%) | 5 (8%) | ns |
| 11q13 | 31 (22%) | 6 (8%) | 25 (40%) | <.001 |
| −13 or del(13) | 75 (54%) | 26 (35%) | 49 (78%) | <.001 |
| 8q24 | 14 (10%) | 5 (7%) | 9 (14%) | –ns |
| del(6q) | 39 (28%) | 20 (27%) | 19 (30%) | ns |
| −8 or del(8p) | 42 (30%) | 23 (31%) | 19 (30%) | ns |
| dup(1q) | 57 (41%) | 26 (35%) | 31 (49%) | ns |
| del(1p) | 46 (33%) | 22 (29%) | 24 (38%) | ns |
| del or add(17p) | 16 (11.5%) | 7 (9%) | 9 (14%) | ns |
| Abnormalities . | Total series (n = 138) . | Hyperdiploid group (n = 75) . | Hypodiploid group (n = 63) . | P4-150 . |
|---|---|---|---|---|
| Ig regions | ||||
| 14q32 | 43 (31%) | 8 (11%) | 35 (56%) | <.001 |
| 22q11 | 10 (7%) | 5 (7%) | 5 (8%) | ns |
| 11q13 | 31 (22%) | 6 (8%) | 25 (40%) | <.001 |
| −13 or del(13) | 75 (54%) | 26 (35%) | 49 (78%) | <.001 |
| 8q24 | 14 (10%) | 5 (7%) | 9 (14%) | –ns |
| del(6q) | 39 (28%) | 20 (27%) | 19 (30%) | ns |
| −8 or del(8p) | 42 (30%) | 23 (31%) | 19 (30%) | ns |
| dup(1q) | 57 (41%) | 26 (35%) | 31 (49%) | ns |
| del(1p) | 46 (33%) | 22 (29%) | 24 (38%) | ns |
| del or add(17p) | 16 (11.5%) | 7 (9%) | 9 (14%) | ns |
ns, indicates statistically not significant.
Hyperdiploid versus hypodiploid group.
Chromosome 13.
Implication of chromosome 13 (13qA) was the most common abnormality. Complete monosomy 13 was found in 66 cases, while partial deletion including the 13q14 region was recorded in only 8 cases. The 13qA's were significantly associated with the hypodiploid group (P < .001).
Chromosome 1.
Total or partial duplications of chromosome 1 long arm were detected in 57 cases and were more frequent in the hypodiploid group (49%) than in the hyperdiploid one (35%). Chromosome 1 short arm was affected in 46 cases, equally distributed in the 2 groups.
The presence of chromosome derivatives resulting from unbalanced translocations of chromosome whole arms was found as a frequent event, especially for 1q but also for other chromosomes.
Breaks at immunoglobulin gene locations.
The 14q32 region was implicated in 43 cases (31%), including 8 cases (11%) in the hyperdiploid and 35 cases (56%) in the hypodiploid group (P < .001).
Rearrangements were mainly translocations of identified chromosome regions leading to t(8;14)(q24;q32) in 2 cases or to t(11;14)(q13;q32) in 25 cases (18%); in 4 cases the partner of 14q32 was identified as 1q12, 12p?, 7q22, and 3p11 (1 case each). In 11 cases the partner was not identified, and the rearrangement was described as add(14)(q32). In 7 cases an interstitial deletion involved the 14q23q32 region. In 3 cases with a t(11;14), the second chromosome 14 was rearranged; it was implicated in a t(8;14) in 1 case and in a common deletion del(14)(q23q32) in 2.
The 22q11 and 2p12 regions.
The 22q11 region was involved in 4 translocations (2 with chromosome 1 and 2 with unknown material) and 6 deletions. The 2p12 region was implicated only once in a Burkitt-type translocation.
Chromosome 8.
Total or partial loss of chromosome 8 was identified as a frequent recurrent change that was found in 42 cases equally distributed in the 2 groups. This abnormality resulted either from complete monosomy of chromosome 8 (20 cases), from translocation on 8p of unknown material (14 cases), or part of identified chromosome (8 cases). In 5 of these cases the translocation was located in centromeric regions, leading to dicentric chromosomes in 4 cases.
Chromosome 11.
An 11q13 involvement was found in 31 cases, corresponding to an 11;14 translocation in 25 cases, mainly in the hypodiploid group (22 cases) and seldom in the hyperdiploid group (3 cases) (P < .001). Noteworthy, the t(11;14) was found in 7 of the 9 pseudodiploid karyotypes.
Breaks in the 8q24 region and 6q deletions.
These were equally involved in the 2 groups.
Survival: correlation with clinical and biological features
The median follow-up of the 159 patients available for analysis was 32 months. The median OS of these patients, whatever the cytogenetic result, was 30.6 months. Previously reported prognostic factors were found to correlate with survival in univariate analysis: an adverse prognostic was observed in patients with stage III (127 of 159, median survival 27.1 months, while the median was not reached for stage I and II patients, P < .01), β2-microglobulin more than 3 mg/L (90 of 159 patients, median survival 21 vs 55 months, P < .0001), bone marrow plasmocytosis more than 20% (113 of 159 patients, median survival 27.1, while the median was not reached for patients with < 20%, P < .04), and creatinine level more than 200 mM/L (22 of 159 patients, median survival 16.7 vs 32.4 months,P < .03).
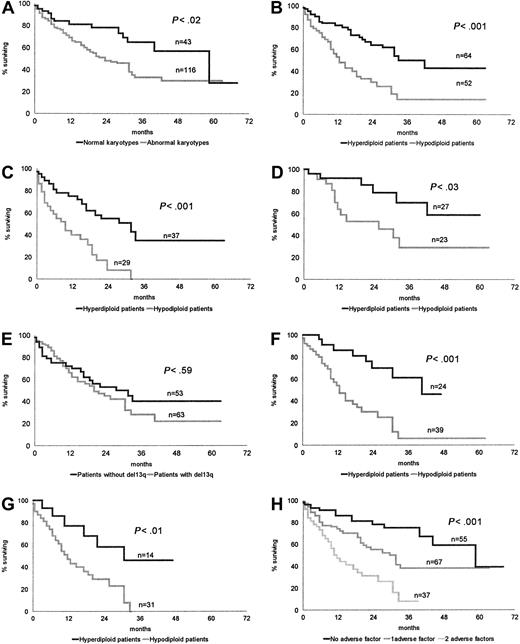
The 43 patients with normal karyotypes had a longer survival than the 116 patients with CCAs: median OS 45.2 months versus 22.7 months, respectively, P < .02 (Figure1A).
OS of patients according to cytogenetic and biological characteristics.
(A) Normal karyotypes versus abnormal karyotypes. (B) Hyperdiploid karyotypes versus hypodiploid karyotypes. (C) Conventional chemotherapy: hyperdiploid patients versus hypodiploid patients. (D) HDC SCT: hyperdiploid patients versus hypodiploid patients. (E) Absence or presence of chromosome 13 loss or deletion. (F) Impact of chromosome number pattern in patients with chromosome 13q abnormality. (G) Impact of chromosome number pattern in patients with both a high β2-microglobulin level and chromosome 13q abnormality. (H) Stratification of patients according to the absence or presence of a hypodiploid karyotype and high β2-microglobulin.
OS of patients according to cytogenetic and biological characteristics.
(A) Normal karyotypes versus abnormal karyotypes. (B) Hyperdiploid karyotypes versus hypodiploid karyotypes. (C) Conventional chemotherapy: hyperdiploid patients versus hypodiploid patients. (D) HDC SCT: hyperdiploid patients versus hypodiploid patients. (E) Absence or presence of chromosome 13 loss or deletion. (F) Impact of chromosome number pattern in patients with chromosome 13q abnormality. (G) Impact of chromosome number pattern in patients with both a high β2-microglobulin level and chromosome 13q abnormality. (H) Stratification of patients according to the absence or presence of a hypodiploid karyotype and high β2-microglobulin.
The median OS of 64 hyperdiploid patients, 33.8 months, was significantly higher than that of 52 patients belonging to the hypodiploid group, 12.5 months, P < .001 (Figure 1B).
This significant difference was maintained whatever the treatment—conventional chemotherapy: hyperdiploid group, 37 patients, median OS 28.3 months; hypodiploid group, 29 patients, median OS 9.2,P < .001 (Figure 1C); and HDC SCT: hyperdiploid group, 27 patients, median OS not reached; hypodiploid group, 23 patients, median OS 20.1 months, P < .03 (Figure 1D).
Whatever the group, hyperdiploid or hypodiploid, an implication of the14q32 region (36 patients) significantly worsened the prognosis (median survival 17.8 months vs 29.9, P < .02).
We failed to find any prognostic value to the presence of an 11q13 abnormality, which was present in 26 patients (median survival 18.4 months vs 25.5 months).
The presence of a deleted chromosome 6q, a complete loss or partial deletion of chromosome 8, a rearrangement of chromosome 8q24 region, or an alteration of chromosome 1p or 1q was not correlated with the outcome.
Comparing OS of CCA patients with (63 patients) or without (53 patients) a chromosome 13 abnormality, we did not find any significant difference (P < .59) (Figure 1E).
In the same way, 13qA did not modify OS in hyperdiploid patients (42 vs 31.7 months, P < .72) or in hypodiploid patients (12.5 vs 12 months, P < .43). Conversely, in patients bearing 13qA, those with a hypodiploid karyotype had a worse outcome (median OS 12.5 vs 42 months, P < .001) (Figure 1F).
However, comparing the patients with 13q abnormalities with all other patients including those with normal cytogenetics (63 of 159 patients), median survival was significantly different: 20.6 and 37.1 months, respectively (P < .04). In the same way, comparing OS of the hypodiploid patients versus all other patients (52 vs 107), we found a highly significant difference:median OS 12.5 versus 44.5 months, respectively (P < .000 01).
Within the high-risk population characterized by both a high β2-microglobulin level and 13qA, we found a significant difference in OS between patients with a hyperdiploid or hypodiploid karyotype; median survival 28.7 versus 11.1 months, respectively (P < .01) (Figure 1G).
A multivariate analysis of several prognostic factors, including Durie and Salmon stage, creatinine, β2-microglobulin level, bone marrow plasmocytosis, chromosome 13qA, hyperdiploid and hypodiploid groups, conventional therapy, and HDC SCT, showed that hypodiploidy was the most important independent factor for OS (P < .0001), followed by the treatment (P < .001), Durie and Salmon stage (P < .04), and β2-microglobulin level (P < .10).
Prognostic model
CCA patients were stratified according to the first unfavorable prognostic factor identified by multivariate analysis, hypodiploidy, and to a high β2-microglobulin level. This led to identification of 3 different risk groups: a low-risk group, including patients without hypodiploidy and showing low β2-microglobulin (31 patients, median OS 44.4 months); a group of patients with intermediate risk, showing either hypodiploidy or a high β2-microglobulin level (42 patients, median OS 23.3 months); and a high-risk group, showing both hypodiploidy and high β2-microglobulin (37 patients, median OS 10.9 months;P < .001). Identical results were obtained when patients with normal karyotypes were included in the analysis (low risk, 55 patients, median OS 51.7 months; intermediate risk, 67 patients, median OS 29.7 months; high risk, 37 patients, OS 10.9 months;P < .001) (Figure 1H).
Discussion
This large independent series including 138 complex myeloma karyotypes in 208 patients (66%) confirms our previous data.18 A 72-hour unstimulated culture allows a rather high detection rate of the abnormal clone. On the whole, conventional cytogenetics allows identification of 2 different groups based on the number of chromosomes in abnormal mitoses. A first group defined by the presence of a hyperdiploid clone with trisomies 3, 5, 7, 9, 11, 15, and 19 associated or not with structural abnormalities; a second group characterized by a hypodiploid, pseudodiploid, or hypotetraploid karyotype always associated with structural abnormalities. The type and frequency of structural abnormalities found in this series are in agreement with previously published data,14,16,29-35except for the total or partial loss of chromosome 8 not yet reported and found here in 42 cases (30%). This loss could be related to the possible localization in 8p21 of a putative tumor suppressor gene.36 37 In our series, 35 of 42 cases showed an 8p21 deleted region. This abnormality was not correlated with outcome.
Patients with normal karyotypes displayed a survival advantage as compared with those with CCAs (Figure 1A). As others, we think that a normal karyotype should be considered as a detection failure of the abnormal clone.14,38-40 In these cases, the normal karyotype could reflect a low labeling index of malignant plasma cells, explaining the longer survival.6 41-43 Thus, the evaluation of the prognostic value of CCAs in MM patients should be based only on abnormal informative karyotypes.
Conventional cytogenetic studies in MM are particularly efficient in stage III MM patients (detection of the abnormal clone in 75% patients), where this technique enables the study of the totality of complex genetic changes present in mitoses. Conversely, interphase fluorescence in situ hybridization techniques (IF)—widely used because they allow the demonstration of abnormalities in nearly 90% of patients irrespective of disease status7,40,44-47—only focus on selected abnormalities regardless of global complexity.48 Nevertheless, IF published results regarding the frequency of chromosome 13q14 (46% and 52%)8 15 are in accordance with our findings using conventional cytogenetics: 54% in 138 abnormal karyotypes.
Using univariate analysis, we found that among the different chromosome abnormalities identified in this series, the chromosome number pattern was the most important prognostic factor: median OS was 33.8 months for hyperdiploid patients compared with 12.5 months for hypodiploid patients (P < .001) (Figure 1B).
The prognostic value of numerical abnormalities, as it is correlated with the DNA content, has mainly been investigated using flow cytometry techniques: Patients with a hypodiploid DNA content had a poor response to chemotherapy and a very short survival,49-52 and a hyperdiploid content was associated with a better prognosis.53 In the same way, in conventional cytogenetic studies hypodiploidy was associated with a poor prognosis,12,17 and on multivariate analysis the absence of hypodiploidy was found to be the most favorable prognosis variable for OS.17 Finally, a study using IF showed that the presence of various trisomies was associated with prolonged survival.7
We did not find any significant difference in OS between patients with or without 11q13 abnormality (18.4 vs 25.5 months, respectively); this is in discordance with previous reports, because this abnormality was considered an important prognostic factor by some authors9-11,35 although not confirmed by others.12 17 However, this abnormality is often associated with the hypodiploid group in our study: 39.5% versus 7% in the hyperdiploid group.
On the other hand, we confirm the reported11 18 negative impact of the presence of 14q32 abnormalities whatever the number of chromosomes (survival 17.8 vs 29.9 months respectively,P < .02).
Our results regarding the prognostic value of chromosome 13q abnormalities are in accordance with those of previous reports.7-13 Taking into account, as in literature,10-13 the whole series including patients with normal karyotypes, we found a significant difference when we compared patients with 13qA with all other patients (P < .04).
However, this study demonstrates that 13qA is not an independent prognostic factor: the comparison of OS of CCA patients with and without chromosome 13qA does not show any significant difference between the 2 populations (Figure 1E). In the same way, the presence of a chromosome 13qA does not modify OS in hyperdiploid patients or in hypodiploid patients. Conversely, in patients with 13qA, the chromosome number pattern conserves an important prognostic value (Figure 1F). Moreover, looking at the impact of hypodiploidy on the whole series, including patients with a normal karyotype, we found that this parameter was the best predictive indicator (P < .000 01). However, considering that normal karyotypes are representative of the tumor probably introduces a bias in the analysis.
Upon multivariate analysis, we found that a hypodiploid karyotype was the most powerful prognostic factor, followed by treatment, whereas chromosome 13qA did not appear as an independent prognostic factor. These findings are different from previously published results in which 13qA was identified as the first independent adverse prognostic factor.8,10 13 Because we found that hypodiploidy was the most powerful prognostic factor and because the presence of a 13qA is significantly associated with hypodiploidy, testing the presence of a 13qA without introducing the chromosome number pattern probably represents an indirect way to test the adverse effect of this parameter.
A very high–risk population was recently individualized by several groups,9 13 characterized by the presence of both 13qA and a high β2-microglobulin level. In this population, our results show that the chromosome number pattern is able to identify 2 subgroups of patients with highly different outcome (Figure 1G). Moreover, we found that hypodiploidy and the β2-microglobulin level allows stratification of these patients in 3 risk groups. In particular, the presence of both hypodiploidy and a high β2-microglobulin level identifies a very poor–risk population (Figure 1H).
On the basis of our results, the chromosome number pattern appears as the most powerful prognostic factor in newly diagnosed MM patients. The presence of a hypodiploid, pseudodiploid, or hypotetraploid karyotype represents a very strong adverse factor.
Looking at the distribution of chromosomal abnormalities in this series, we observed that the hyperdiploid group was particularly homogeneous, identified by the presence of identical trisomies, while the hypodiploid group was more heterogeneous, showing an important variety of structural aberrations. Pseudodiploid karyotypes appeared more homogeneous: most of them (7 of 9) showed an 11;14 translocation.
In conclusion, this independent series confirms our previous results,18 and a large study measuring DNA content of malignant plasma cells should be undertaken to evaluate the prognostic value of hypodiploidy in all patients with MM.
We thank Professor Hervé Tilly and Professor Albert Najman for critical reading of this manuscript and Sylvie Dumont and Catherine Arnould for their technical assistance.
Participants of GFCH are listed with the locations of centers and the number of cases in parentheses.
N. Smadja, M. J. Grange, A. Najman, F. Isnard, J. L. Dutel, B. Varet, C. Belanger, J. P. Vernant, V. Leblond, M. Amar, N. Atkhen, J. P. Fermand, M. Janvier (Paris St Antoine 1; 85); C. Brigaudeau, V. Praloran, F. Trimoreau, P. Turlure, D. Bordessoule, A. Jaccard, M. Touati, L. Remenieras (Limoges; 30); D. Leroux, M. Callanan, B. Pégourié-Bandelier, F. Garban, C. Dascalescu, J. J. Sotto (Grenoble; 21); L. Michaux, J. M. Libouton, C. H. Verellen-Dumoulin, J. M. Scheiff, V. Deneys, A. Ferrant (Brussels; 13); F. Mugneret, B. Favre, I. Luquet, F. Girodon, M. Maynadie, D. Caillot (Dijon; 11); C. Bastard, M. Monconduit, C. Fruchart, V. Izydorczyk (Rouen; 8); P. Talmant (Nantes; 8); C. Terre, S. Castaigne, J. N. Bastie, F. Suzan, I. Garcia (Versailles; 7); C. Barin, L. Benbouker, P. Colombat, A. Petit, J. L. Bremond (Tours; 7); E. Jeandidier, P. Hénon, G. Jung (Mulhouse; 5); A. Daudignon, J. P. Pollet (Valenciennes; 4); M. Lafage-Pochitaloff, M. J. Mozziconacci, M. Merlin, C. Arnoulet, D. Sainty, D. Blaise (Marseille; 3); J. Van Den Akker, M. Divine, J. L. Taillemite (Paris St Antoine 2; 2); J. Lespinasse, M. F. Pedron (Chambery; 2); and C. Charin, I. Tigaud (Lyon; 1); F. Desangles, C. Fouet (Paris Val de grace; 1).
A complete list of the members of GFCH who participated in this study is given in an at the end of this article.
Submitted November 30, 2000; accepted June 4, 2001.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
N. V. Smadja, Laboratoire de Recherche en Cytogénétique Hématologique, Pavillon Moı̈ana, Service Pr JL. Taillemite, Hôpital Saint Antoine, 184 rue du Faubourg St Antoine, 75571 Paris, France.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal