The zinc finger transcription factor GATA-2 plays a critical role in the survival and proliferation of hematopoietic stem cells. This study examined the interaction of GATA-2 with histone deacetylases (HDACs) to define the involvement of HDACs in the regulation of GATA-2 function. GATA-2 directly associates with HDAC3 but not with HDAC1. Consistent with this, HDAC3 suppressed the transcriptional potential of GATA-2, whereas HDAC1 did not affect GATA-2–dependent transcription. Results further demonstrated that GATA-2 and HDAC3 colocalized in the nucleus. These results identify GATA-2 as a nuclear target for HDAC3-mediated repression. Furthermore, GATA-2 also directly associated with HDAC5 but not with other class II HDACs examined, that is, HDAC4 and HDAC6. This is the first demonstration that a tissue-specific transcription factor directly and selectively interacts with HDAC3 and HDAC5 among HDAC family members.
Introduction
The members of the GATA family of DNA-binding proteins contribute to the transcriptional regulation of cell lineage commitment and differentiation.1-3 Six vertebrate GATA factors have been described. Each recognizes a DNA consensus sequence motif (T/A)GATA(A/G) through a highly conserved DNA-binding domain comprised of 2 zinc fingers of the Cys-X2-Cys-X17-Cys-X2-Cys type.4 Three members of the GATA family, GATA-1, GATA-2, and GATA-3, have been identified as important regulators of gene expression in hematopoietic cells.5 GATA-1, the founding member of this family,4,6 is highly expressed in erythroid cells, mast cells, and megakaryocytes, and its expression is required for primitive and definitive erythropoiesis.7-9 GATA-2 is highly expressed in pluripotent hematopoietic stem cells.10,11 Analysis of a targeted disruption of theGATA2 gene in mice and GATA-2–deficient embryonic stem cells suggests the necessity of GATA-2 for the survival of early hematopoietic stem cells.12-14 GATA-3, which is expressed in T lymphocytes, is essential for T-lymphoid cell development.15 16
The organization of chromatin is a fundamentally important element of gene regulation in all eukaryotic cells. The modification of nucleosome histones determines whether chromatin is transcriptionally active or repressed.17-19 The transcriptional coactivators, such as CREB-binding protein (CBP) and p300, have been identified as histone acetyltransferases (HATs).20,21 Almost in parallel with the discovery of HATs, a number of histone deacetylase (HDAC) enzymes have been identified. Class I HDACs (HDAC1, HDAC2, HDAC3, and HDAC8) are homologous to yeast RPD3.22-26 Various transcriptional repressors recruit these complexes to inhibit transcription. The nuclear receptor corepressor N-CoR, for example, interacts with HDAC1 and HDAC2 through the mSin3 complex.27-31 Class II HDACs (HDAC4, HDAC5, HDAC6, and HDAC7) contain domains significantly similar to the catalytic domain of yeast HDA1.32-34 HDAC5 and HDAC7 directly interact with silencing mediator for retinoid and thyroid receptors (SMRT)/N-CoR and also bind to mSin3A through a region different from the one where HDAC1 binds.35 At present, members of class I HDACs are distinguished solely on the basis of sequence, with HDAC1 and HDAC2 being more closely related to each other than to HDAC3.24This closer relationship suggests that HDAC3 may play a unique role, but the question of whether any HDAC3-specific binding proteins exist remains open.
Several molecules that bind GATA proteins and possibly regulate their transcriptional activity have been identified. GATA-1 has been shown to bind to other zinc finger–containing transcription factors such as Sp-1,36 EKLF,36 a multiple zinc finger protein FOG-1 (friend of GATA-1),37 and most recently a lineage-specific transcription factor PU.1.38,39 We have shown that promyelocytic leukemia (PML) protein associates with and potentiates GATA-2.40 The transcriptional activator CBP binds to GATA-1 and enhances GATA-1 transactivation.41Cooperation of CBP with GATA-1 is further required for GATA-1–dependent erythroid differentiation. In contrast, GATA-1 has been revealed to interact with the myeloid PU.1 transcription factor and repress PU.1-dependent transcription.42,43 However, the repression mechanisms of not only GATA family members but also other hematopoietic-specific transcription factors are poorly understood. Induction of GATA-2 activity in the interleukin-3 (IL-3)–dependent multipotential hematopoietic progenitor cell model FDCP mix blocks factor-dependent self-renewal and cells undergo cell cycle arrest and cease proliferating but do not apoptose,44 indicating that the repression of GATA-2 activity may be critical for keeping the cell cycle going in hematopoietic stem cells. In accordance with this, enforced expression of GATA-2 causes a block in the proliferation of primitive murine bone marrow cells.14 These facts prompted us to investigate whether HDACs associate with GATA-2 transcription factor and repress its transcriptional activity. We show here that HDAC3 uniquely associates with GATA-2 and represses GATA-2–dependent transcriptional potential.
Materials and methods
Expression plasmids
The full-length human GATA-2 complementary DNA (cDNA) was generously provided by S. H. Orkin (Harvard Medical School, Boston, MA). The full-length human HDAC1 and HDAC3 cDNAs are kind gifts from K. Tamai and T. Miyazaki (MBL, Nagano, Japan). Flag-hGATA-2-pCMV, Myc-HDAC1, and Myc-HDAC3 were constructed by using appropriate restriction sites in either pFlag-CMV2 expression vector (Eastman Kodak, New Haven, CT) or pcDNA3.1-Myc/His expression vector (Invitrogen, Carlsbad, CA). The Flag-tagged expression vectors for human class II HDACs (HDAC4, HDAC5, and HDAC6) were kindly provided by S. L. Shreiber (Harvard University, Cambridge, MA). The full-length HDAC5 was excised and introduced in pcDNA 3.1 expression vector.
Cells
COS and 293T cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum (FCS). A human hematopoietic cell line KG-1 was maintained in RPMI 1640 medium supplemented with 10% FCS.
Protein interaction assay in cells
COS cells (5 × 105) grown in 10-cm diameter plates were transfected with the indicated expression plasmids. The total amount of plasmids was equalized by addition of the corresponding empty vectors. Transfection was carried out using the Lipofectamine 2000 reagent (Gibco BRL, Rockville, MD) in accordance with the manufacturer's instruction. Forty-eight hours after transfection, cells were lysed in a buffer (50 mM Tris pH 7.6, 150 mM NaCl, 1 mM EGTA, 0.5% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride plus protease inhibitors). After preclear, immunoprecipitation assays were performed at 4°C by using anti-Flag antibody M2 in combination with avidin-agarose beads (Sigma Chemical, St Louis, MO), anti–c-Myc antibody (Rosche, Indianapolis, IN), anti–GATA-2 antibody (H116, Santa Cruz Biotechnology, Santa Cruz, CA) or anti–X-press antibody (Invitrogen). After 4 washes with the lysis buffer, immune complexes were analyzed by Western blotting using the indicated antibodies as described previously.45 46 Antibody against HDAC3 was purchased from Transduction Laboratories (Lexington, KY).
Pull-down assay
Fragments of cDNA encoding hGATA-2 were produced using convenient restriction enzymes and polymerase chain reaction (PCR) methods and then cloned into the glutathione S-transferase (GST) fusion vector pGEX 5X-1 (Pharmacia, Uppsala, Sweden). The GST constructs were transformed into the Escherichia coli strain, BL21, and the GST fusion proteins were obtained according to the manufacturer's instructions. HDAC1, HDAC3, and HDAC5 were transcribed and translated in vitro in the presence of [35S]-methionine by using the T7-coupled reticulocyte lysate system (Promega, Madison, WI) in accordance with the manufacturer's instructions. For pull-down assay, equal amounts of the GST fusion proteins were incubated with the HDAC in vitro–transcribed and translated reaction mixture in phosphate-buffered saline (PBS). After 1 hour at 4°C, the beads were washed 4 times with PBS containing 0.5% Nonidet P-40 and resuspended in 2 × sodium dodecyl sulfate (SDS) sample buffer. Proteins were then separated by SDS–polyacrylamide gel electrophoresis (PAGE) before autoradiography.
DNA-binding assay
For electrophoretic mobility assays (EMSAs), nuclear extract from transfected COS cells was incubated in 10 μL binding buffer (10 mM Tris-HCl, pH 7.5, 75 mM KCl, 1 mM EDTA, 5 mM dithiothreitol, 4% Ficoll, 0.5 μg poly [dI-dC])6 and a 32P-labeled double-strand oligonucleotide probe (CACTTGATAACAGAAAGTGATAACTCT). After 20 minutes at room temperature (RT), the protein-DNA complex was resolved on a 4% native polyacrylamide gel and visualized by autoradiography. A 2000-fold molar excess of unlabeled oligonucleotide was used for competition experiments, and 1 μL of each antibody was used for supershift assay.
Transactivation assays
A luciferase reporter plasmid in which a murine GATA-1 promoter (position −798 to −574) containing a double GATA site was arrayed upstream of the β-globin minimal promoter (designated as GATA-1/Luc), a gift from M. Yamamoto (Tsukuba University, Tsukuba, Japan). A luciferase reporter plasmid in which 2 copies of back-to-back double GATA sites in the mouse CD34 promoter were placed upstream of the β-globin minimal promoter driving the luciferase gene (designated CD34 × 2/Luc.) was generated as we described.40 The mutant reporter in which core recognition sites were mutated from GATA to TTTA (mutant CD34 × 2/Luc.) was also used. The 293T cells (2 × 105/35-mm diameter plate) were transfected with the indicated expression plasmids. Cell lysates were prepared 48 hours after transfection and assayed for luciferase activity using a luciferase assay system (Promega) according to the manufacturer's instructions. Total amounts of plasmids used for transfection were equalized by the addition of the corresponding empty vectors. Transfection efficiency was normalized on the basis of β-galactosidase activity expressed from cotransfected pCMV/β-gal plasmids (Promega). The relative luciferase activities presented reflect triplicate values from a representation of 4 independent experiments.
Immunofluorescence and confocal microscopy
KG-1 cells were cytospun onto glass slides and fixed in methanol for 5 minutes at RT. After blocking for 60 minutes at RT with 1% bovine serum albumin in PBS, anti–GATA-2 rabbit antibody and anti-HDAC3 mouse antibody were applied for 60 minutes at RT. Subsequently, they were washed 3 times with PBS and incubated with antirabbit fluorescein isothiocyanate (FITC)–conjugated secondary antibody (Santa Cruz Biotechnology) and antimouse immunoglobulin G conjugated with Alexa 568 (Molecular Probes, Eugene, OR) for 60 minutes at RT. Images were acquired using a 1024MRC Bio-Rad microscope (Bio-Rad, Hercules, CA) equipped with a krypton-argon laser and exported to a PowerMac computer for further processing with Adobe Photoshop.
Results
GATA-2 interacts with HDAC3 but not with HDAC1
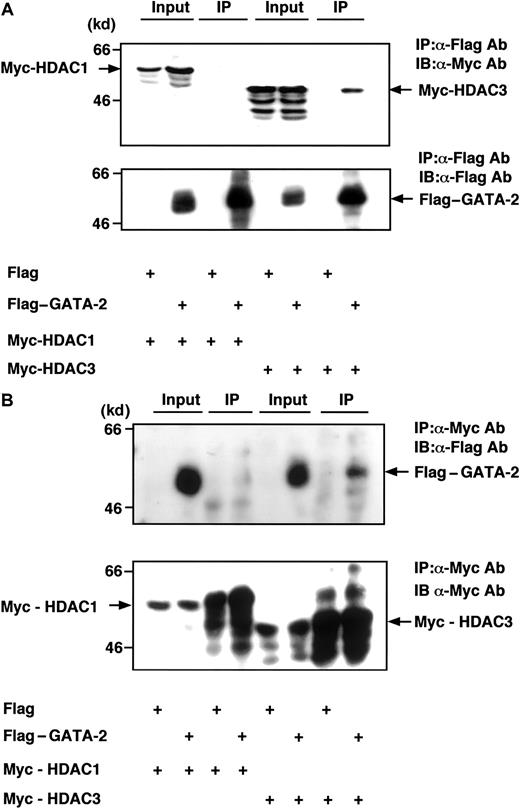
We first examined whether GATA-2 interacts with HDACs in mammalian cells. Flag-tagged GATA-2 and Myc-tagged HDAC1 or HDAC3 were coexpressed in COS cells and we measured their association using immunoprecipitation assays. Whole cell extracts were immunoprecipitated with anti–Flag M2 beads, followed by immunoblot analysis with anti-Myc antibody. HDAC3 was coimmunoprecipitated with GATA-2 in this system, whereas HDAC1 was not detected in the GATA-2 immunoprecipitates (Figure1A). Reciprocally, the same whole cell extracts as used above were immunoprecipitated with anti-Myc antibody followed by immunoblotting with anti-Flag antibody, suggesting that Flag–GATA-2 was coimmunoprecipitated with HDAC3 but not with HDAC1 (Figure 1B). Consistent with this, we confirmed the interaction by a mammalian 2-hybrid assay and found that GATA-2 showed much higher affinity with HDAC3 than HDAC1 (data not shown).
GATA-2 interacts with HDAC3 in vivo.
(A) Flag–GATA-2 (5 μg) was cotransfected with Myc-HDAC1 (5 μg) or Myc-HDAC3 (5 μg) into COS cells. Whole cell lysates were prepared and immunoprecipitated with anti–Flag M2 beads. Immunoprecipitates were subjected to immunoblot analysis with anti-Myc antibody (upper panel). The blot was reprobed with anti-Flag antibody to confirm that GATA-2 was successfully immunoprecipitated (lower panel). Input represents 5% of whole cell lysates used for each immunoprecipitation applied to confirm whether each HDAC or GATA-2 was highly expressed. (B) The same lysates described above were immunoprecipitated with anti-Myc antibody followed by immunoblot analysis with anti-Flag antibody (upper panel). Successful immunoprecipitation was confirmed by reprobing with anti-Myc antibody.
GATA-2 interacts with HDAC3 in vivo.
(A) Flag–GATA-2 (5 μg) was cotransfected with Myc-HDAC1 (5 μg) or Myc-HDAC3 (5 μg) into COS cells. Whole cell lysates were prepared and immunoprecipitated with anti–Flag M2 beads. Immunoprecipitates were subjected to immunoblot analysis with anti-Myc antibody (upper panel). The blot was reprobed with anti-Flag antibody to confirm that GATA-2 was successfully immunoprecipitated (lower panel). Input represents 5% of whole cell lysates used for each immunoprecipitation applied to confirm whether each HDAC or GATA-2 was highly expressed. (B) The same lysates described above were immunoprecipitated with anti-Myc antibody followed by immunoblot analysis with anti-Flag antibody (upper panel). Successful immunoprecipitation was confirmed by reprobing with anti-Myc antibody.
GATA-2 binds to HDAC3 in vitro
To investigate the possibility of direct interaction between GATA-2 and HDAC3, we prepared bacterially expressed fusion proteins containing GST fused to GATA-2 and examined the interaction with in vitro transcribed/translated HDAC1 and HDAC3. In pull-down assays, GST–GATA-2 exhibited a binding preference to HDAC3, whereas it failed to show any detectable binding to HDAC1 (Figure2A). These results suggest that GATA-2 directly associates with HDAC3 but not with HDAC1.
GATA-2 interacts with HDAC3 in vitro.
(A) [35S]-methionine-labeled HDAC1 or HDAC3 translated in vitro was incubated with GST–GATA-2 fusion protein or GST protein alone and pulled down with GST-agarose beads. After 4 washes, the precipitates were dissolved by SDS-PAGE and visualized by autoradiography. Input represents 4% of in vitro–translated HDAC proteins used for the pull-down assay. (B) [35S]-labeled full-length HDAC3 was incubated with GST fusion proteins containing the various portions of GATA-2 as indicated in the upper panel. The precipitates were visualized by autoradiography following SDS-PAGE. Input represents 4% of in vitro–translated HDAC3 protein used for the pull-down assay. N-Zf indicates amino-terminal zinc finger; C-Zf, carboxyl-terminal zinc finger. (C) Various HDAC3 deletion constructs as indicated in the upper panel were produced by in vitro translation reaction and subjected to pull-down analysis by GST–GATA-2. Input represents 4% of each in vitro– translated HDAC3 protein used for the pull-down assay. (D) Various HDAC3 deletion constructs were transfected in COS cells. Whole cell lysates were extracted and subjected to pull-down by GST–GATA-2. After 4 washes the precipitates were subjected to immunoblot analysis with anti–X-press antibody. Inputs represent 5% of whole cell lysates used for each pull-down assay applied to confirm whether each HDAC3 was highly expressed.
GATA-2 interacts with HDAC3 in vitro.
(A) [35S]-methionine-labeled HDAC1 or HDAC3 translated in vitro was incubated with GST–GATA-2 fusion protein or GST protein alone and pulled down with GST-agarose beads. After 4 washes, the precipitates were dissolved by SDS-PAGE and visualized by autoradiography. Input represents 4% of in vitro–translated HDAC proteins used for the pull-down assay. (B) [35S]-labeled full-length HDAC3 was incubated with GST fusion proteins containing the various portions of GATA-2 as indicated in the upper panel. The precipitates were visualized by autoradiography following SDS-PAGE. Input represents 4% of in vitro–translated HDAC3 protein used for the pull-down assay. N-Zf indicates amino-terminal zinc finger; C-Zf, carboxyl-terminal zinc finger. (C) Various HDAC3 deletion constructs as indicated in the upper panel were produced by in vitro translation reaction and subjected to pull-down analysis by GST–GATA-2. Input represents 4% of each in vitro– translated HDAC3 protein used for the pull-down assay. (D) Various HDAC3 deletion constructs were transfected in COS cells. Whole cell lysates were extracted and subjected to pull-down by GST–GATA-2. After 4 washes the precipitates were subjected to immunoblot analysis with anti–X-press antibody. Inputs represent 5% of whole cell lysates used for each pull-down assay applied to confirm whether each HDAC3 was highly expressed.
To determine the region of GATA-2 required for interaction with HDAC3, various deletion constructs of GATA-2 fused to GST were produced in bacteria and tested for binding to [35S]-labeled HDAC3. A pull-down assay by GST beads was performed for the interaction with various deletion mutants of GATA-2. The result clearly indicated that the portion comprising amino acids 270 to 393, which contains whole 2 zinc fingers, was required for HDAC3 binding (Figure 2B). We next performed reciprocal pull-down experiments to determine the region in HDAC3 required for the binding to GATA-2. Various HDAC3 deletion constructs were produced by an in vitro transcription/translation reaction and were subjected to pull-down by full-length GST–GATA-2. The truncated form of HDAC3 comprising amino acids 1 to 180 and longer forms of HDAC3 exhibited the equivalent binding to GATA-2, whereas a shorter truncated form of HDAC3 encompassing amino acids 1 to 132 (designated as HDAC3 1-132) failed to show any binding to GATA-2 (Figure 2C). We further examined the interaction between recombinant GATA-2 protein and various HDAC3 deletion constructs expressed in vivo. His-tagged HDAC3 deletion constructs were transfected in COS cells. Forty-eight hours later whole cell lysates were extracted and subjected to pull-down by GST–GATA-2. Consistent with the interaction in vitro, HDAC3 (1-132) expressed in COS cells could not bind to GST–GATA-2, whereas HDAC3 (1-180) successfully bound to GST–GATA-2 as effectively as full-length HDAC3 (Figure 2D). These results suggest that the portion comprising amino acids 132 to 180 in human HDAC3 is involved mainly in GATA-2 binding.
GATA-2 associates with HDAC3 in hematopoietic cells
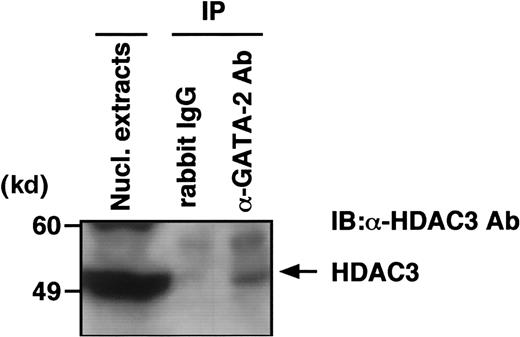
Given that HDAC3 coimmunoprecipitated with GATA-2 in extracts from COS cells overexpressing both proteins, the interaction would be expected also in hematopoietic progenitors cells that express GATA-2 at a high level. Among human hematopoietic cell lines, GATA-2 is expressed at relatively high levels in KG-1 cells. The nuclear extract from KG-1 cells was used for immunoprecipitation with an antibody to GATA-2 followed by immunoblot analysis with anti-HDAC3 antibody. As expected, the complex immunoprecipitated with GATA-2 antibody, but not that selected with preimmune immunoglobulin, and also contained HDAC3 protein (Figure 3), suggesting that GATA-2 associated with HDAC3 in KG1 cells. Successful immunoprecipitation of GATA-2 was confirmed by reprobing the blot with anti–GATA-2 antibody (data not shown).
GATA-2 interacts with HDAC3 in hematopoietic cells.
Nuclear extracts from KG-1 cells were immunoprecipitated with either preimmune rabbit serum or anti–GATA-2 antibody (H116). Immunoprecipitates were subjected to immunoblot analysis with anti-HDAC3 antibody.
GATA-2 interacts with HDAC3 in hematopoietic cells.
Nuclear extracts from KG-1 cells were immunoprecipitated with either preimmune rabbit serum or anti–GATA-2 antibody (H116). Immunoprecipitates were subjected to immunoblot analysis with anti-HDAC3 antibody.
HDAC3 does not alter DNA-binding activity of GATA-2
We next asked whether HDAC3 could alter DNA-binding activity of GATA-2 using EMSAs. We transiently expressed Flag-tagged GATA-2 in the presence or absence of HDAC3 in COS cells, and nuclear lysates were extracted 48 hours later. EMSAs using an oligonucleotide containing GATA recognition sites (Figure 4, lane 1) revealed a protein-DNA complex (lane 2). Competitive experiments were performed using a 2000-fold excess of the unlabeled oligonucleotide (lane 3). Supershift assays using α-Flag antibody indicated the presence of GATA-2 proteins in the complex (lane 5). Coexpression of GATA-2 and HDAC3 did not diminish DNA binding (lane 7).
HDAC3 does not alter DNA-binding activity of GATA-2.
Nuclear extracts from COS cells transfected with GATA-2 expression vector (Flag–GATA-2) in the presence or absence of HDAC3 expression vector (Myc-HDAC3) were used in EMSAs. A 32P-labeled double-stranded oligonucleotide containing a GATA consensus recognition site was used as a probe. The protein-DNA complex was revealed in lanes 2 and 4. Competitive experiments were performed using a 2000-fold excess of the unlabeled oligonucleotide (lane 3). Supershift experiments were performed by addition of α-Flag antibody, with the combination α-GATA-1 antibody serving as a control, as indicated. Specific GATA-2–DNA complexes and the supershifted complexes are indicated. The supershifted band by α-Flag antibody is denoted with an asterisk.
HDAC3 does not alter DNA-binding activity of GATA-2.
Nuclear extracts from COS cells transfected with GATA-2 expression vector (Flag–GATA-2) in the presence or absence of HDAC3 expression vector (Myc-HDAC3) were used in EMSAs. A 32P-labeled double-stranded oligonucleotide containing a GATA consensus recognition site was used as a probe. The protein-DNA complex was revealed in lanes 2 and 4. Competitive experiments were performed using a 2000-fold excess of the unlabeled oligonucleotide (lane 3). Supershift experiments were performed by addition of α-Flag antibody, with the combination α-GATA-1 antibody serving as a control, as indicated. Specific GATA-2–DNA complexes and the supershifted complexes are indicated. The supershifted band by α-Flag antibody is denoted with an asterisk.
HDAC3 represses transcriptional activity of GATA-2
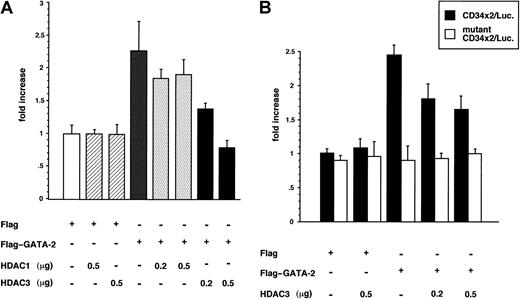
We further asked whether the association of HDAC3 with GATA-2 had any functional consequences for GATA-2 activity. Because HDAC3 is capable of repressing transcription, we examined the effect of HDAC3 on GATA-2–directed transcriptional activity of a luciferase reporter gene linked in cis to a GATA recognition motif. We have shown that GATA-2 transactivated a luciferase reporter containing a double GATA element derived from the murine GATA-1 promoter. Using this reporter system in 293T cells, GATA-2 showed a modestly increased level of luciferase activity (approximately 2.3-fold) as we reported previously.40 The addition of HDAC3 repressed the transactivation potential of GATA-2 in a dose-dependent manner (Figure5A). We also compared the effects of HDAC3 with HDAC1, which lacks the binding affinity to GATA-2. Consistent with this, HDAC1 did not significantly suppress GATA-2 transactivating potential, further sustaining our suggestion that HDAC3 represses GATA-2 activity through direct interaction. To further confirm the repressing effects of HDAC3 on GATA-2 transcriptional activity, we performed another luciferase reporter assay using a different GATA reporter gene in which 2 copies of back-to-back double GATA sites in the mouse CD34 promoter were placed upstream of the β-globin minimal promoter driving the luciferase gene (CD34 × 2/Luc.). This reporter showed GATA-2–dependent activity (approximately 2.4-fold), and the activity was again repressed by HDAC3 (Figure 5B). The GATA-2–dependent activity and its repression by HDAC3 were abrogated when the GATA sites in the reporter were disrupted (mutant CD34 × 2/Luc.).
HDAC3 represses GATA-2–directed transcriptional activity.
(A) The 293T cells were transfected with a luciferase reporter containing GATA consensus motif (GATA-1/Luc, 0.5 μg), together with expression plasmid for human GATA-2 (Flag–GATA-2, 0.5 μg) and human HDAC1 and HDAC3, (pcDNA3.1-HDAC1 or pcDNA3.1-HDAC3, 0.2 μg or 0.5 μg, as indicated). Luciferase activity is standardized against β-galactosidase activity from cotransfected control vector (pCMV/β-gal). The relative luciferase activities presented reflect triplicate value from a representation of at least 3 independent experiments. (B) Experiments similar to those represented in panel A were conducted using a luciferase reporter containing 2 copies of back-to-back double GATA sites from the mouse CD34 promoter (CD34 × 2/Luc., 0.5 μg; solid bar) or its mutant in which GATA sites were disrupted (mutant CD34 × 2/Luc., 0.5 μg; open bar), together with GATA-2 (Flag–GATA-2, 0.5 μg) and human HDAC3, (pcDNA3.1-HDAC3, 0.2 μg or 0.5 μg, as indicated).
HDAC3 represses GATA-2–directed transcriptional activity.
(A) The 293T cells were transfected with a luciferase reporter containing GATA consensus motif (GATA-1/Luc, 0.5 μg), together with expression plasmid for human GATA-2 (Flag–GATA-2, 0.5 μg) and human HDAC1 and HDAC3, (pcDNA3.1-HDAC1 or pcDNA3.1-HDAC3, 0.2 μg or 0.5 μg, as indicated). Luciferase activity is standardized against β-galactosidase activity from cotransfected control vector (pCMV/β-gal). The relative luciferase activities presented reflect triplicate value from a representation of at least 3 independent experiments. (B) Experiments similar to those represented in panel A were conducted using a luciferase reporter containing 2 copies of back-to-back double GATA sites from the mouse CD34 promoter (CD34 × 2/Luc., 0.5 μg; solid bar) or its mutant in which GATA sites were disrupted (mutant CD34 × 2/Luc., 0.5 μg; open bar), together with GATA-2 (Flag–GATA-2, 0.5 μg) and human HDAC3, (pcDNA3.1-HDAC3, 0.2 μg or 0.5 μg, as indicated).
GATA-2 and HDAC3 colocalize in hematopoietic cells
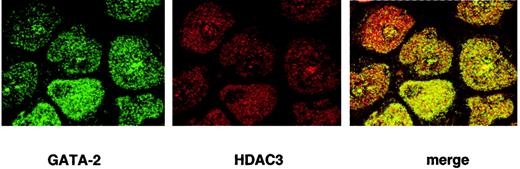
To verify the interaction between GATA-2 and HDAC3 in vivo, we further examined their colocalization in the nuclei using confocal microscopy. HDAC3 localizes to specific areas in the nucleus by immunofluorescence using anti-HDAC3 antibody. GATA-2 showed a similar localization in the nucleus by immunofluorescence using anti–GATA-2 antibody. The colocalization of HDAC3 and GATA-2 is clearly observed when the 2 confocal images are merged (Figure6, right).
GATA-2 and HDAC3 colocalize in the nucleus.
KG-1 cells were cytospun onto glass slides and fixed in methanol and stained with anti–GATA-2 and anti-HDAC3 antibodies followed by FITC-conjugated and Alexa 568–conjugated secondary antibody. Cells were analyzed by confocal microscopy. GATA-2 is immunostained in green and HDAC3 is immunostained in red.
GATA-2 and HDAC3 colocalize in the nucleus.
KG-1 cells were cytospun onto glass slides and fixed in methanol and stained with anti–GATA-2 and anti-HDAC3 antibodies followed by FITC-conjugated and Alexa 568–conjugated secondary antibody. Cells were analyzed by confocal microscopy. GATA-2 is immunostained in green and HDAC3 is immunostained in red.
HDAC5 interacts with GATA-2 in vivo and in vitro
The interaction between GATA-2 and class II HDACs (HDAC4, HDAC5, and HDAC6) was also examined by similar immunoprecipitation analysis. Among the class II HDACs examined, HDAC5 was exclusively coimmunoprecipitated (Figure 7A). Furthermore, in the pull-down assay, GST–GATA-2 interacted with in vitro transcribed/translated HDAC5 (Figure 7B). As shown in Figure 2A, HDAC3 was consistently pulled down by GATA-2, whereas any detectable binding to HDAC1 was not observed. These results suggest that HDAC5 also directly associates with GATA-2.
GATA-2 interacts with HDAC5 in vivo and in vitro.
(A) pcDNA3.1/His–GATA-2 (5 μg) was cotransfected with 5 μg of either Flag-HDAC4, Flag-HDAC5, or Flag-HDAC6 into COS cells. Whole cell lysates were prepared and immunoprecipitated with anti–X-press antibody. Immunoprecipitates were subjected to immunoblot analysis with anti-Flag antibody (upper panel). The blot was reprobed with anti–X-press antibody to confirm that GATA-2 was successfully immunoprecipitated (lower panel). Input represents 5% of whole cell lysates used for each immunoprecipitation applied to confirm whether each class II HDAC was highly expressed. (B) GATA-2 interacts with HDAC3 and 5 in vitro. [35S]-methionine-labeled HDAC1, HDAC3, and HDAC5 translated in vitro were incubated with GST–GATA-2 fusion protein or GST protein alone and pulled down with GST-agarose beads. After 4 washes, the precipitates were dissolved by SDS-PAGE and visualized by autoradiography. Input represents 4% of in vitro–translated HDAC proteins used for the pull-down assay.
GATA-2 interacts with HDAC5 in vivo and in vitro.
(A) pcDNA3.1/His–GATA-2 (5 μg) was cotransfected with 5 μg of either Flag-HDAC4, Flag-HDAC5, or Flag-HDAC6 into COS cells. Whole cell lysates were prepared and immunoprecipitated with anti–X-press antibody. Immunoprecipitates were subjected to immunoblot analysis with anti-Flag antibody (upper panel). The blot was reprobed with anti–X-press antibody to confirm that GATA-2 was successfully immunoprecipitated (lower panel). Input represents 5% of whole cell lysates used for each immunoprecipitation applied to confirm whether each class II HDAC was highly expressed. (B) GATA-2 interacts with HDAC3 and 5 in vitro. [35S]-methionine-labeled HDAC1, HDAC3, and HDAC5 translated in vitro were incubated with GST–GATA-2 fusion protein or GST protein alone and pulled down with GST-agarose beads. After 4 washes, the precipitates were dissolved by SDS-PAGE and visualized by autoradiography. Input represents 4% of in vitro–translated HDAC proteins used for the pull-down assay.
Discussion
The SMRT and N-CoR large protein complex contains HDAC1/HDAC2 and class II HDACs including HDAC4, HDAC5, and HDAC7.34,47,48In addition, HDAC3 is also contained in the SMRT/N-CoR complex.49,50 These findings raise the question as to how the specificity of each HDAC family member for the repression mediated by SMRT/N-CoR, if any, is preserved. To date, little is known about the substrate specificity of the respective HDACs. Only a few transcription factors have been shown to directly bind to HDACs without intermediate corepressor. Retinoblastoma protein is revealed to interact with HDAC1 directly51,52 and Sp1 also interacts with HDAC1.53 YY1-induced transcriptional repression is due to direct interaction with HDAC2.23 HDAC1 and HDAC2 also interact directly with DNA topoisomerase II and modify topoisomerase II activity.54 Recently, myocyte enhancer factor 2 (MEF2) was shown to interact directly with HDAC4 and HDAC5.55 56 In the present study, we succeed in presenting evidence that HDAC3 and HDAC5 interact with GATA-2. This is the first time that GATA transcription family members were seen to directly bind to the HDAC family.
More importantly, we show that HDAC3 and HDAC5 but not other HDACs examined (HDAC1, HDAC4, and HDAC6) directly interact with tissue-specific transcription factors to repress the transcription. Among class II HDACs, HDAC5 but not HDAC4 nor HDAC6 was consistently coimmunoprecipitated with GATA-2. Although sharing a similar domain organization and sequence homology, HDAC4 and HDAC5 are probably able to execute different functions in hematopoietic cells partly by associating with different partners. The putative binding region in human HDAC3 (amino acids 132-180) are completely conserved at amino acid sequence level among human, mouse, and chicken HDAC3. However, in the binding region in HDAC3, there exist no sites, which are conserved with HDAC5 but not with HDAC4, as revealed by amino acid sequence homology search (MacVector version 7.0). It is likely that the binding regions to GATA-2 are different between HDAC3 and HDAC5. The homology search also unveils that several amino acid residues in the putative binding domain of HDAC3 are different from those in the corresponding domain of HDAC1 and HDAC2 (data not shown). To identify the minimal binding site of HDAC3, we are currently generating mutant HDAC3 that lacks binding affinity to GATA-2 by mutagenesis. Interestingly it was reported that the active domain for the deacetylation was contained in the region comprising amino acids 78 to 187 of chicken HDAC3.57 They also indicated that histidine residue at amino acid 135 was critical for the deacetylation activity. The putative domain of HDAC3 for GATA-2 binding, which we have identified, completely falls in the active domain for deacetylation described above. The relevance of GATA-2 binding to the active domain not only for GATA-2 activity but also for HDAC3 activity may be an interesting issue.
What are the consequences of these interactions? The binding of GATA-2 to HDAC3 presumably means the recruitment of HDAC3 and HDAC5 to the GATA-2 target promoters, thereby deacetylating target chromatin and repressing GATA-2–dependent target genes. Very recently, GATA-1 was speculated to be a repressor of antimullerian hormone expression58 and a repressor of enhancer activity of the β-globin locus control region.59 However, no direct evidence has so far been shown that indicates that GATA-2 is a repressor of specific gene expressions. Identifying the repressed target genes of GATA-2 in hematopoiesis may unveil whether the repressed transcription is associated with the binding of HDAC3 to GATA-2 on the target promoters/enhancers. Of relevance to human disease is the finding that GATA-2 is down-regulated in hematopoietic stem cells of aplastic anemia.60 We have recently reported a point mutation of GATA-2 in human acute myeloid leukemia blasts.61 Understanding the mechanisms involved in the specific interaction between HDAC3 and GATA-2 will not only contribute to our understanding of normal hematopoiesis regulated by GATA-2, but also could lead to novel therapeutic strategies against hematopoietic disorders that involve GATA-2.
It is also possible that the binding may modify the acetylation status of GATA-2 itself, along with target histones. The founding GATA family member GATA-1 has been shown to be acetylated by CBP/p300 HATs, and GATA-1 acetylation is essential for erythroid differentiation.62,63 GATA-3 is also acetylated by HATs, thereby affecting T-cell survival and homing to secondary lymphoid organs.64 The other hematopoietic GATA transcription factor GATA-2 is also acetylated by HATs both in vitro and in vivo (F. Hayakawa and M. Towatari, unpublished data, June 1999). The mechanisms of deacetylation of GATA factors, however, are poorly understood. Only a very few transcriptional regulators are shown to be regulated by deacetylation; the function of transcription factor E2F, essential for cell cycle control, is reversibly regulated through the balance of its acetylation and deacetylation.65 It is of particular importance to determine whether GATA-2 is specifically deacetylated by HDAC3 but not by other HDAC family members. Elucidating mechanisms of GATA-2 regulation by the interactive HATs and HDACs should be a good model for studying how tissue-specific and lineage-specific gene expressions are controlled in the context of acetylation/deacetylation of histone/nonhistone proteins on the chromatin.
The authors are grateful to Dr S. L. Schreiber for providing human class II HDACs (HDAC4, HDAC5, and HDAC6) clones. We also thank S. Suzuki and C. Wakamatsu for technical assistance.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Masayuki Towatari, First Department of Internal Medicine, Nagoya University School of Medicine, 65 Tsurumai-cho, showa-ku, Nagoya 466-8550, Japan; e-mail: towatari@med.nagoya-u.ac.jp.

![Fig. 2. GATA-2 interacts with HDAC3 in vitro. / (A) [35S]-methionine-labeled HDAC1 or HDAC3 translated in vitro was incubated with GST–GATA-2 fusion protein or GST protein alone and pulled down with GST-agarose beads. After 4 washes, the precipitates were dissolved by SDS-PAGE and visualized by autoradiography. Input represents 4% of in vitro–translated HDAC proteins used for the pull-down assay. (B) [35S]-labeled full-length HDAC3 was incubated with GST fusion proteins containing the various portions of GATA-2 as indicated in the upper panel. The precipitates were visualized by autoradiography following SDS-PAGE. Input represents 4% of in vitro–translated HDAC3 protein used for the pull-down assay. N-Zf indicates amino-terminal zinc finger; C-Zf, carboxyl-terminal zinc finger. (C) Various HDAC3 deletion constructs as indicated in the upper panel were produced by in vitro translation reaction and subjected to pull-down analysis by GST–GATA-2. Input represents 4% of each in vitro– translated HDAC3 protein used for the pull-down assay. (D) Various HDAC3 deletion constructs were transfected in COS cells. Whole cell lysates were extracted and subjected to pull-down by GST–GATA-2. After 4 washes the precipitates were subjected to immunoblot analysis with anti–X-press antibody. Inputs represent 5% of whole cell lysates used for each pull-down assay applied to confirm whether each HDAC3 was highly expressed.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/98/7/10.1182_blood.v98.7.2116/5/m_h81911590002.jpeg?Expires=1767733448&Signature=tBQdqaCNBu69FyLbILH7vj2-bs3olbGIkXeZWeakyL~Ax6VMuJiK4eKqGsda6VYSJZJWpliRuhUzr5Kx8tkhRJzVpn684HoI7CRt1ns1RkFiNmrDIx4vZwSMs~Q-JN~F1xKFpBp8H7AaAhKuqZ674wvIaoakqeF6opc8T2u5ytJQH0ZLUrdldxTzNHoKosqnz1H~xBd2ogrC0ra16UFNPcC8S6-x0TYOqXxS7WwsUNly5sxy9Heye6zi6Hx-UIfLt45Kwc-qPMlr8NHAYtBzhGFI-2f3OYeAeOY91dHAB9vDMXOFRcjuKF5VnFf~hp8ErXPNB0QSBE2pdt6t6nIPqw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 7. GATA-2 interacts with HDAC5 in vivo and in vitro. / (A) pcDNA3.1/His–GATA-2 (5 μg) was cotransfected with 5 μg of either Flag-HDAC4, Flag-HDAC5, or Flag-HDAC6 into COS cells. Whole cell lysates were prepared and immunoprecipitated with anti–X-press antibody. Immunoprecipitates were subjected to immunoblot analysis with anti-Flag antibody (upper panel). The blot was reprobed with anti–X-press antibody to confirm that GATA-2 was successfully immunoprecipitated (lower panel). Input represents 5% of whole cell lysates used for each immunoprecipitation applied to confirm whether each class II HDAC was highly expressed. (B) GATA-2 interacts with HDAC3 and 5 in vitro. [35S]-methionine-labeled HDAC1, HDAC3, and HDAC5 translated in vitro were incubated with GST–GATA-2 fusion protein or GST protein alone and pulled down with GST-agarose beads. After 4 washes, the precipitates were dissolved by SDS-PAGE and visualized by autoradiography. Input represents 4% of in vitro–translated HDAC proteins used for the pull-down assay.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/98/7/10.1182_blood.v98.7.2116/5/m_h81911590007.jpeg?Expires=1767733448&Signature=rKY9BzdrvLJ-f50nKhLns6I3IGAXHU2WFfVB7q2Pyc4B2Nv8-ESEWWJ6tVCOKdAMq5z74cXXxqdUoAWb~GgMwQjGNQa50AZasvgo9QcPlsL9D5RW41cZDeesFYuwlt5estT-2TMs3-IHzsCL~1Ew2tQzdXGitY3xxhAy4MszliFzJHp4A4uFjP~1B02Y175DfRuDQJgHoeVagc~KOYVbhUoxxRepiFyjGYEXKW5DF~fAPlEf5cl5A4~Kfoza7bm7TdRcFPnc7y9LyRZu-sPuRXOHQplRSJl~69wyYP-hTAokxHFXIlegVlE1ldg5eHXgqu3PaVFK3kbcW7bfocf6kg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal