Abstract
Complementary and genomic DNA for the murine transferrin receptor 2 (TfR2) were cloned and mapped to chromosome 5. Northern blot analysis showed that high levels of expression of murine TfR2 occurred in the liver, whereas expression of TfR1 in the liver was relatively low. During liver development, TfR2 was up-regulated and TfR1 was down-regulated. During erythrocytic differentiation of murine erythroleukemia (MEL) cells induced by dimethylsulfoxide, expression of TfR1 increased, whereas TfR2 decreased. In MEL cells, expression of TfR1 was induced by desferrioxamine, an iron chelator, and it was reduced by ferric nitrate. In contrast, levels of TfR2 were not affected by the cellular iron status. Reporter assay showed that GATA-1, an erythroid-specific transcription factor essential for erythrocytic differentiation at relatively early stages, enhanced TfR2 promoter activity. Interestingly, FOG-1, a cofactor of GATA-1 required for erythrocyte maturation, repressed the enhancement of the activity by GATA-1. Also, CCAAT-enhancer binding protein, which is abundant in liver, enhanced the promoter activity. Thus, tissue distribution of TfR2 was consistent with the reporter assays. Expression profiles of TfR2 were different from those of TfR1, suggesting unique functions for TfR2, which may be involved in iron metabolism, hepatocyte function, and erythrocytic differentiation.
Introduction
Iron, one of the essential elements of life, is carried by transferrin (Tf) in the serum and absorbed by the cells through transferrin receptor (TfR1)-mediated mechanisms.1Diferric Tf in the serum binds to TfR1 on the cell surface, followed by endocytosis of the receptor-ligand complex. HFE is an atypical major histocompatibility complex (MHC) class I molecule that can form a complex with TfR1 on the cell surface and interfere with binding of Tf to TfR1.2-4 Mutations of HFE are thought to be related to hereditary hemochromatosis. After internalization of the Tf-TfR1 complex, iron is released from the Tf-TfR1 complex in the endosome and is transported to the cytoplasm and mitochondria by DMT1/Nramp2, a transmembrane iron transporter.5-8
Recently, we cloned human TfR2, another transferrin receptor gene. Two alternatively spliced forms of human TfR2 transcripts were identified: α and β. TfR2-α was highly expressed in K562, an erythroid leukemia cell line; in liver; and in HepG2, a hepatoma cell line.9 Increased Tf binding and iron uptake were observed in Chinese hamster ovary cells that had been stably transfected with human TfR2-α. Although human TfR2-α and TfR1 are very similar in their primary structure and Tf-binding property, the expression pattern of TfR2 mRNA is quite different from that of TfR1. We have found that in K562 cells, iron chelation up-regulates TfR1 and down-regulates TfR2, and iron overload down-regulates TfR1 and slightly up-regulates TfR2.10 TfR1 knockout mice did not survive beyond embryonic day 12.5 because of severe anemia and neurologic abnormalities, which indicates that murine TfR2 cannot fully compensate for the functions of TfR1.11 Recently, a homozygous nonsense mutation of the TfR2 gene, Y250X, was found in some patients with hereditary hemochromatosis that was not associated with altered HFE.12 This observation indicates that the main function of TfR2 may not be direct iron uptake, but may be regulation of iron uptake through indirect mechanisms.
In this paper, we describe the molecular cloning, chromosomal mapping, and characterization of expression of murine TfR2. Tissue distribution of TfR2 was clearly different from that of TfR1. We compared expression profiles of TfR1 and TfR2 mRNAs in murine erythroleukemia (MEL) cells in response to iron status and during their erythrocytic differentiation. We also examined the regulation of the murine TfR2 promoter by transcription factors that may be related to its tissue-specific expression. These results will help to elucidate the physiologic function of TfR2.
Materials and methods
Cell culture and differentiation
NIH-3T3 cells were obtained from American Type Culture Collection (Manassas, VA), and MEL cells, clone DS19, were provided by Dr Murate.13 NIH-3T3 cells were maintained in Dulbecco modified Eagle medium (Life Technologies, Gaithersburg, MD) supplemented with 10% adult bovine serum, and MEL cells were maintained in RPMI 1640 (Life Technologies) supplemented with 10% fetal bovine serum. To differentiate MEL cells toward erythrocytic lineage, we added 75 μM hemin (Sigma, St Louis, MO) and/or 2% dimethylsulfoxide (DMSO) to the medium. In some experiments, 10−7 M dexamethasone was also added. The cells were harvested on days 1, 3, and 5. Erythrocytic differentiation was monitored by hemoglobin concentration determined by the benzidine method, as described elsewhere.14 Protein concentrations were determined by the Bio-Rad protein assay kit (Bio-Rad, Hercules, CA).
Molecular cloning of complementary and genomic DNA of murine TfR2
A murine EST clone, IMAGE 1450755, which has significant homology to human TfR2-α cDNA (GenBank accession number AF067864), was purchased from Research Genetics (Huntsville, AL) and sequenced. The 3′ and 5′ cDNA ends for murine TfR2 were amplified from a cDNA library of MEL cells with primers 5′-AGGAGCAGCCAAGTCTGCAGTGGGGAC-3′ and 5′-GAAGGTCGAACCACTCCGCAGGATGGCA-3′, respectively, using the Marathon cDNA Amplification Kit (Clontech, Palo Alto, CA). The putative full-length coding sequence of the murine TfR2 was amplified from the same library using primers 5′-CACAAGCATGGAGCAACGTTGG-3′ and 5′-AGGGAGAAAGGAGAATCACGTGG-3′. Genomic DNA clones for murine TfR2 were isolated from a murine genomic library (Lambda FIX II Library; Stratagene, La Jolla, CA) using murine TfR2 cDNA fragments as probes.
Chromosomal mapping
T31 Mouse Radiation Hybrid Panel, RH04.02 (Research Genetics), was used to determine the chromosomal location of the TfR2 gene, as described previously.15 The primers 5′-TGGTAGACCACCTGCGGATG-3′ and 5′-AGGGAGAAAGGAGAATCACGTGG-3′ amplified a 227-bp fragment. The polymerase chain reaction (PCR) products were subjected to electrophoresis, Southern blotting, and hybridization with a 32P-labeled probe of murine TfR2. The results were analyzed by the Jackson Laboratory Mapping Panels (www.http://jax.org/resources/documents/cmdata).
Reverse transcriptase–PCR and Northern blot analyses
Reverse transcriptase–PCR (RT-PCR) and Northern blot analyses were performed essentially as described previously.9 As a template of RT-PCR, Mouse Multiple Tissue cDNA Panel I (Clontech) was used. To amplify murine TfR2 cDNA, we used primers 5′-TGCACAAGATGCTGCGAGGT-3′ and 5′-GTTCCGCTCCGAGCTGTAA-3′. Conditions for amplification were 32 cycles of 94°C for 30 seconds, 56°C for 40 seconds, and 72°C for 1 minute. As a control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was amplified in a separate reaction for 24 cycles using primers coming with the cDNA panel. The products were subjected to electrophoresis, Southern blotting, hybridization with radiolabeled probes, and autoradiography. For Northern blots, various murine tissues were prepared from C3H mice, and total RNA was extracted. Intestinal mucosa was prepared by scraping the inner surface of small intestines. As probes, 0.5 kb of the murine TfR1, the 3′ part of 1.4-kb murine TfR2, and 0.9-kb murine GAPDH cDNA fragments were used. In some experiments, relative expression levels of TfRs were calculated by an image analyzer, standardized by the expression levels of GAPDH, and shown as bar graphs.
Reporter assay
Reporter constructs pGL-mTfR2-pro-0.3 kb, -0.4 kb, -1.0 kb, and -2.1 kb were created by subcloning fragments of the murine TfR2 promoter from the transcription starting site to either the 0.3-kb upstream McsI site, the nucleotide at −392, the approximately 1-kb upstream BglII site, or the 2.1-kb upstream NheI site, respectively, into pGL3-basic vector plasmid (Promega, Madison, WI). Expression plasmids for GATA-1 and FOG-1, pXM-GATA1, and pMT2-FOG were kindly provided by Drs A. Tsang and S. Orkin.16,17 An expression plasmid for murine CCAAT/enhancer binding protein (C/EBP-α) was provided by Dr Xanthropoulos.18 An expression plasmid for erythroid Kruppel-like factor (EKLF) was created as follows. The complete coding sequence of EKLF was amplified by RT-PCR using K562 cDNA as template, tagged with FLAG at the 5′-end, and subcloned into pcDNA3.1(+) (Invitrogen, Carlsbad, CA). Expression of EKLF protein at a proper size was confirmed by Western blotting after transient transfection of NIH-3T3 cells. NIH-3T3 cells were transfected by GenePorter system (Gene Therapy Systems, San Diego, CA) with either empty pGL3-basic or pGL-mTfR2 promoter plasmids together with expression plasmids for GATA-1, FOG, EKLF, and/or C/EBP-α. To normalize the transfection efficiency, we cotransfected pCMV · SPORT-β Gal (Life Technologies). Cells were harvested after 60 hours, and luciferase and β-galactosidase activities were measured with a luminometer and with the β-galactosidase Enzyme Assay System (Promega), respectively.
Results
Homology between human and murine TfR2
Approximately 2.8 kb of cDNA sequences for murine TfR2 was obtained from the cDNA library of MEL cells by assembling sequences of 5′ and 3′ rapid amplification of cDNA ends (RACE) products (GenBank accession number AF207741). As Fleming et al19reported previously analyzing an EST clone, putative amino acid sequences of murine TfR2 were similar to those of human TfR2-α, sharing 85% identical and 92% similar amino acids. Murine TfR2 had a putative internalization motif, YRRV, and a transmembrane domain (amino acids 78-102) consisting of a hydrophobic amino acid stretch. We also cloned genomic DNAs including the promoter region (GenBank accession number AF207742). When we compared this genomic sequence with the human counterpart, promoter sequences from −360 to 0 were considerably conserved (Figure 1). Both human and murine TfR2 promoters contained 2 typical GATA-1 (an erythroid-specific transcription factor) consensus sequences, (W)GATA(R) at −52 to −57 and −23 to −19 for mouse, and −61 to −56 and −26 to −22 for man, respectively (underlined in Figure 1). Also, putative C/EBP binding sites, a doublet of CAAT at around −240, and a CCAAT sequence in the reverse direction at around −190, were seen in both human and murine TfR2 promoters (double underlines). In addition, several CACCC sequences, which can be EKLF consensus sequences, were found both in the human and murine TfR2 promoters within the upstream 1 kb from the transcription starting sites (GenBank accession number AF207742). We also found an interesting palindrome structure at −978 to −997, CAGTTTCCCAGGGAAACTG, although its meaning and function are unknown. Our cDNA sequence of murine TfR2 is identical to that reported by Fleming et al19 except for the 5′ portion; Fleming's sequence has an additional noncoding exon before our exon 1 (exon 1a, −248 to −189 base; Figure 1) and lacks 17 nucleotides at the 5′ portion of our exon 1. Because of these differences, putative GATA-1 and C/EBP sites are located in intron 1 and exon 1a of Fleming's sequences, respectively.
DNA sequences of the promoter region of murine TfR2 aligned with those of human TfR2.
Identical residues are shaded. Sequences for exon 1 of murine TfR2 and human TfR2-α are encircled by a box. Putative GATA-1 and C/EBP sites are underlined and double-underlined, respectively. The murine TfR2 promoter sequence up to −1150 base is available in GenBank (accession number AF207742).
DNA sequences of the promoter region of murine TfR2 aligned with those of human TfR2.
Identical residues are shaded. Sequences for exon 1 of murine TfR2 and human TfR2-α are encircled by a box. Putative GATA-1 and C/EBP sites are underlined and double-underlined, respectively. The murine TfR2 promoter sequence up to −1150 base is available in GenBank (accession number AF207742).
Alternative forms of murine TfR2 transcripts
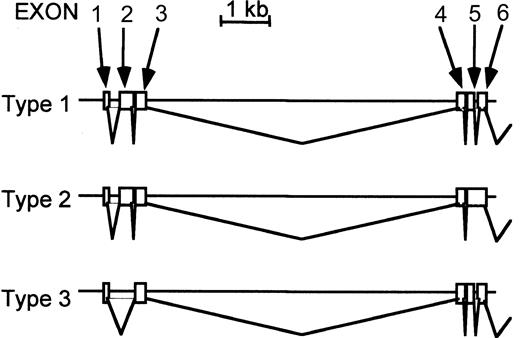
When we amplified the putative coding sequence of murine TfR2 by RT-PCR, we obtained clones of 3 different lengths (Figure2). The sequences of the middle-length form (type 1 in Figure 2) were identical to clones that we obtained from RACE; thus, this is probably the dominant form of murine TfR2 transcripts. The longer clone (type 2 in Figure 2) had an additional sequence between exons 5 and 6, which was the sequence of intron 5, and the shorter clone (type 3) had a deletion of exon 2. The former splicing variation caused a 14–amino acid addition after amino acid 237, followed by a stop codon (VRFPGWGAHHVLIGX). The latter caused an in-frame deletion of amino acids 12 to 93, which includes most of the putative intracellular and part of the transmembrane domains. Thus, the cDNA library from MEL cells contains at least 3 different lengths of TfR2 transcripts. We did not find murine TfR2 clones corresponding to the β-form of human TfR2 when we performed 5′-RACE using the MEL cDNA library. However, we did see at least 3 bands with different sizes on Northern analysis when we used the 1.4-kb 3′-portion of the TfR2 cDNA as a probe, indicating the presence of alternative forms of murine TfR2 transcripts (Figure3B). These 3 bands, however, do not correspond to the 3 forms shown in Figure 2 because the sizes of the latter 3 forms are too close to separate in Northern blots (type 1, 2.8 kb; type 2, 2.9 kb; type 3, 2.5 kb). On the other hand, when we used the 1.4-kb 5′-portion of TfR2 cDNA as a probe, only one band with the size of approximately 2.9 kb was observed. Probably variations occur in the 5′-portion of the transcripts coding for TfR2, and those corresponding to the β-form may exist although they might not be abundant, explaining why we could not isolate them.
Alternative forms of murine TfR2 transcripts.
Using primers for 5′ and 3′ ends of the murine TfR2 coding region, the transcripts were amplified by RT-PCR, cloned, sequenced, and compared with the genomic sequences. The sequences of the primers are 5′-CACAAGCATGGAGCAACGTTGG-3′ and 5′-AGGGAGAAAGGAGAATCACGTGG-3′. Type 1 is the representative form, which is identical to that cloned by 5′ and 3′ RACE. Type 2 is a longer form that has additional sequences (intron 5 in the type 1) between exons 5 and 6. Type 3 is a shorter form that lacks exon 2. Sequences after exon 6 were the same in all 3 types, so only the 5′ portion of this gene (exons 1-6) is shown in this figure.
Alternative forms of murine TfR2 transcripts.
Using primers for 5′ and 3′ ends of the murine TfR2 coding region, the transcripts were amplified by RT-PCR, cloned, sequenced, and compared with the genomic sequences. The sequences of the primers are 5′-CACAAGCATGGAGCAACGTTGG-3′ and 5′-AGGGAGAAAGGAGAATCACGTGG-3′. Type 1 is the representative form, which is identical to that cloned by 5′ and 3′ RACE. Type 2 is a longer form that has additional sequences (intron 5 in the type 1) between exons 5 and 6. Type 3 is a shorter form that lacks exon 2. Sequences after exon 6 were the same in all 3 types, so only the 5′ portion of this gene (exons 1-6) is shown in this figure.
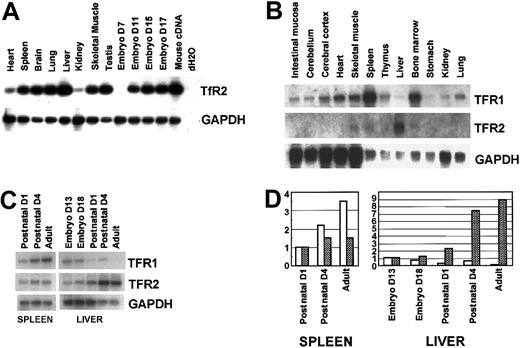
Tissue distribution of murine TfR2.
(A) RT-PCR analyses. RT-PCR was performed with primers for murine TfR2 (32 cycles) as well as GAPDH (24 cycles). Mouse multiple cDNA panels were used as templates. Whole-mouse cDNA and dH2O were used as positive and negative controls, respectively (right 2 lanes). The products were subjected to electrophoresis, Southern blotting, hybridization with radiolabeled probes, and autoradiography. D indicates day. (B) Northern blot analysis of various murine tissues. Various murine tissues were prepared from C3H mice, and total RNA (5 μg/lane) was extracted and Northern blotted. (C) Expression profiles of TfR1 and TfR2 in the liver and spleen during development. Spleens and livers were excised from day 13 and day 18 embryos, from postnatal day 1 and day 4 mice, and adult mice. (B-C) Total RNA was extracted, and expression levels of TfR1, TfR2, and GAPDH were compared by Northern analysis. (D) Relative expression levels of TfRs in the liver and spleen during development are calculated by an image analyzer, standardized by the values of GAPDH, and shown as bar graphs (■, TfR1;  , TfR2). The values of postnatal day 1 for spleen and day 13 embryo for liver are adjusted to 1.0.
, TfR2). The values of postnatal day 1 for spleen and day 13 embryo for liver are adjusted to 1.0.
Tissue distribution of murine TfR2.
(A) RT-PCR analyses. RT-PCR was performed with primers for murine TfR2 (32 cycles) as well as GAPDH (24 cycles). Mouse multiple cDNA panels were used as templates. Whole-mouse cDNA and dH2O were used as positive and negative controls, respectively (right 2 lanes). The products were subjected to electrophoresis, Southern blotting, hybridization with radiolabeled probes, and autoradiography. D indicates day. (B) Northern blot analysis of various murine tissues. Various murine tissues were prepared from C3H mice, and total RNA (5 μg/lane) was extracted and Northern blotted. (C) Expression profiles of TfR1 and TfR2 in the liver and spleen during development. Spleens and livers were excised from day 13 and day 18 embryos, from postnatal day 1 and day 4 mice, and adult mice. (B-C) Total RNA was extracted, and expression levels of TfR1, TfR2, and GAPDH were compared by Northern analysis. (D) Relative expression levels of TfRs in the liver and spleen during development are calculated by an image analyzer, standardized by the values of GAPDH, and shown as bar graphs (■, TfR1;  , TfR2). The values of postnatal day 1 for spleen and day 13 embryo for liver are adjusted to 1.0.
, TfR2). The values of postnatal day 1 for spleen and day 13 embryo for liver are adjusted to 1.0.
Chromosomal mapping
The radiation hybrid panel analysis revealed that the murine TfR2 was on the distal end of chromosome 5, between the D5Mit168 and D5Mit326 markers. This location is syntenic to human chromosome 7q22, to which human TfR2 was mapped.9
Tissue distribution of murine TfR2
By RT-PCR, murine TfR2 was detected in all the tissues that we tested, although the expression was low in the heart and kidney (Figure3A). Relatively high levels of expression were seen in liver and testis when normalized with GAPDH. In the embryo, expression of TfR2 was not detectable until day 11. By Northern blot analysis, expression of TfR2 was the highest in liver and the second highest in spleen (Figure 3B). Expression of TfR2 was not detectable in the intestinal mucosa, where iron absorption takes place. In contrast, expression of TfR1 was ubiquitous, but it was relatively low in the liver. In the embryo, expression of TfR2 increased in the liver during development, whereas expression of TfR1 decreased (Figure 3C-D). In the spleen, expression of TfR1 increased and levels of TfR2 were constant during development (Figure 3C-D).
Expression of TfR2 mRNA in MEL cells during erythrocytic differentiation
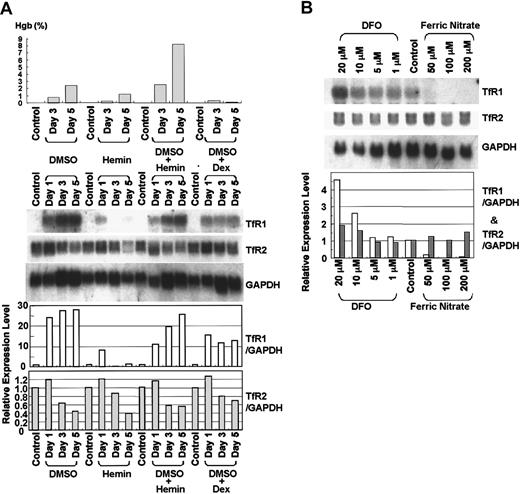
To induce erythrocytic differentiation, we cultured MEL cells with hemin and/or DMSO for 5 days. After 5 days, hemoglobin concentration in the cell lysates increased from 0.03% to 2.46%, 1.21%, or 8.24% in the presence of 2% DMSO, 75 mM hemin, or the combination of 2% DMSO and 75 mM hemin, respectively (Figure 4A, upper panel). The increase in hemoglobin concentration by DMSO was blocked by simultaneous addition of 10−7 M dexamethasone. Northern blot analysis showed that expression of TfR1 mRNA increased dramatically and that of TfR2 decreased during either DMSO or DMSO plus hemin–induced erythrocytic differentiation of these cells (Figure 4A, lower panel). These changes were blocked by simultaneous addition of 10−7 M dexamethasone. In contrast, expression of TfR1 mRNA was not induced and that of TfR2 decreased slightly during erythrocytic differentiation of MEL cells cultured with hemin alone (Figure 4A).
Expression of TfR2 mRNA in MEL cells during erythrocytic differentiation and in response to iron status.
(A) Expression of TfR2 mRNA during differentiation of MEL cells. MEL cells were cultured in the presence of 2% DMSO, 75 μM hemin, 2% DMSO + 75 μM hemin, or 2% DMSO + 10−7 M dexamethasone (Dex) for 5 days. Upper panel shows the hemoglobin concentration in the cell lysates. Hemoglobin concentration was determined by the benzidine method and shown as a percentage of total cellular lysate. Lower panel shows the results of Northern blot analysis. Approximately 20 μg total RNA was loaded in each lane. The membrane was sequentially hybridized with32P-labeled murine TfR1, TfR2 (3′-part, 1.4 kb), and murine GAPDH cDNA probes. (B) Effects of cellular iron status on expression of murine Tf receptors in MEL cells. MEL cells were cultured with various concentrations of Fe2(NO3)3 or DFO for 2 days, and Northern blot analysis was performed. The membrane was sequentially hybridized with 32P-labeled murine TfR1, TfR2, and GAPDH cDNA probes. (A-B) Relative expression levels of TfRs are calculated by an image analyzer, standardized by the values of GAPDH, and shown as bar graphs (■, TfR1; , TfR2).
, TfR2).
Expression of TfR2 mRNA in MEL cells during erythrocytic differentiation and in response to iron status.
(A) Expression of TfR2 mRNA during differentiation of MEL cells. MEL cells were cultured in the presence of 2% DMSO, 75 μM hemin, 2% DMSO + 75 μM hemin, or 2% DMSO + 10−7 M dexamethasone (Dex) for 5 days. Upper panel shows the hemoglobin concentration in the cell lysates. Hemoglobin concentration was determined by the benzidine method and shown as a percentage of total cellular lysate. Lower panel shows the results of Northern blot analysis. Approximately 20 μg total RNA was loaded in each lane. The membrane was sequentially hybridized with32P-labeled murine TfR1, TfR2 (3′-part, 1.4 kb), and murine GAPDH cDNA probes. (B) Effects of cellular iron status on expression of murine Tf receptors in MEL cells. MEL cells were cultured with various concentrations of Fe2(NO3)3 or DFO for 2 days, and Northern blot analysis was performed. The membrane was sequentially hybridized with 32P-labeled murine TfR1, TfR2, and GAPDH cDNA probes. (A-B) Relative expression levels of TfRs are calculated by an image analyzer, standardized by the values of GAPDH, and shown as bar graphs (■, TfR1; , TfR2).
, TfR2).
Expression of murine TfR2 in response to cellular iron status
MEL cells were cultured with various concentrations of the iron chelator desferrioxamine (DFO) or ferric nitrate for 2 days, and expression levels of TfR1 and TfR2 mRNA were examined by Northern blot analysis. As shown in Figure 4B, expression of TfR1 was up-regulated by DFO and down-regulated by ferric nitrate. In contrast, expression of TfR2 was largely unchanged by the identical conditions.
Transactivation of TfR2 promoter by GATA-1 and C/EBP-α
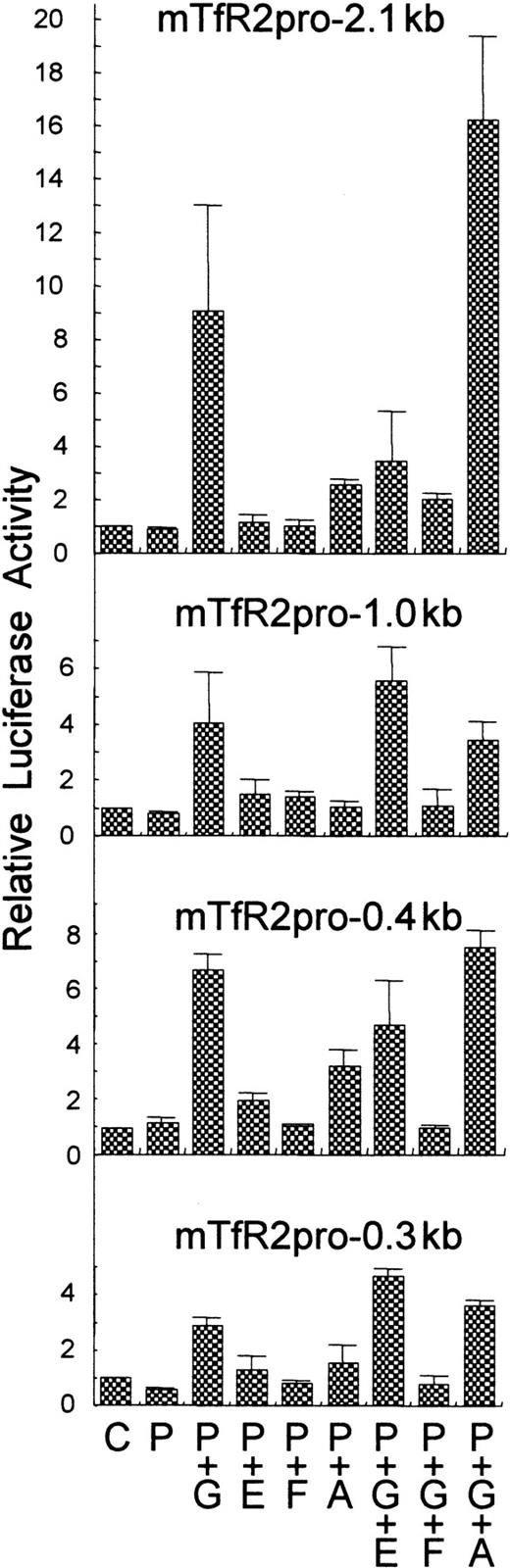
Murine TfR2 promoter activity was analyzed by transfecting NIH-3T3 cells with pGL3-mTfR2 reporter plasmids: pGL-mTfR2-pro-0.3 kb, -0.4 kb, -1.0 kb, and -2.1 kb, containing approximately 0.3 kb, 0.4 kb, 1.1 kb, and 2.1 kb TfR2 promoter, respectively. Almost no promoter activity was observed when these promoter constructs alone were transfected (Figure5). However, when cotransfected with expression plasmids for GATA-1, an erythroid-specific transcription factor, clear induction of luciferase activity (3- to 9-fold) was observed (Figure 5). To a much lesser extent, EKLF, another erythroid-specific transcription factor, induced the promoter activity. C/EBP-α also slightly enhanced the activity when using mTfR2-pro-0.3 kb, -0.4 kb, and -2.1 kb (Figure 5). On the other hand, FOG-1, a cofactor of GATA-1, did not enhance the activity but rather abrogated the enhancement of the activity by GATA-1. EKLF combined with GATA-1 showed a variable response, inhibiting GATA-1 stimulation with the longest TfR2 promoter (2.1 kb) and slightly stimulating GATA-1 activity with the shortest promoter (0.3 kb). The strongest TfR2 promoter activity (16-fold) occurred with the longest TfR2 promoter (2.1 kb) when stimulated by GATA-1 and C/EBP-α (Figure 5).
Reporter assay for murine TfR2 promoter.
NIH-3T3 cells were transfected by Geneporter system (Gene Therapy Systems) with either empty pGL3-basic (C) or mTfR2 promoter reporter plasmids (P) pGL-mTfR2-pro-0.3 kb, -0.4 kb, -1.0 kb, or -2.1 kb together with expression plasmids for GATA-1 (G), FOG-1 (F), EKLF (E), and/or C/EBP-α (A). To normalize the transfection efficiency, pCMV · SPORT-β-Gal (Life Technologies) was cotransfected. Cells were harvested after 60 hours, and luciferase and β-galactosidase activities were measured with a luminometer and with the β-galactosidase Enzyme Assay System (Promega), respectively. Experiments were performed in triplicate and were repeated twice, with very similar results. Mean values of relative luciferase activities and SDs of a representative experiment are shown.
Reporter assay for murine TfR2 promoter.
NIH-3T3 cells were transfected by Geneporter system (Gene Therapy Systems) with either empty pGL3-basic (C) or mTfR2 promoter reporter plasmids (P) pGL-mTfR2-pro-0.3 kb, -0.4 kb, -1.0 kb, or -2.1 kb together with expression plasmids for GATA-1 (G), FOG-1 (F), EKLF (E), and/or C/EBP-α (A). To normalize the transfection efficiency, pCMV · SPORT-β-Gal (Life Technologies) was cotransfected. Cells were harvested after 60 hours, and luciferase and β-galactosidase activities were measured with a luminometer and with the β-galactosidase Enzyme Assay System (Promega), respectively. Experiments were performed in triplicate and were repeated twice, with very similar results. Mean values of relative luciferase activities and SDs of a representative experiment are shown.
Discussion
We have previously cloned human TfR2 as the second receptor for transferrin. We identified 2 alternatively spliced forms of human TfR2 transcripts, α and β.9 TfR2-α protein is a type-2 membrane protein, and TfR2-β protein may be a soluble form lacking both intracellular and transmembrane domains of the α-form. The function of the β-form is unknown. In this study, we identified 3 different transcripts for murine TfR2, types 1, 2, and 3 (our Northern blot analyses suggest that additional variations also occur at the 5′-portion of TfR2 transcripts). Type 1 transcript is the most common form, encoding a type-2 membrane protein corresponding to human TfR2-α. TfR2 type-2 protein, if it exists, lacks most of the extracellular domain. In contrast, TfR2 type-3 protein lacks most of the transmembrane domain, so theoretically, this may be a soluble protein similar to the β-form of human TfR2.
Our previous studies suggested that molecular properties of human TfR1 and TfR2-α are similar. First, both can bind to Tf on the cell surface.9 Second, both can mediate cellular iron uptake.9 Third, binding of Tf to both TfR1 and TfR2-α is pH dependent; namely, holo-Tf binds to these receptors at basic pH and apo-Tf binds to them at acidic pH.10 Fourth, both support growth of TfR1-deficient cells in vitro and in vivo when stably transfected with them.10
However, tissue distribution of these receptors was considerably different. Expression of TfR1 occurs almost ubiquitously, but levels are particularly low in the liver19 (Figure 3B). In contrast, expression of TfR2 occurs almost exclusively in the liver19 (Figure 3B). Expression of TfR1 decreased in the liver during development, from embryonic day 13 to postnatal day 1, whereas levels of TfR2 increased dramatically during the same period (Figure 3C). During murine development, hematopoietic cells, especially erythroid cells, appear first at embryonic day 7 to 8 in the yolk sac, and then move to the liver starting on embryonic day 10. Our RT-PCR results shown in Figure 3A indicate that expression of TfR2 starts between embryonic days 8 and 11, which may reflect the development of the liver, where erythropoiesis occurs.20
Expression of TfR1 is regulated at the levels of transcription and posttranscription by factors such as cellular status of iron, oxygen, proliferation, and differentiation.21-26 Expression profiles of TfR1 and TfR2 in response to cellular iron status were also different. Intracellular iron levels regulate expression of TfR1 mainly at a posttranscriptional level. If iron is scarce, iron-regulatory proteins (IRPs) bind to iron-responsive elements (IREs) of the 3′-untranslated region of TfR1 mRNA and stabilize the transcript. In the presence of excess iron, IRPs are released from IREs of the TfR1 mRNA, resulting in degradation of the transcript. Previous reports have shown that in K562 and other cells, a cell membrane–permeable iron chelator (DFO) up-regulates and an excess of iron down-regulates levels of TfR1 mRNA, consistent with our results from Northern blot analysis shown in Figure 4.10,21,27-31 In contrast, expression of murine TfR2 mRNA was not clearly changed by either DFO or iron overloading in MEL cells (Figure 4), which is similar to the findings of human TfR2 in K562 cells.10
TfR1 has been reported to be up-regulated during erythrocytic differentiation in MEL cells.31-33 This is consistent with our results of DMSO-induced differentiation shown in Figure 4. In the experiments with hemin alone, up-regulation of TfR1 during differentiation was not obvious, probably because iron in the hemin may diminish the up-regulation of TfR1. On the other hand, TfR2 mRNA was down-regulated during erythrocytic differentiation of MEL cells. If our differentiation system reflects gene expression in normal erythropoiesis, our results suggest that TfR2 is a dominantly expressed receptor in early erythroid precursors and TfR1 becomes the dominant receptor during differentiation.
Together, tissue distribution and expression profiles of TfR1 and TfR2 were quite different in response to iron and during erythrocytic differentiation. These results call into question whether TfR2 is just a second receptor for Tf and functions similarly to TfR1. Inappropriate iron accumulation in the body causes hemochromatosis, and to date, 3 types of hereditary hemochromatosis have been identified. Type 1 is the most common type that is related to mutations of the HFEgene.2 Type-2 hemochromatosis is the juvenile type, the locus of which was recently determined to be chromosome 1q, but the responsible gene is not yet determined.34Recently, type-3 hemochromatosis was found in 2 families, who had a nonsense mutation at codon 250 of the human TfR2 gene, with a chromosomal locus at 7q22.12 This observation strongly suggests that defects of TfR2 can cause hemochromatosis, suggesting a relation between TfR2 and iron metabolism, but the main function of TfR2 does not appear to be cellular iron uptake. How then might TfR2 regulate iron metabolism?
TfR1 can form a complex with HFE, an atypical MHC class I molecule. TfR1 can also physically associate with other MHC class I and II molecules on the cell surface.35 MHC molecules, some of which are presenting self- and/or nonself antigens, are recognized by T lymphocytes; thus, they are involved in the immune response. The primary structure of the extracellular domain of both human and murine TfR2 is similar to that of TfR1, so TfR2 may also be able to bind to some of the MHC molecules, which may affect immune function or iron metabolism. Although the affinity of TfR2 for HFE is low compared with that of TfR1,36 TfR2 may bind to other MHC molecules. Once T lymphocytes recognize TfR1 or TfR2 together with MHC molecules on the cell surface, these cells may produce cytokines that may change the iron status of the body. TfR2 may regulate iron metabolism through such a mechanism. An association between iron homeostasis and the immune response has been proposed by Salter-Cid et al.37
In both man and mouse, elevated levels of expression of TfR2 were observed in the liver. C/EBP-α is highly abundant in normal liver, and it is down-regulated during proliferation of hepatocytes.38 This is consistent with our reporter assay, which indicated that C/EBP-α enhanced TfR2 promoter activity (Figure5). Another tissue in which high expression of TfR2 occurs is the erythroid compartment. Two consensus sequences for GATA-1 are present in the proximal portion of the promoter of mTfR2. The GATA-1 erythroid-specific transcription factor enhanced TfR2 promoter activity dramatically (Figure 5), probably through the GATA-1 consensus sequences on the promoter of mTfR2. GATA-1 is essential for erythroid development at the relatively immature proerythroblast stage,39 although over-expression of GATA-1 inhibits erythroid differentiation.40 Several CACCC sequences are found in the TfR2 promoter region; these sequences can be binding sites for EKLF, another erythroid-specific transcription factor. EKLF alone slightly enhanced the TfR2 promoter activity (Figure 5), suggesting that expression of TfR2 is regulated by the erythroid-specific transcriptional machinery. Intriguingly, FOG-1, a cofactor of GATA-1, repressed the GATA-1 enhancement of the promoter activity (Figure 5). FOG-1 is essential for erythroid cell maturation.16Down-regulation of TfR2 during erythrocyte maturation may be due to up-regulation of FOG-1. Thus, transactivation of the TfR2 by GATA-1, EKLF, and C/EBP-α is consistent with tissue distribution of expression of TfR2 as shown in Figure 3.
In this study, we described molecular cloning of murine TfR2 and its expression profile compared with that of TfR1. Further studies defined the transcription factors required for a robust transcription of TfR2. Accumulating evidence suggests that the physiologic function of TfR2 seems to be different from that of TfR1. Murine knockout models for TfR2 will help to clarify the function of TfR2; it is probably involved in iron metabolism, hepatic function, and erythropoiesis.
We thank Dr T. Murate (Nagoya University, Japan) for providing us with MEL cells, Drs A. Tsang and S. Orkin for plasmids pXM-GATA1 and pMT2-FOG, and Dr Xanthopoulos for the C/EBP-α expression plasmid.
Supported in part by grants from the National Institutes of Health, C. and H. Koeffler Fund, Parker Hughes Trust, and Horn Foundation. H.P.K. holds the Mark Goodson Endowed Chair of Oncology and is a member of the Jonsson Cancer Center.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
H. Phillip Koeffler, Division of Hematology/Oncology, Cedars-Sinai Medical Center, UCLA School of Medicine, 8700 Beverly Blvd, B-208, Los Angeles, CA 90048; e-mail:koeffler@cshs.org.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal