Abstract
Cytokines, such as interleukin-2 (IL-2), activate intracellular signaling pathways via rapid tyrosine phosphorylation of their receptors, resulting in the activation of many genes involved in cell growth and survival. The deubiquitinating enzyme DUB-2 is induced in response to IL-2 but as yet its function has not been determined. The results of this study show that DUB-2 is expressed in human T-cell lymphotropic virus-I(HTLV-1)–transformed T cells that exhibit constitutive activation of the IL-2 JAK/STAT (signal transducers and activators of transcription) pathway, and when expressed in Ba/F3 cells DUB-2 markedly prolonged IL-2–induced STAT5 phosphorylation. Although DUB-2 did not enhance IL-2–mediated proliferation, when withdrawn from growth factor, cells expressing DUB-2 had sustained STAT5 phosphorylation and enhanced expression of IL-2–induced genescis and c-myc. Moreover, DUB-2 expression markedly inhibited apoptosis induced by cytokine withdrawal allowing cells to survive. Taken together these data suggest that DUB-2 can enhance signaling through the JAK/STAT pathway, prolong lymphocyte survival, and, when constitutively expressed, may contribute to the activation of the JAK/STAT pathway observed in some transformed cells.
Introduction
The survival, proliferation, and differentiation of lymphocytes are controlled by cytokines that act through specific transmembrane receptors.1 Interleukin-2 (IL-2) regulates T-cell survival through a receptor composed of 3 chains, IL-2Rα, IL-2Rβ, and the common gamma chain (γc). Binding of IL-2 to its receptor results in activation of the cytoplasmic tyrosine kinases JAK1 and JAK3, which, in turn, induce phosphorylation of the receptor and associated signaling molecules including the signal transducers and activators of transcription (STATs).2 The model predicts that JAK kinases phosphorylate specific tyrosine residues in the cytoplasmic domain of the receptors and thus provide docking sites for SH2-containing adaptor molecules such as SHC, SHP-2, and the transcription factor STAT5. This results in the initiation of signaling cascades and the expression of genes including c-myc, cd25, cis, socs-3, pim-1,c-Fos, and vcp.3 Although many of these gene products are involved in proliferation and differentiation of lymphocytes, some act in a feedback loop to modulate signaling, presumably by regulating JAK and STAT activation.
Although the activation of the receptor signaling pathways has been studied in detail, how these processes are regulated is unclear. One important mechanism of signal transduction regulation is the dephosphorylation of signaling intermediates by protein tyrosine phosphatases such as SHP-1 and phospholipid phosphatases like SHIP.4 Recent studies have identified 2 new families of negative regulatory molecules termed suppressors of cytokine signaling (SOCS/JAB/SSIs) and protein inhibitors of activated STATs (PIAS), which suppress signal transduction via the JAK-STAT pathway possibly by directly inhibiting JAK kinases and receptor-mediated STAT activation.5 6
Additionally, proteolytic degradation is also a suggested mechanism for the regulation of receptor turnover, and a number of receptors can be targeted for endocytosis and proteolytic degradation by ubiquitination.7 Recently, the ubiquitin (Ub)-mediated proteolytic pathway has been implicated in many intracellular processes such as transcriptional activation, cell cycle regulation, and the turnover of plasma membrane receptors such as the c-Kit receptor. Proteins that are targeted for degradation are conjugated with a 76-amino acid Ub polypeptide tag. This is accomplished via a series of enzymes termed the Ub-conjugating complex consisting of components including Ub-activating (E1), Ub-conjugating (E2), and Ub-ligating (E3) enzymes.8 Polyubiquitinated proteins are then targeted to the proteasome where degradation can occur. Deubiquitinating enzymes are a family of cysteine proteases that cleave Ub from Ub-conjugated protein substrates and hence also control ubiquitination.9Deubiquitinating enzymes can act at many points in the Ub pathway including polyubiquitin chain processing, removing Ub from substrates to rescue from degradation, or by removing residual Ub to assist in proteasomal degradation. Many novel candidate deubiquitinating enzymes have been identified indicating that these enzymes may be the largest group involved in the Ub system.10 These deubiquitinating enzymes or Ub-processing proteases (UBPs) have considerable sequence diversity and are therefore likely to have a broad range of specificities. However, their substrate specificities and function have not been determined.
Recently 2 cytokine-inducible genes have been described that belong to a novel class of deubiquitinating enzymes termed DUB-1 and DUB-2.10 They were identified as hematopoietic-specific immediate-to-early genes that are rapidly induced after cytokine stimulation. DUB-1 is induced by IL-3, and granulocyte-macrophage colony-stimulating factor (GM-CSF) and is expressed in a variety of hematopoietic cells types, but DUB-2 appears to be specifically regulated by IL-2 and its expression is restricted to T cells.11-13 The function of these enzymes is not known, but the fact that both DUB-1 and DUB-2 are induced after cytokine stimulation suggests that they may play a role in cytokine-mediated signal transduction.
To determine DUB-2 function we expressed the enzyme in a Ba/F3 B-lymphocyte cell line and found that it enhanced IL-2 receptor and STAT5 tyrosine phosphorylation in response to IL-2. We also detected DUB-2 expression in human T-cell lymphotropic virus I (HTLV-1)–transformed T cells that exhibit constitutive activation of the IL-2 JAK/STAT pathway. When DUB-2 was expressed in IL-2Rβ+ Ba/F3 cells, it delayed apoptosis induced by cytokine withdrawal. Therefore, DUB-2 can enhance signaling specifically in response to IL-2 and can contribute to constitutive activation of the JAK/STAT pathway and enhanced lymphocyte survival.
Materials and methods
Plasmids
DUB-2 wild-type (WT) and DUB-2 C60S complementary DNAs (cDNAs) were tagged with the FLAG epitope at their C-termini by standard polymerase chain reaction (PCR)–based methods. Each cDNA was amplified by PCR, using a 5′ primer containing an EcoRI site and an ATG codon, and a 3′ primer containing a ClaI site. The EcoRI/ClaI PCR fragment was subcloned betweenEcoRI and ClaI sites of a modified pME18S vector in frame with the FLAG epitope. The pUHD 10-3 plasmid expressing DUB-2 was constructed by subcloning the DUB-2 cDNA containing a FLAG tag. The human IL-2Rβ was expressed from the pNA plasmid (gift from Dr Brad Nelson).
Cell culture and transfections
The transformed T-cell lines MT-2 and Molt-4β were grown in RPMI 1640/10% fetal bovine serum (FBS). The IL-2–dependent human T-cell line KIT-225 was grown in RPMI 1640/10% FBS containing 100 IU IL-2/mL. The IL-3–dependent pro-B cell line Ba/F3 was grown in RPMI 1640/10% FBS containing 10 ng/mL IL-3. Ba/F3 cells expressing the IL-2Rβ were generated by stable transfection; these cells are able to grow in response to either IL-3 or IL-2. Ba/F3/IL-2Rβ (Ba/F3β) cell lines expressing WT DUB-2 were also generated by stable transfection. Transfectants were generated by coelectroporating 107 cells with linearized plasmids containing the desired genes and a plasmid encoding neomycin or puromycin resistance, using a Gene Pulser (Biorad, Hercules, CA; 300 V, 960 μF). After 24 hours, cells were plated and selected in medium containing either neomycin 2 mg/mL (Gibco BRL, Rockville, MD) or 200 μg/mL Puromycin (Biorad). Resistant clones were tested for IL-2Rβ and DUB-2 expression by Western blotting, using IL-2Rβ antiserum (Santa Cruz Biotechnology, Santa Cruz, CA) and FLAG antiserum (Zymed, South San Francisco, CA), respectively. Ba/F3 cells expressing the tetracycline-responsive transcriptional activator (tTA) protein were generated by electroporation with pUHD15-1 modified to contain a puromycin resistance gene.24 A Ba/F3β/tTA cell line expressing the IL-2Rβ was generated as described above. Ba/F3β/tTA cells were transfected with pUHD10-3 containing DUB-2 by electroporation as above to generate the Ba/F3β/tTA DUB-2 cell lines and selected using 1.2 mg/mL hygromycin. Tetracycline 1 μg/mL was replaced every 48 hours. To induce expression of DUB-2, cells were grown in the absence of tetracycline for 24 to 48 hours. For cytokine stimulation, cells were washed twice, incubated in cytokine-free media for 4 to 9 hours in RPMI 1640/10% FBS, and then treated with 100 U/mL IL-2 for the indicated period.
Apoptosis assays
The Ba/F3β/tTA DUB-2 cells were grown out of tetracycline for 48 hours, rested for 12 hours, stimulated with IL-2 for 4 to 5 hours, and then withdrawn from IL-2 for the indicated period. Apoptotic cells were quantified by standard trypan blue exclusion assay using a 0.4% trypan blue stain solution (Gibco BRL), or by flow cytometry after propidium iodine (PI) staining as follows. Cells were washed once in phosphate-buffered saline (PBS), resuspended in a hypotonic buffer containing 0.1% nonylphenoxy polyethoxy ethanol (NP-40), 0.1% sodium citrate, and 50 μg/mL PI (Sigma, St Louis, MO), and subjected to flow cytometry analysis using FACSCalibur and CellQuest software (Becton Dickinson, Franklin Lakes, CA).
Immunoprecipitations and Western blotting
Cells were washed in PBS, lysed in 10 mM Tris pH 7.5, 150 mM NaCl, 2 mM EDTA, 0.875% Brij 96, 0.125% NP-40, 1 mM Na3VO4, 5 mM NaF, 10 μg/mL leupeptin, 10 μg/mL aprotinin, and 1 mM p-nitrophenyl p′-guanidine benzoate (NPGB), and centrifuged at 18 000g at 4°C for 15 minutes. Lysates were immunoprecipitated using either monoclonal antibody (mAb) IL-2Rβ,24 polyclonal DUB,12antibodies to STAT5b, monoclonal M2 anti-FLAG (Sigma), or mAb to ERK2 (Santa Cruz Biotechnology). The immunoprecipitates were washed 5 times in lysis buffer, and separated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA), and immunoblotted. Antibodies used were as follows: FLAG epitope (Octapeptide epitope, Zymed), DUB,12 DUB-2 (rabbit polyclonal no. 305), Bcl-x (Transduction Laboratories, San Diego, CA), phosphotyrosine mAb (4G10, UBI), phospho-Akt and Akt (New England Biolabs, Beverly, MA), STAT5b, IL-2Rβ, c-Myc, phospho-ERK and ERK-2 mAb (Santa Cruz Biotechnology), and Cis (a gift from Dr A. Yoshimura). The blots were visualized by enhanced chemoluminescence (ECL; Pierce, Rockford, IL).
Results
DUB-2 is constitutively expressed in HTLV-1–transformed MT2 cells
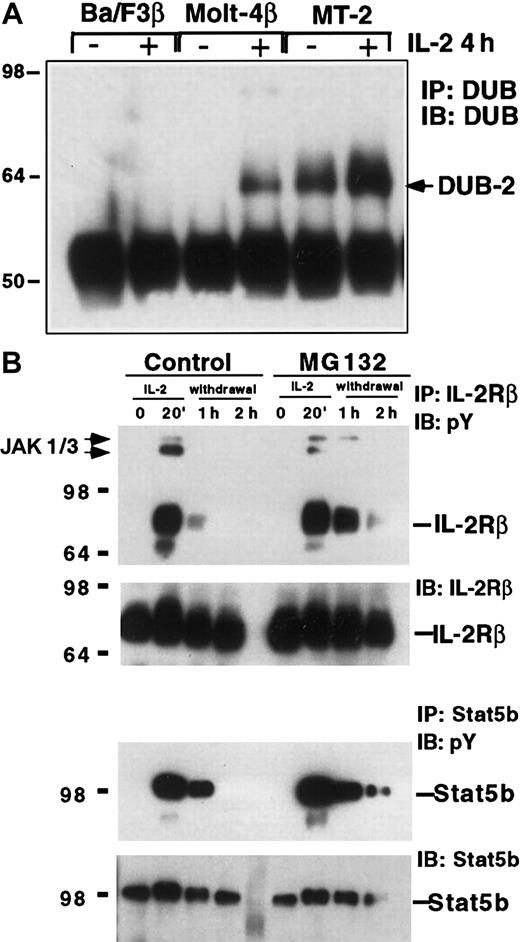
Considerable evidence suggests that the JAK/STAT pathway plays a positive role in leukemia.14 Indeed oncogenic transformation of T cells by HTLV-1 can result in constitutive tyrosine phosphorylation of the IL-2 receptor complex and STAT5.15-17 To explore IL-2–induced genes that were expressed in HTLV-1–transformed T cells, we examined DUB-2 expression in MT-2 cells using an antibody that recognizes DUB-1 and DUB-2.12 We compared DUB protein expression in MT2 cells, Ba/F3 cells (pro-B cell), and MOLT-4β cells (T-cell leukemia) before and after 4 hours of IL-2 stimulation (Figure1A). Although no induction of DUB protein was detected in Ba/F3 cells, a protein of 64 to 68 kd was recognized by the DUB-specific antibody in MOLT-4β cells after IL-2 treatment. The apparent molecular weight of the protein (64 kd) and the fact that it was seen in an IL-2–responsive T-cell line such as MOLT-4β suggested that it was human DUB-2. Interestingly, MT-2 showed a protein of the same size that was constitutively expressed prior to IL-2 treatment and further induced by IL-2. Although the IL-2 signaling pathway was constitutively active in MT2 cells, expression of other IL-2–induced genes including SOCS3 andCIS was not observed (results not shown). Therefore in an HTLV-1–transformed T-cell line DUB-2 is uniquely stably expressed, perhaps suggesting that it may play a role in transformation.
DUB expression and ubiquitination may regulate IL-2 signaling.
(A) Constitutive expression of DUB-2 in HTLV-1–transformed T cells. Ba/F3β, Molt-4β, and MT-2 cells were stimulated with IL-2 for 4 hours and lysates were immunoprecipitated with antisera to DUB,12 followed by Western blotting with the same antibody. (B) The proteasome inhibitor MG132 stabilizes IL-2–induced tyrosine phosphorylation of IL-2Rβ and STAT5b. KIT-225 cells were pretreated with 5 μM MG132 or control DMSO and stimulated with IL-2 for 20 minutes. The cells were then incubated in cytokine-free media containing MG132 or DMSO for the indicated times. Cells lysates were immunoprecipitated with anti–IL-2Rβ or anti-STAT5b, and immunoblotted with antiphosphotyrosine (pY), IL-2Rβ, or STAT5b antisera as indicated.
DUB expression and ubiquitination may regulate IL-2 signaling.
(A) Constitutive expression of DUB-2 in HTLV-1–transformed T cells. Ba/F3β, Molt-4β, and MT-2 cells were stimulated with IL-2 for 4 hours and lysates were immunoprecipitated with antisera to DUB,12 followed by Western blotting with the same antibody. (B) The proteasome inhibitor MG132 stabilizes IL-2–induced tyrosine phosphorylation of IL-2Rβ and STAT5b. KIT-225 cells were pretreated with 5 μM MG132 or control DMSO and stimulated with IL-2 for 20 minutes. The cells were then incubated in cytokine-free media containing MG132 or DMSO for the indicated times. Cells lysates were immunoprecipitated with anti–IL-2Rβ or anti-STAT5b, and immunoblotted with antiphosphotyrosine (pY), IL-2Rβ, or STAT5b antisera as indicated.
Proteasome inhibitors block dephosphorylation of the IL-2 receptor and STAT5
Deubiquitinating enzymes such as DUB-2 can block Ub- and proteasome-mediated proteolytic degradation.10 Two recent reports have suggested that signaling via the EPO receptor18 and the IL-3 receptor19 can be regulated in a proteasome-dependent manner. To explore whether the Ub system was involved in the regulation of IL-2 signaling we tested the effect of the proteasome inhibitor MG132. KIT-225 cells were preincubated in media containing MG132 (5 μM) or control dimethyl sulfoxide (DMSO) for 1 hour, and then treated with IL-2 for 20 minutes prior to washing and resuspension in media without IL-2. Cell lysates were immunoprecipitated with an antibody to the IL-2Rβ, run on SDS-PAGE, and blotted for antiphosphotyrosine. IL-2–induced rapid tyrosine phosphorylation of the IL-2Rβ chain was evident at 20 minutes, and following withdrawal from IL-2 the level of IL-2Rβ phosphorylation returned to background within 1 hour in cells not treated with proteasome inhibitor (Figure 1B, upper panel). However, in the presence of MG132 the level of IL-2Rβ tyrosine phosphorylation had not decreased by 1 hour but remained elevated up to 2 hours following cytokine withdrawal (Figure 1B, upper panel). Interestingly the JAK kinases JAK1 and JAK3 also coprecipitated with the IL-2R and in the presence of MG132 remained associated and phosphorylated up to 1 hour following IL-2 removal. Reblotting with anti–IL-2Rβ antibody showed equivalent protein in each lane. This finding suggested that turnover of the phosphorylated IL-2R complex required proteasome-mediated degradation. To determine whether proteasome inhibitors affected other components of the IL-2 signaling pathway, we examined the phosphorylation of the STAT5b using the same lysates. In DMSO-treated cells IL-2–induced STAT5 tyrosine phosphorylation was not detected 2 hours after cytokine withdrawal, but remained elevated up to 2 hours in the presence of MG132 (Figure 1B, lower panels). This effect on IL-2–induced STAT5 has been reported previously and coupled with our data suggest that proteasome inhibitors can stabilize both IL-2Rβ and STAT5 phosphorylation, suggesting that their dephosphorylation was dependent on the proteasome pathway and perhaps the Ub system.
STAT5 phosphorylation is prolonged in cells expressing DUB-2
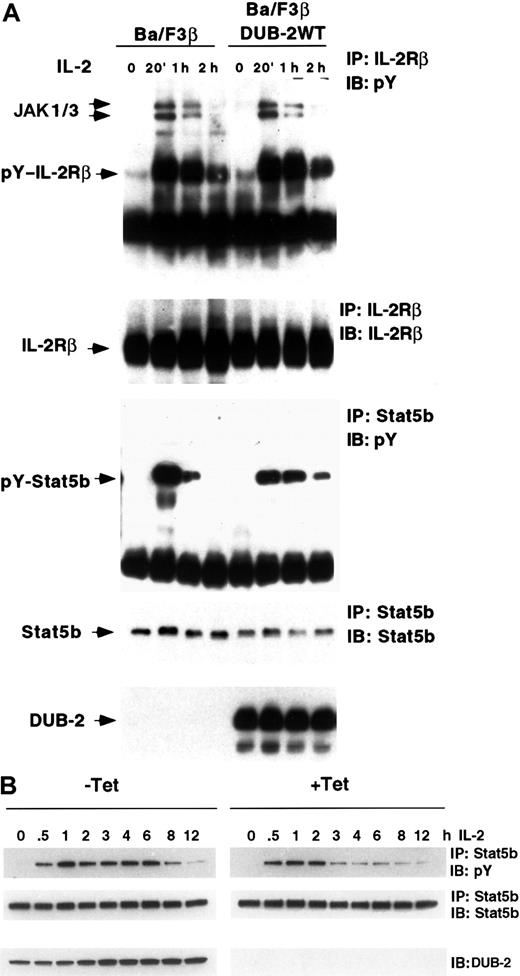
Having found that IL-2R and STAT5 dephosphorylation can be inhibited by a proteasome-dependent mechanism, and that DUB-2 was constitutively expressed in adult T-cell leukemia (ATL) cells, we wondered whether IL-2–induced phosphorylation might be affected by DUB-2. Because neither DUB-2 nor DUB-1 was detected in Ba/F3 cells following IL-2 treatment (Figure 1A), we made Ba/F3 cells that stably expressed DUB-2 (and IL-2Rβ) and explored IL-2–induced STAT5 and IL-2Rβ phosphorylation. The cells were treated with IL-2 for the periods shown and the lysates immunoprecipitated with anti–IL-2Rβ, run on SDS-PAGE, and blotted with antiphosphotyrosine. IL-2–induced phosphorylation of the IL-2R was not significantly altered in cells expressing DUB-2 (Figure 2A, upper panel). We then examined IL-2–mediated STAT5 phosphorylation, by immunoprecipitating with anti-STAT5b and blotting for phosphotyrosine. In control BA/F3β cells IL-2–induced tyrosine phosphorylation of STAT5b returned to almost basal levels by 2 hours (Figure 2A, lower panels). However, in cells expressing DUB-2, STAT5b phosphorylation was markedly prolonged with phosphorylation still detected at 2 hours. These findings suggest that when DUB-2 is expressed it can prolong IL-2–mediated STAT5 phosphorylation.
STAT5 phosphorylation is prolonged in cells expressing DUB-2.
(A) Ba/F3β (Ba/F3 cells expressing the IL-2Rβ) cells were stimulated with IL-2 for 20 minutes, then removed from cytokine and incubated in cytokine-free medium as indicated. Lysates were immunoprecipitated with anti–IL-2Rβ, anti-STAT5b, or anti-FLAG M2, and immunoblotted with antiphosphotyrosine (pY), anti–[IL-2Rβ, anti-STAT5b, or anti-FLAG as indicated. (B) Ba/F3β/tTA DUB-2 were grown with or without tetracycline (+/− Tet) for 48 hours and stimulated with IL-2 for the indicated times. Lysates were immunoprecipitated with anti-STAT5b and immunoblotted with antiphosphotyrosine (pY) or STAT5b antisera as indicated. Whole cell lysates were also run on SDS-PAGE and immunoblotted with DUB-2 antiserum.
STAT5 phosphorylation is prolonged in cells expressing DUB-2.
(A) Ba/F3β (Ba/F3 cells expressing the IL-2Rβ) cells were stimulated with IL-2 for 20 minutes, then removed from cytokine and incubated in cytokine-free medium as indicated. Lysates were immunoprecipitated with anti–IL-2Rβ, anti-STAT5b, or anti-FLAG M2, and immunoblotted with antiphosphotyrosine (pY), anti–[IL-2Rβ, anti-STAT5b, or anti-FLAG as indicated. (B) Ba/F3β/tTA DUB-2 were grown with or without tetracycline (+/− Tet) for 48 hours and stimulated with IL-2 for the indicated times. Lysates were immunoprecipitated with anti-STAT5b and immunoblotted with antiphosphotyrosine (pY) or STAT5b antisera as indicated. Whole cell lysates were also run on SDS-PAGE and immunoblotted with DUB-2 antiserum.
As previously reported, DUB-2 is induced in response to IL-2 and is not expressed in resting lymphocytes. Thus it is possible that sustained expression of DUB-2 may affect many proteasome-dependent events. We therefore created Ba/F3 cells containing tTA such that DUB-2 expression was induced on removal from tetracycline, and examined tyrosine phosphorylated STAT5 following IL-2 treatment. Cells grown for 48 hours either in the presence or absence of tetracycline were stimulated with IL-2 for up to 12 hours, and tyrosine phosphorylated STAT5 examined. In cells not expressing DUB-2 (Figure 2B, +Tet) IL-2–induced tyrosine phosphorylation of STAT5 returned to background levels by 3 hours. However, in cells expressing DUB-2 (Figure 2B, −Tet) strong STAT5 phosphorylation was detected up to 6 to 8 hours after IL-2 treatment. Equivalent amounts of STAT5 were present in each lane and DUB-2 expression was evident only in the absence of tetracycline (Figure 2B, bottom panels). These findings were observed in more than 5 experiments and suggest that when DUB-2 is induced in lymphocytes it can prolong IL-2–mediated STAT5 phosphorylation.
DUB-2 inhibits STAT5 dephosphorylation
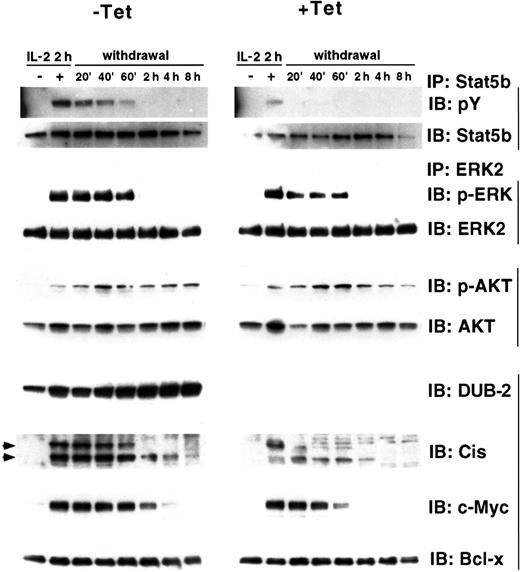
Dephosphorylation of signaling intermediates occurs rapidly after removal of cytokine. We therefore wanted to determine whether STAT5 phosphorylation would remain elevated after cytokine withdrawal in cells expressing DUB-2. Cells were stimulated with IL-2 for 2 hours, washed, and incubated in cytokine-free medium for the indicated times and tyrosine- phosphorylated STAT5 was examined both in cells removed from or maintained in tetracycline. In cells not expressing DUB-2 IL-2–induced tyrosine phosphorylation of STAT5b was not detected 20 minutes following cytokine withdrawal (Figure3, +Tet, top panel). However, in cells expressing DUB-2, STAT5b phosphorylation was still observed at 1 hour after cytokine removal (Figure 3, −Tet, top panel). To examine whether DUB-2 could prolong phosphorylation of other signaling intermediates we checked IL-2–mediated ERK-2 and AKT phosphorylation. ERK-2 phosphorylation was observed up to1 hour after cytokine withdrawal whether or not DUB-2 was expressed (Figure 3, third panel). The level of ERK-2 expression in both cells was comparable (Figure 3, fourth panel). No significant difference in AKT phosphorylation was observed between cells expressing and not expressing DUB-2 (Figure 3, fifth panel). Thus, DUB-2 expression can markedly inhibit dephosphorylation of STAT5, but not ERK-2 or AKT, following cytokine withdrawal, suggesting that DUB-2 may specifically enhance JAK/STAT signaling.
DUB-2 enhances expression of STAT5-dependent genes.
Ba/F3β/tTA DUB-2 cells were grown with or without tetracycline for 48 hours, stimulated with IL-2 for 2 hours, and incubated in cytokine-free medium for the indicated times. Cells lysates were immunoprecipitated with anti-STAT5b or ERK2, and immunoblotted with antiphosphotyrosine (pY), antiphospho-ERK (p-ERK), anti-STAT5b, or anti-ERK2 antisera, as indicated. Whole cell lysates were also run and immunoblotted with antisera for phospho-AKT (p-AKT), AKT, DUB-2, Cis, c-Myc, and Bcl-x, as indicated.
DUB-2 enhances expression of STAT5-dependent genes.
Ba/F3β/tTA DUB-2 cells were grown with or without tetracycline for 48 hours, stimulated with IL-2 for 2 hours, and incubated in cytokine-free medium for the indicated times. Cells lysates were immunoprecipitated with anti-STAT5b or ERK2, and immunoblotted with antiphosphotyrosine (pY), antiphospho-ERK (p-ERK), anti-STAT5b, or anti-ERK2 antisera, as indicated. Whole cell lysates were also run and immunoblotted with antisera for phospho-AKT (p-AKT), AKT, DUB-2, Cis, c-Myc, and Bcl-x, as indicated.
DUB-2 enhances the expression of STAT5-induced gene products
We next wished to examine whether the prolonged activation of STAT5 would result in extended expression of the IL-2–induced STAT5-dependent genes such as the cytokine-induced SH2-containing protein (CIS). CIS can be strongly induced by many cytokines that activate STAT5, including IL-2, and its expression had been reported to be dependent on STAT5.20 CIS expression was examined in the lysates described above by immunoblotting with an antibody specific for CIS (Figure 3, eighth panel). In the presence of DUB-2 CIS expression was markedly prolonged up to 8 hours following cytokine withdrawal, whereas it was not detected beyond 2 hours in the absence of DUB-2. We also observed markedly enhanced expression ofc-Myc, a gene also known to be activated by STAT5 (Figure 3, ninth panel).21 However, Bcl-x protein levels were not affected in cells expressing DUB-2 (Figure 3, tenth panel). Bcl-x was shown to be STAT5 regulated,22 but in these cells Bcl-x protein was expressed in unstimulated cells and its expression was not further enhanced by IL-2 stimulation. Taken together these findings suggest that DUB-2 expression can prolong the expression of STAT5-regulated genes in lymphocytes.
DUB-2 inhibits apoptosis following cytokine withdrawal
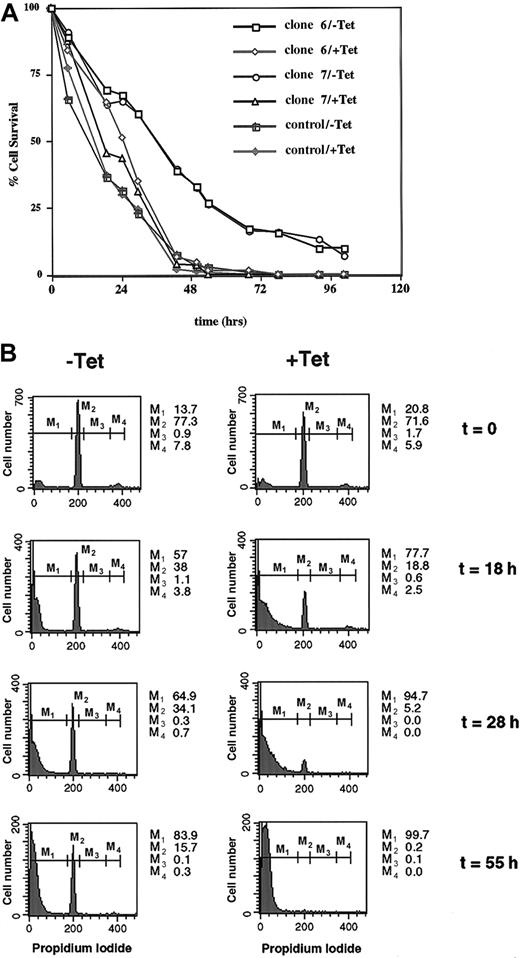
Although DUB-2 could prolong IL-2–mediated signaling, its expression in Ba/F3 cells did not enhance cytokine-mediated proliferation (results not shown). We therefore wondered whether DUB-2 might improve the survival of cells withdrawn from growth factor. Tetracycline-responsive cells were stimulated with IL-2 for 4 hours, withdrawn from cytokine, and survival-monitored using standard assays. In Figure 4A, cell death was monitored by trypan blue exclusion at different periods following removal from cytokine. Two days after cytokine withdrawal 100% loss of cell viability was evident in cells not expressing DUB-2. By contrast, at 2 days 35% of cells that expressed DUB-2 were resistant to cell death induced by cytokine withdrawal. Even 5 days after cytokine withdrawal 10% of the cells expressing DUB-2 still did not take up trypan blue (Figure 4A). This effect was evident in over 10 experiments in 3 separate DUB-2 clones and the data suggest that DUB-2 can strongly inhibit apoptosis induced by cytokine withdrawal.
DUB-2 prolongs survival following cytokine withdrawal.
(A) Ba/F3β/tTA DUB-2 (clones 6 and 7) were incubated with or without tetracycline for 48 hours, stimulated with IL-2 for 4 hours, and incubated in cytokine-free medium for the indicated times. Cell death was monitored by trypan blue exclusion assay, and data expressed as percentage of cells excluding trypan blue. (B) Ba/F3β/tTA DUB-2 cells (clone 6, +/−Tet) were stimulated with IL-2 for 4 hours, and withdrawn from cytokine as above. At the indicated times, cells were stained with PI and analyzed by flow cytometry. Apoptotic cells correspond to cells with sub-G1 DNA content (M1). Numbers indicate the percentage of cells in the different phases of the cell cycle (M2: G0/G1; M3: S; M4: G2/M).
DUB-2 prolongs survival following cytokine withdrawal.
(A) Ba/F3β/tTA DUB-2 (clones 6 and 7) were incubated with or without tetracycline for 48 hours, stimulated with IL-2 for 4 hours, and incubated in cytokine-free medium for the indicated times. Cell death was monitored by trypan blue exclusion assay, and data expressed as percentage of cells excluding trypan blue. (B) Ba/F3β/tTA DUB-2 cells (clone 6, +/−Tet) were stimulated with IL-2 for 4 hours, and withdrawn from cytokine as above. At the indicated times, cells were stained with PI and analyzed by flow cytometry. Apoptotic cells correspond to cells with sub-G1 DNA content (M1). Numbers indicate the percentage of cells in the different phases of the cell cycle (M2: G0/G1; M3: S; M4: G2/M).
Apoptosis was also monitored by PI staining and flow cytometry analysis (Figure 4B). Apoptotic cells correspond to the cell population localized in the sub-G1 peak (M1). Twenty-eight hours after IL-2 withdrawal 95% of Ba/F3β cells not expressing DUB-2 were in the sub-G1 peak (M1), whereas 35% of the cells expressing DUB-2 were still alive (M2+M3+M4). DUB-2 expression appeared to enhance the survival of these cells by keeping them in the G1 phase of the cell cycle (Figure 4B, M2). This is interesting because Zhu and coworkers11 observed that DUB-1 could block cell cycle progression by maintaining cells in G1. These results further support the idea that DUB-2 can significantly block the onset of apoptosis induced by cytokine withdrawal. Apoptosis was also measured by the TUNEL assay, which detects double-stranded DNA breaks that are a hallmark of apoptosis, and confirmed that DUB-2 strongly inhibited apoptosis induced by cytokine withdrawal (results not shown). Altogether these results suggest that DUB-2 can act not only by enhancing IL-2–mediated signaling, but also by inhibiting apoptosis in Ba/F3 cells expressing the IL-2 receptor.
Discussion
In the present study, we show that the deubiquitinating enzyme DUB-2 can enhance IL-2–mediated signaling and can serve to suppress apoptosis in lymphocytes following withdrawal of growth factor. We found that DUB-2 was constitutively expressed in HTLV-1–transformed T cells that exhibit STAT5 activation and growth factor independence. Cells that expressed DUB-2 had prolonged phosphorylation of STAT5 and enhanced expression of STAT5-dependent genes. It would be interesting to examine whether serine phosphorylation of STAT5 and tyrosine phosphorylation of STAT3 are affected by DUB-2 expression. Unfortunately, we did not detect STAT3 tyrosine phosphorylation in response to IL-2 in Ba/F3 cells expressing IL-2Rβ. These findings suggest that DUB-2, which is specifically regulated by IL-2 in T cells, may play an important role in controlling T- lymphocyte survival and perhaps contribute to T-cell transformation.
We clearly found that DUB-2 could prolong STAT5 phosphorylation induced by IL-2 and maintained STAT5 activation on cytokine withdrawal. However, we did not observe STAT5 constitutive activation in DUB-2–expressing cells without prior cytokine stimulation. This suggests that although the activation of STAT5 was prolonged in the presence of DUB-2, it required cytokine-induced receptor activation. In these experiments, no clear effect of DUB-2 on IL-2R phosphorylation was detected, suggesting that the receptor may not be a direct target for DUB-2. We have also observed enhanced IL-3–mediated STAT5 phosphorylation in cells expressing DUB-2, but only at times earlier than 2 hours following IL-3 treatment (data not shown). This IL-3 result is complicated by DUB-1, the DUB-2 homologue, which is known to be expressed in Ba/F3 cells 2 hours after IL-3 treatment.12 Because DUB-1 and DUB-2 are close homologues, this result is difficult to interpret, but perhaps both deubiquitinating enzymes can enhance STAT5 activation and therefore the expression of STAT5-dependent genes and hence improve survival in more physiologic concentrations of cytokine.
How DUB-2 enhances IL-2–mediated phosphorylation is unknown, but our data suggest that regulation of the JAK/STAT pathway likely involves the Ub system and several mechanisms could be proposed. First, DUB-2 may inhibit dephosphorylation by blocking the activity of a phosphatase and this would be supported by the observation that proteasome inhibitors prolong STAT5 phosphorylation (Callus and Mathey-Prevot,19 Verdier et al,20 and this study), and also by the observation that STAT5 phosphorylation is maintained on IL-2 withdrawal (Figure 3). Second, STAT phosphorylation is clearly controlled by phosphatases such as SHP-123 and other proteins such as SOCS3 and CIS,24-26 and it is possible that deubiquitinating enzymes regulate these STAT-inhibitory molecules. However, CIS expression is up-regulated in cells expressing DUB-2 (Figure 3), making it unlikely that DUB-2 prolongs Stat5 phosphorylation through this family of STAT inhibitors. Third, DUB-2 may inhibit STAT turnover. However, this seems unlikely because ubiquitination of STAT5 has not been detected, and because Wang and colleagues27 reported that proteasome inhibitors have no effect on STAT5 half-life. Fourth, receptor turnover may be affected by DUB-2. Interestingly, the γc component of the IL-2R can be cleaved in a calpain-dependent manner suggesting that the IL-2Rβ partner chain can be targeted for degradation by protease cleavage.28However, no PEST (single-letter amino acid codes) sequence has been identified in IL-2Rβ suggesting that this receptor chain must not be directly targeted for cleavage in a similar way. We show that dephosphorylation of the IL-2Rβ (and STAT5) is dependent on the proteasome pathway. However, we did not observe a clear effect of DUB-2 on receptor phosphorylation, suggesting that DUB-2 might not directly target IL-2R, but rather acts downstream by inhibiting STAT5 dephosphorylation. These findings suggest that the dephosphorylation of both the IL-2Rβ and STAT5 occurs by a proteasome-dependent mechanism, and that DUB-2 might counteract or inhibit STAT5 dephosphorylation and thus prolong signaling. Clearly, identification of DUB-2 targets will help elucidate the mechanism by which DUB-2 regulates the IL-2 signaling.
Whether the antiapoptotic effect observed with DUB-2 can be mimicked by other DUB family members is unclear. Zhu and coworkers11 reported that DUB-1 can force cells into the G1 phase of the cell cycle and hence has proposed that DUB-1 may function to inhibit cell proliferation. We have also found that DUB-2 can inhibit cell proliferation and, following IL-2 withdrawal, cells that express DUB-2 survive longer and accumulate in G1. Because DUB-1 is induced by IL-3 and GM-CSF and is expressed in a variety of hematopoietic cell types, it may be important for survival of other cell types such as hematopoietic stem cells and the myeloid lineage. It should be pointed out that a strong survival effect was also observed in DUB-2–expressing cells on IL-3 withdrawal (data not shown), which may be the result of not only DUB-2 but also DUB-1 expression. Our data support the idea that DUBs do not enhance proliferation, but rescue from programmed cell death, and may therefore be important in directing cell survival in vivo.
Many forms of leukocyte transformation are associated with activation of the JAK/STAT pathway and STAT5, in particular, contributes to transformation mediated by v-ABL29 and BCR/ABL.30 31 BCR/ABL-dependent STAT5 activation has been shown to be important for protection from apoptosis and for growth factor independence. Indeed an oncogenic mutation of STAT5A is known to induce growth factor independence in Ba/F3 cells. In our hands DUB-2 can prolong STAT5 phosphorylation but was not sufficient to induce proliferation because these cells did not multiply independent of growth factor, but we have found that some cells that express DUB-2 can become growth factor–independent (results not shown).
Human T-cell lymphotropic virus I is the etiologic agent for ATL and tropical spastic paraparesis (TSP/HAM [HTLV-1–associated myelopathy]).32,33 HTLV-1–transformed T-cell lines that proliferate independently of IL-2 have constitutively activated IL-2 signaling.16 The transcriptional transactivator Tax, encoded by HTLV-1, can enhance expression of genes such as IL2Rα chain and c-myc, and modulation of these genes is likely to play a role in the development of HTLV-1–associated disease.34 We had set out to examine which, if any, of the known IL-2–induced gene products would be expressed in ATL cells. Despite the active IL-2 signaling pathway, other gene products induced by IL-2 such as SOCS3 were not expressed. DUB-2 exhibited an altered expression profile being constitutively expressed in these HTLV-1–transformed T-cell lines, but because the expression of DUB-2 alone did not lead to factor-independent proliferation other signals from IL-2R must be required. This is not surprising because other IL-2–induced genes such as pim-1and c-myc can play an important role in lymphomagenesis,35 and because a cooperation between c-Myc, Pim-1 and Vcp have been implicated in cytokine- independent growth.36
Deubiquitinating enzymes some such as FAF (fat facets in Drosophila) can cleave Ub from specific targets regulating cellular pools of free Ub. Other enzymes remove Ub from cellular proteins and rescue from Ub-mediated degradation, whereas yet others accelerate proteasome-mediated degradation by removing Ub from degradation remnants.10,11 It will be important to explore whether deubiquitinating enzymes such as Fam, which can interact with and stabilize the oncogene β-catenin, can influence survival.37 Interestingly, a mutant form of the deubiquitinating enzyme tre-2 encodes an oncogene indicating a role for the Ub system in aberrant growth control. The mechanism underlying the ability of DUB-2 to inhibit programmed cell death is not understood, but because it can enhance survival, it may act along with other oncogenic mutations to induce transformation. Clearly transformation and survival are tightly linked phenomena and there are important proto-oncogenes, including Akt, Bcl-2, andWT1, that act by maintaining cell viability.36 38-40
In conclusion, the observation that DUB-2 is expressed in ATL cells and may prolong cell survival suggests that it may be important for transformation. Key remaining questions include the mechanisms responsible for the prolonged phosphorylation of STAT5 and the mechanism by which DUB-2 may inhibit cell death. Nonetheless, this is the first report that a deubiquitinating enzyme can enhance survival, and although the mechanism will require further investigation, the findings clearly show an important role for the Ub pathway in the regulation of cell survival.
We thank Dr Bruno Amati for critical comments and Maribel Andonian for graphics.
The DNAX Research Institute is fully supported by Schering-Plough.
T.-S.M., M.H., and A.R. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
James A. Johnston, Department of Immunology, Whitla Medical Building, Queen's University Belfast, 97 Lisburn Rd, Belfast BT9 7BL, Northern Ireland; e-mail: jim.johnston@qub.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal