Abstract
Antigenic peptides recognized by virus-specific cytotoxic T lymphocytes (CTLs) are useful tools for studying the CTL responses exclusively among those who own the major histocompatibility complex (MHC) class I molecules that present the peptides. For widening the application, an efficient strategy to determine such epitopes in the context of a given MHC is highly desirable. A rapid and efficient strategy is presented for the determination of CTL epitopes in the context of given MHC molecules of interest through multiple screenings consisting of a computer-assisted algorithm and MHC stabilization and enzyme-linked immunospot assays. A major cytomegalovirus (CMV)–specific CTL epitope, QYDPVAALF, in the amino acid sequence of its lower matrix 65 kd phosphoprotein (pp65) presented by HLA-A*2402 molecules was identified from 83 candidate peptides. The results indicate that the CMV-specific CTL response is highly focused to pp65 in the context of HLA-A*2402. Endogenous processing and presentation was confirmed using a peptide-specific CD8+ T-cell clone as the effectors and autologous fibroblast cells infected with recombinant vaccinia virus expressing pp65 gene or CMV as antigen-presenting cells. Flow cytometric analysis of intracellular interferon-γ production revealed 0.04% to 0.27% of CD8+ T cells in peripheral blood of HLA-A24+ and CMV-seropositive donors to be specific for the peptide. The tetrameric MHC-peptide complexes specifically bound to the reactive T-cell clone and 0.79% of CD8+ T cells in peripheral blood from a seropositive donor. The peptide could be a useful reagent to study CTL responses to CMV among populations positive for HLA-A*2402.
Introduction
Human cytomegalovirus (CMV) is a ubiquitous β-herpesvirus. Primary infection in healthy hosts is usually asymptomatic, and the virus persists in CD33+progenitors expressing markers of dendritic and myeloid lineage as major reservoirs of latent infection virus1,2 without any apparent clinical symptoms. However, in immunocompromised hosts, such as patients with advanced human immunodeficiency virus (HIV) infection and those who have undergone bone marrow transplantation (BMT), CMV is frequently reactivated and disease resulting from the progression of CMV infection is a major cause of infectious morbidity and mortality.3-5
CD8+ cytotoxic T lymphocytes (CTLs) are essential to the control of CMV infection. In patients undergoing allogeneic BMT, the delayed regeneration of CMV-specific CD8+ T lymphocytes following transplantation has been correlated with CMV disease.5,6 In addition, adoptive transfer of CMV-specific CD8+ T-cell clones has led to restoration of virus-specific immunity in BMT patients with relatively low frequencies of CMV-related disease.7,8 In addition, recent data indicate that there is an inverse correlation between the CMV-specific CTL response and CMV antigenemia after renal transplantation.9 This observation underlines the potential benefit of testing for CMV-specific CTL responses in immunocompromised patients in order to monitor and control CMV infection.
Recently, we have shown that monitoring of the Epstein-Barr virus (EBV) load and EBV-specific CD8+ T-cell frequencies, determined by multiparameter flow cytometric analysis to detect rapid accumulation of interferon γ (IFN-γ) in antigen-reacting CD8+ T cells, is useful for care of EBV-related post-BMT lymphoproliferation.10-12 Antigen-presenting cells for the detection of EBV-specific CD8+ T cells are easily obtained and propagated by transforming peripheral blood B lymphocytes with a laboratory strain of EBV. However, detection of CMV-specific CD8+ T cells using the same approach is practically hampered by difficulties in obtaining the dermal fibroblast cells necessary for presenting CMV antigens to relevant CTLs.
Virus-specific CD8+ T cells recognize short peptides—8 to 10 amino acids in most cases—presented by major histocompatibility complex (MHC) class I molecules. Identification of CMV-specific CD8+ T-cell epitopes has brought advantages for studies of many aspects of the T-cell response. These peptides can directly stimulate the T cells for rapid IFN-γ production13-16 or directly stain the T cells after being assembled into fluorescent-labeled tetrameric MHC–peptide complexes.17-21 Both techniques are sufficiently sensitive and rapid for monitoring the CMV-specific CD8+ T-cell immunity in the clinical setting. In addition, such peptides have the potential of eliciting CMV-specific CTL responses in vivo when used as vaccines preceding CMV challenge.22 Staining with tetramers carrying the antigenic peptides could make it possible to sort fluorescent-labeled CTLs from peripheral blood mononuclear cells (PBMCs), and the sorted T cells could be expanded in vitro for large-scale production of reagents for adoptive transfer treatment.23 However, the antigenic peptides need to be identified in the context of HLA, and application is always restricted to patients who possess the allele. For widening the application, an efficient strategy to determine CMV-specific CD8+ T-cell epitopes in the context of a given HLA is highly desirable.
We present here a rapid and efficient strategy for determination of CTL epitopes in the context of HLA-A*2402 molecules through multiple screenings consisting of a computer-assisted algorithm, an in vitro MHC stabilization assay, and an enzyme-linked immunospot (ELISPOT) assay. In many ethnic groups, HLA-A24 is one of the most common alleles24,25 and, especially in our country, HLA-A24 is the most frequently encountered HLA class I allele and the genotype is almost exclusively A*2402.26 We here introduce an amino acid sequence from the lower matrix 65-kd phosphoprotein (pp65) of CMV, QYDPVAALF, as one of the the major CMV-specific CTL epitopes presented by HLA-A*2402 molecules.
Materials and methods
Donors
The study design and purpose, which had been approved by the institutional review board of Aichi Cancer Center, were fully explained to all donors. Skin biopsies and peripheral blood were obtained after informed consent was confirmed. Standard serologic HLA typing was performed using peripheral blood for some donors.
Virus and cell lines
Dermal fibroblast lines were generated from skin biopsies obtained from the volunteers and propagated in Dulbecco modified Eagle medium (Gibco, Grand Island, NY) supplemented with 10% fetal calf serum (FCS), 2 mM L-glutamine, 50 U/mL penicillin, and 50 μg/mL streptomycin.
AD169 strain CMV (VR-538, American Type Culture Collection, Manassas, VA) was propagated in fibroblast cells infected at a multiplicity of infection 0.1. Whole infected cultures were harvested when a 100% cytopathic effect was evident. They were frozen, thawed once, and spun at 3000 rpm. Aliquots of the supernatant with a titer of 5 × 106 plaque-forming units per milliliter were stocked at −80°C until used.
The peptide transporter-negative B × T hybrid cell line 174 CEM.T2 (referred to as T2) was transfected with a plasmid expressing HLA-A*2402,27 a kind gift from Dr K. Itoh, Kurume University, Japan. The transfected cell line was cloned by limiting dilution and an A24+ clone screened by indirect immunofluorescence using an anti-A24 monoclonal antibody (mAb) (clone 0041HA, One Lambda, Canoga Park, CA), and antimouse fluorescein isothiocyanate (FITC)-labeled antibodies, named T2-A24, were further cultured in Iscoves modified Dulbecco medium (Gibco) supplemented with 2 mM L-glutamine, 50 U/mL penicillin, 50 μg/mL streptomycin, 5 × 10−5 M β-mercaptoethanol, 10% FCS, and 0.8 mg/mL G418 (Gibco).
Peptides
To identify the potential HLA-A24–binding peptides within CMV proteins (accession code X17403),28 a computer-based program was employed by access through the World Wide Web site BioInformatics and molecular analysis section (BIMAS) HLA peptide bonding predictions.29,30 Most peptides were synthesized with a Cleaved PepSet from Mimotope (Melbourne, Australia) dissolved in 100 μL dimethyl sulfoxide and further diluted in 40% acetonitrile, 0.1 M HEPES (pH 7.4), where necessary. The yield of each peptide was assumed to be 1 μM. HLA-A24–binding peptides RYLRDQQLL, derived from the HIV envelope protein,31 and TYGPVFMSL, derived from EBV latent membrane protein 2A,32 were synthesized (Sawady, Tokyo, Japan) and used as control peptides. The octamer QYDPVAAL of CMV pp65 used in the CTL assay was also synthesized (Sawady). Peptides used in this paper are listed in Table1.
Characters of synthetic oligopeptides and results with the MHC stabilization assay
| Peptide no. . | Amino acid sequence . | ORF name . | Character . | Start position . | Amino acid length . | Score* . | % MFI increase† . |
|---|---|---|---|---|---|---|---|
| 1 | RYTAESLRL | UL69 | IE | 259 | 9 | 400 | 227 |
| 2 | DYEPVPRKF | 33 | 9 | 277 | 141 | ||
| 3 | VYNAPNLHTL | 329 | 10 | 300 | 196 | ||
| 4 | AYCPFDEQSL | 517 | 10 | 288 | 92 | ||
| 5 | DYLLHIRQQF | 301 | 10 | 252 | 123 | ||
| 6 | TYRCGAVSDL | 477 | 10 | 200 | 58 | ||
| 7 | VYLFSLVVL | US3 | IE | 164 | 9 | 300 | 175 |
| 8 | VYDLANNTL | IRS1 | IE | 218 | 9 | 240 | 194 |
| 9 | RYDDESWRPL | 640 | 10 | 480 | 36 | ||
| 10 | FYQIRKPPWL | 598 | 10 | 300 | 217 | ||
| 11 | VYDLANNTL | TRS1 | IE | 218 | 9 | 240 | 136 |
| 12 | LYLALGAGF | 206 | 9 | 180 | 79 | ||
| 13 | RYTRPTEPPL | 631 | 10 | 400 | 17 | ||
| 14 | AYVCCQEYL | UL36 | Teg | 69 | 9 | 300 | 172 |
| 15 | LYDAITENL | 350 | 9 | 280 | 202 | ||
| 16 | CYHQLLGAL | 231 | 9 | 240 | 114 | ||
| 17 | LYRGDVESL | 156 | 9 | 200 | 155 | ||
| 18 | KYAESDYIF | UL37 | IE | 353 | 9 | 240 | 177 |
| 19 | KYAESDYIFL | 353 | 10 | 480 | 145 | ||
| 20 | SYRWIQRKRL | 21 | 10 | 200 | −20 | ||
| 21 | QYHLEGWFPL | 262 | 10 | 200 | 109 | ||
| 22 | QYILGADPL | UL123 | IE | 353 | 9 | 300 | 189 |
| 23 | KYTQTEEKF | 107 | 9 | 220 | 142 | ||
| 24 | AYAQKIFKIL | 248 | 10 | 336 | 152 | ||
| 25 | AYLGLSKKL | UL48 | Teg | 433 | 9 | 396 | 146 |
| 26 | DYLRFPTRL | 2026 | 9 | 300 | 104 | ||
| 27 | EYHGVYEHL | 563 | 9 | 280 | 144 | ||
| 28 | TYRDRDIDL | 223 | 9 | 240 | −21 | ||
| 29 | TYSTLDRAL | 470 | 9 | 240 | 165 | ||
| 30 | FYDLRDLKL | 662 | 9 | 220 | 121 | ||
| 31 | TYAQMVKKL | 880 | 9 | 220 | 212 | ||
| 32 | GYQKKVQQDL | 1016 | 10 | 504 | −29 | ||
| 33 | LYPEYIYTVL | 1753 | 10 | 504 | 161 | ||
| 34 | TYVLVTVNSL | 136 | 10 | 360 | 203 | ||
| 35 | YYEALFLYML | 197 | 10 | 300 | 215 | ||
| 36 | DYGNVAFKYL | 1609 | 10 | 240 | 47 | ||
| 37 | KYCDPNVLL | UL56 | Teg | 27 | 9 | 480 | 149 |
| 38 | LYLLLYRHL | 756 | 9 | 360 | 157 | ||
| 39 | TYNKEWPLL | 735 | 9 | 300 | 202 | ||
| 40 | IYELCNGPL | 617 | 9 | 300 | 133 | ||
| 41 | MYTKIKHIL | 397 | 9 | 280 | 155 | ||
| 42 | VYHQLCRAL | 320 | 9 | 240 | 148 | ||
| 43 | LYLEMLLKAL | 72 | 10 | 432 | 36 | ||
| 44 | LYNETFGKQL | 708 | 10 | 360 | 34 | ||
| 45 | SYIQEAEEAL | 269 | 10 | 300 | −9 | ||
| 46 | TYNKEWPLLL | 735 | 10 | 300 | 238 | ||
| 47 | AYIYTTYLL | UL55 | gB | 154 | 9 | 300 | 239 |
| 48 | SYENKTMQL | 205 | 9 | 300 | 116 | ||
| 49 | AYEYVDYLF | 624 | 9 | 210 | 225 | ||
| 50 | DYLFKRMIDL | 629 | 10 | 300 | −2.4 | ||
| 51 | IYNKPIAARF | 531 | 10 | 210 | 169 | ||
| 52 | SYAYIYTTYL | 152 | 10 | 200 | 228 | ||
| 53 | QYRSYKRSL | UL100 | gM | 174 | 9 | 200 | −16 |
| 54 | GYHLGAFFGL | 268 | 10 | 240 | 157 | ||
| 55 | YYHVVDFERL | 46 | 10 | 240 | 196 | ||
| 56 | KYESWLRPL | UL115 | gL | 92 | 9 | 600 | 103 |
| 57 | YYAGLPPEL | 277 | 9 | 264 | 196 | ||
| 58 | KYYAGLPPEL | 276 | 10 | 528 | 172 | ||
| 59 | LYNAVKEFCL | 253 | 10 | 300 | 185 | ||
| 60 | NYLDLSALL | UL75 | gH | 306 | 9 | 432 | 192 |
| 61 | SYVVTNQYL | 589 | 9 | 360 | 209 | ||
| 62 | SYLKDSDFL | 291 | 9 | 300 | 174 | ||
| 63 | TYALVSKDL | 125 | 9 | 280 | 178 | ||
| 64 | SYRSFSQQL | 135 | 9 | 240 | 82 | ||
| 65 | TYGRPIRFL | 45 | 9 | 200 | 169 | ||
| 66 | YYVFHMPRCL | 88 | 10 | 300 | 227 | ||
| 67 | MYMHDSDDVL | 668 | 10 | 300 | 133 | ||
| 68 | HYFTTLKQYL | UL18 | Env | 344 | 10 | 288 | 182 |
| 69 | EYTRLGVWL | US27 | Env | 31 | 9 | 240 | 91 |
| 70 | EYAKHHPKL | 89 | 9 | 220 | 33 | ||
| 71 | IYQLFEYTRL | 26 | 10 | 300 | 169 | ||
| 72 | QYLLDHNSL | US28 | Env | 91 | 9 | 360 | 161 |
| 73 | FYVAMFASL | 111 | 9 | 360 | 172 | ||
| 74 | LYVFVGTKF | 290 | 9 | 231 | 71 | ||
| 75 | RYRPVKQACL | 137 | 10 | 480 | 117 | ||
| 76 | SYPIILNVEL | 183 | 10 | 396 | 183 | ||
| 77 | TYNCTYNRL | UL4 | Env | 110 | 9 | 360 | 122 |
| 78 | HYRYEVANL | 183 | 10 | 200 | −22 | ||
| 79 | TYNRLTLLNL | 114 | 10 | 300 | 59 | ||
| 80 | VYALPLKML | UL83 | Teg | 100 | 9 | 200 | 151 |
| 81 | QYDPVAALF | 328 | 9 | 168 | 215 | ||
| 82 | QYVKVYLESF | 209 | 10 | 150 | 26 | ||
| 83 | QYDPVAALFF | 328 | 10 | 120 | 185 | ||
| 84 | RYLRDQQLL‡ | 584 | 9 | 720 | 141 | ||
| 85 | TYGPVFMSL1-153 | 287 | 9 | 403 | 229 |
| Peptide no. . | Amino acid sequence . | ORF name . | Character . | Start position . | Amino acid length . | Score* . | % MFI increase† . |
|---|---|---|---|---|---|---|---|
| 1 | RYTAESLRL | UL69 | IE | 259 | 9 | 400 | 227 |
| 2 | DYEPVPRKF | 33 | 9 | 277 | 141 | ||
| 3 | VYNAPNLHTL | 329 | 10 | 300 | 196 | ||
| 4 | AYCPFDEQSL | 517 | 10 | 288 | 92 | ||
| 5 | DYLLHIRQQF | 301 | 10 | 252 | 123 | ||
| 6 | TYRCGAVSDL | 477 | 10 | 200 | 58 | ||
| 7 | VYLFSLVVL | US3 | IE | 164 | 9 | 300 | 175 |
| 8 | VYDLANNTL | IRS1 | IE | 218 | 9 | 240 | 194 |
| 9 | RYDDESWRPL | 640 | 10 | 480 | 36 | ||
| 10 | FYQIRKPPWL | 598 | 10 | 300 | 217 | ||
| 11 | VYDLANNTL | TRS1 | IE | 218 | 9 | 240 | 136 |
| 12 | LYLALGAGF | 206 | 9 | 180 | 79 | ||
| 13 | RYTRPTEPPL | 631 | 10 | 400 | 17 | ||
| 14 | AYVCCQEYL | UL36 | Teg | 69 | 9 | 300 | 172 |
| 15 | LYDAITENL | 350 | 9 | 280 | 202 | ||
| 16 | CYHQLLGAL | 231 | 9 | 240 | 114 | ||
| 17 | LYRGDVESL | 156 | 9 | 200 | 155 | ||
| 18 | KYAESDYIF | UL37 | IE | 353 | 9 | 240 | 177 |
| 19 | KYAESDYIFL | 353 | 10 | 480 | 145 | ||
| 20 | SYRWIQRKRL | 21 | 10 | 200 | −20 | ||
| 21 | QYHLEGWFPL | 262 | 10 | 200 | 109 | ||
| 22 | QYILGADPL | UL123 | IE | 353 | 9 | 300 | 189 |
| 23 | KYTQTEEKF | 107 | 9 | 220 | 142 | ||
| 24 | AYAQKIFKIL | 248 | 10 | 336 | 152 | ||
| 25 | AYLGLSKKL | UL48 | Teg | 433 | 9 | 396 | 146 |
| 26 | DYLRFPTRL | 2026 | 9 | 300 | 104 | ||
| 27 | EYHGVYEHL | 563 | 9 | 280 | 144 | ||
| 28 | TYRDRDIDL | 223 | 9 | 240 | −21 | ||
| 29 | TYSTLDRAL | 470 | 9 | 240 | 165 | ||
| 30 | FYDLRDLKL | 662 | 9 | 220 | 121 | ||
| 31 | TYAQMVKKL | 880 | 9 | 220 | 212 | ||
| 32 | GYQKKVQQDL | 1016 | 10 | 504 | −29 | ||
| 33 | LYPEYIYTVL | 1753 | 10 | 504 | 161 | ||
| 34 | TYVLVTVNSL | 136 | 10 | 360 | 203 | ||
| 35 | YYEALFLYML | 197 | 10 | 300 | 215 | ||
| 36 | DYGNVAFKYL | 1609 | 10 | 240 | 47 | ||
| 37 | KYCDPNVLL | UL56 | Teg | 27 | 9 | 480 | 149 |
| 38 | LYLLLYRHL | 756 | 9 | 360 | 157 | ||
| 39 | TYNKEWPLL | 735 | 9 | 300 | 202 | ||
| 40 | IYELCNGPL | 617 | 9 | 300 | 133 | ||
| 41 | MYTKIKHIL | 397 | 9 | 280 | 155 | ||
| 42 | VYHQLCRAL | 320 | 9 | 240 | 148 | ||
| 43 | LYLEMLLKAL | 72 | 10 | 432 | 36 | ||
| 44 | LYNETFGKQL | 708 | 10 | 360 | 34 | ||
| 45 | SYIQEAEEAL | 269 | 10 | 300 | −9 | ||
| 46 | TYNKEWPLLL | 735 | 10 | 300 | 238 | ||
| 47 | AYIYTTYLL | UL55 | gB | 154 | 9 | 300 | 239 |
| 48 | SYENKTMQL | 205 | 9 | 300 | 116 | ||
| 49 | AYEYVDYLF | 624 | 9 | 210 | 225 | ||
| 50 | DYLFKRMIDL | 629 | 10 | 300 | −2.4 | ||
| 51 | IYNKPIAARF | 531 | 10 | 210 | 169 | ||
| 52 | SYAYIYTTYL | 152 | 10 | 200 | 228 | ||
| 53 | QYRSYKRSL | UL100 | gM | 174 | 9 | 200 | −16 |
| 54 | GYHLGAFFGL | 268 | 10 | 240 | 157 | ||
| 55 | YYHVVDFERL | 46 | 10 | 240 | 196 | ||
| 56 | KYESWLRPL | UL115 | gL | 92 | 9 | 600 | 103 |
| 57 | YYAGLPPEL | 277 | 9 | 264 | 196 | ||
| 58 | KYYAGLPPEL | 276 | 10 | 528 | 172 | ||
| 59 | LYNAVKEFCL | 253 | 10 | 300 | 185 | ||
| 60 | NYLDLSALL | UL75 | gH | 306 | 9 | 432 | 192 |
| 61 | SYVVTNQYL | 589 | 9 | 360 | 209 | ||
| 62 | SYLKDSDFL | 291 | 9 | 300 | 174 | ||
| 63 | TYALVSKDL | 125 | 9 | 280 | 178 | ||
| 64 | SYRSFSQQL | 135 | 9 | 240 | 82 | ||
| 65 | TYGRPIRFL | 45 | 9 | 200 | 169 | ||
| 66 | YYVFHMPRCL | 88 | 10 | 300 | 227 | ||
| 67 | MYMHDSDDVL | 668 | 10 | 300 | 133 | ||
| 68 | HYFTTLKQYL | UL18 | Env | 344 | 10 | 288 | 182 |
| 69 | EYTRLGVWL | US27 | Env | 31 | 9 | 240 | 91 |
| 70 | EYAKHHPKL | 89 | 9 | 220 | 33 | ||
| 71 | IYQLFEYTRL | 26 | 10 | 300 | 169 | ||
| 72 | QYLLDHNSL | US28 | Env | 91 | 9 | 360 | 161 |
| 73 | FYVAMFASL | 111 | 9 | 360 | 172 | ||
| 74 | LYVFVGTKF | 290 | 9 | 231 | 71 | ||
| 75 | RYRPVKQACL | 137 | 10 | 480 | 117 | ||
| 76 | SYPIILNVEL | 183 | 10 | 396 | 183 | ||
| 77 | TYNCTYNRL | UL4 | Env | 110 | 9 | 360 | 122 |
| 78 | HYRYEVANL | 183 | 10 | 200 | −22 | ||
| 79 | TYNRLTLLNL | 114 | 10 | 300 | 59 | ||
| 80 | VYALPLKML | UL83 | Teg | 100 | 9 | 200 | 151 |
| 81 | QYDPVAALF | 328 | 9 | 168 | 215 | ||
| 82 | QYVKVYLESF | 209 | 10 | 150 | 26 | ||
| 83 | QYDPVAALFF | 328 | 10 | 120 | 185 | ||
| 84 | RYLRDQQLL‡ | 584 | 9 | 720 | 141 | ||
| 85 | TYGPVFMSL1-153 | 287 | 9 | 403 | 229 |
ORF indicates open reading frame; IE, immediate early genes; Teg, (possible) tegument proteins; g, glycoproteins; Env, (possible) envelope proteins.
Estimated half-time of dissociation from the HLA-A24 allele (min).
Percent MFI increase of HLA-A*2402 molecules on T2-A24 cells. Percent MFI increase = (MFI with sample peptide − MFI without peptide)/MFI without peptide) × 100. See the MHC stabilization assay in “Materials and methods.”
Derived from the HIV envelope protein.31
Derived from the EBV latent membrane protein 2A.32
MHC stabilization assay
The synthesized peptides were used in an MHC stabilization assay using T2-A24 cells as described earlier.33 Briefly, T2-A24 cells (2 × 105) were incubated with 200 μL RPMI 1640 (Sigma, St Louis, MO) containing 0.1% FCS, 5 × 10−5 M β-mercaptoethanol, and each of the peptides at a concentration of 10 μM at 26°C for 16 hours, followed by incubation at 37°C for 3 hours. After the incubation, surface HLA-A24 molecules were stained with the anti-A24 mAb and antimouse FITC-labeled antibodies. Expression was measured by FACScan (Becton Dickinson, San Jose, CA), and mean fluorescence intensity (MFI) was recorded. Percent MFI increase was calculated as follows: Percent MFI increase = (MFI with the given peptide − MFI without peptide)/(MFI without peptide) × 100.
Generation of polyclonal and clonal CMV-specific CTLs
CMV-specific polyclonal CTLs were generated as described earlier.34 35 Briefly, heparinized peripheral blood was obtained by venipuncture from a CMV-seropositive volunteer, and PBMCs were isolated by density gradient centrifugation over Ficoll-Hypaque (Pharmacia Biotech, Uppsala, Sweden). The PBMCs (1 × 107) were cultured with 5 × 105autologous fibroblast cells infected 2 hours previously with the AD169 strain of CMV at a multiplicity of infection of 5 in individual wells of a 6-well plate in a final volume of 6 mL RPMI 1640 supplemented with 2 mM L-glutamine, 50 U/mL penicillin, 50 μg/mL streptomycin, 5 × 10−5 M β-mercaptoethanol, and 10% FCS (referred to as culture medium). After 7 days, the cultures were restimulated with both autologous CMV-infected fibroblast cells and γ-irradiated (33 Gy) allogeneic PBMCs, and they were then supplemented with recombinant IL-2 (20 U/mL) 2 days thereafter. The CTL line was further stimulated weekly with autologous CMV-infected fibroblast cells, γ-irradiated allogeneic PBMCs, and IL-2.
To generate CMV-specific CTL clones, CD8+ T cells were enriched with the aid of magnetic beads (Dynal, Oslo, Norway) and plated in wells of 96-well round bottom plates at 0.3 and 1 cell per well with 5 × 104 γ-irradiated allogeneic PBMCs (33 Gy), 1 × 104 γ-irradiated EBV-transformed B-lymphoblastoid cells (55 Gy), and anti-CD3 mAb (Ortho Diagnostics, Raritan, NJ) at a concentration of 30 ng/mL.8 36 The next day, IL-2 was added to a final concentration of 50 U/mL. Fourteen days after the stimulation, T cells in positive wells were tested by ELISPOT assay for their specificity. The cloning efficiency was less than 5% in all plates.
ELISPOT assay
Flat-bottom, 96-well MultiScreen-HA plates with a nitrocellulose base (Millipore, Millipore Corporation, Bedford, MA) were coated with 10 μg/mL anti–IFN-γ mAb (R & D Systems, Minneapolis, MN) and incubated overnight at 4°C. After washing with phosphate-buffered saline (PBS), plates were blocked with the culture medium for 1 hour at 37°C. T2-A24 (5 × 104) cells were pulsed with each peptide in 100 μL RPMI 1640 with 0.1% FCS and 5 × 10−5 M β-mercaptoethanol in each well of the plates for 30 minutes at room temperature. A total of 10 000 polyclonal CD8+ T cells suspended in culture medium supplemented with additional FCS to 20% and 20 U/mL IL-2 were seeded in each well. When the spots were too many to count, 1000 CD8+ T cells were used as responder cells, and the numbers of spots were multiplied by 10 (Figure1). For testing the specificity of CMV-specific T-cell clones, indicated numbers of the T cells in figures were used as responders. The peptide concentrations described in the text and figures indicate those in the final assay volume. In some experiments, HLA-A24+ fibroblast cells were infected 2 hours previously with CMV AD169 strain, recombinant vaccinia virus expressing glycoprotein B, or pp65 of CMV (kind gifts from Dr S. R. Riddell, Fred Hutchinson Cancer Research Center, Seattle, WA) at a multiplicity of infection of 5 and used as antigen-presenting cells (10 000 cells/well). All assays were performed in duplicate. The plates were incubated in a 5% CO2 incubator at 37°C for 20 hours and extensively washed with PBS containing 0.05% Tween 20. A polyclonal rabbit anti–IFN-γ antibody (Genzyme, Cambridge, MA) was added to individual wells and left for 90 minutes at room temperature, followed by exposure to peroxidase-conjugated goat antirabbit immunoglobulin G (Genzyme) for an additional 90 minutes. For visualization of IFN-γ–specific spots, 100 μL of 0.1 M sodium acetate buffer (pH 5.0) containing 3-amino-9-ethylcarbazole (Sigma) and 0.015% H2O2 was added to each well. After 40 minutes, the reaction was stopped by washing with water and the plates were dried. Diffuse large spots were counted under a dissecting microscope.
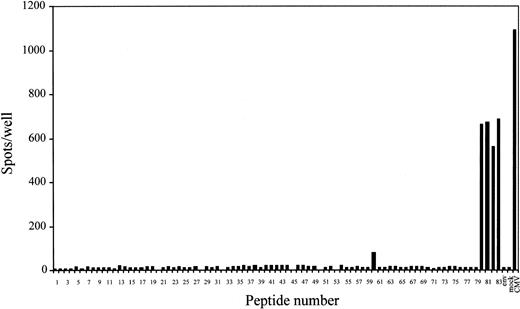
Screening of HLA-A*2402–binding peptides from amino acid sequences of CMV proteins for CD8+ T-cell stimulation by ELISPOT assay.
A total of 10 000 polyclonal CMV-specific CD8+ T cells established from PBMCs of a CMV-seropositive and HLA-A24+donor were cocultured with 50 000 T2-A24 cells in each well in the presence of each peptide at a concentration of 10 μM. When the spots were too many to count, 1000 CD8+ T cells were used as responder cells and the numbers of spots were shown after being multiplied by 10. Only peptides that had been shown to bind to HLA-A*2402 molecules by MHC stabilization assay were tested by ELISPOT. An HLA-A24–binding peptide, RYLRDQQLL, derived from the HIV envelope protein was used as control peptide (env). In some wells, autologous fibroblast cells that had been infected 2 hours previously with CMV AD169 strain (CMV) or mock-infected (mock) were used as antigen-presenting cells (10 000 cells/well). Each bar represents the average number of spots in duplicate wells.
Screening of HLA-A*2402–binding peptides from amino acid sequences of CMV proteins for CD8+ T-cell stimulation by ELISPOT assay.
A total of 10 000 polyclonal CMV-specific CD8+ T cells established from PBMCs of a CMV-seropositive and HLA-A24+donor were cocultured with 50 000 T2-A24 cells in each well in the presence of each peptide at a concentration of 10 μM. When the spots were too many to count, 1000 CD8+ T cells were used as responder cells and the numbers of spots were shown after being multiplied by 10. Only peptides that had been shown to bind to HLA-A*2402 molecules by MHC stabilization assay were tested by ELISPOT. An HLA-A24–binding peptide, RYLRDQQLL, derived from the HIV envelope protein was used as control peptide (env). In some wells, autologous fibroblast cells that had been infected 2 hours previously with CMV AD169 strain (CMV) or mock-infected (mock) were used as antigen-presenting cells (10 000 cells/well). Each bar represents the average number of spots in duplicate wells.
CTL assay
CTL assays were performed employing 51Cr release as previously described.10 11 Briefly,51Cr-labeled T2 or T2-A24 cells (2000 cells/well) were pulsed with each peptide in 100 μL RPMI 1640 with 0.1% FCS and 5 × 10−5 M β-mercaptoethanol in each well of a V-bottom 96-well plate for 30 minutes at room temperature. Desired numbers of CTLs suspended in 100 μL of the culture medium supplemented with additional FCS to 20% were seeded in each well. The peptide concentrations described in the text and figures indicate those in the final assay volume. Each assay was performed in triplicate. After 5 hours of incubation, the supernatants were harvested and radioactivity counted with a γ-counter. Percent specific lysis was calculated as follows: Percent specific lysis = (experimental lysis − minimum lysis) × 100/(maximum lysis − minimum lysis). Minimum lysis was obtained by incubating the target cells with the culture medium alone. Maximum lysis was obtained by exposing the target cells to 1% Nonidet P-40.
Detection of IFN-γ–producing CD8+ T cells in response to the peptide by flow cytometry
Peptide-specific CD8+ T-cell frequencies were measured as previously described with slight modifications.11 13 Briefly, 4 × 106 PBMCs were incubated with 10 μM peptides in 1 mL culture medium supplemented with 20 U/mL IL-2 in a culture tube at 37°C for 6 hours in the presence of brefeldin A (Sigma) during the last 5 hours. After the incubation, the cell suspensions were fixed with 4% paraformaldehyde in PBS for 10 minutes at room temperature. After washing with PBS, cells were permeabilized with IC Perm (BioSource International, Camarillo, CA) and stained with phycoerythrin-cyanin-5.1–labeled anti-CD8, phycoerythrin-labeled anti-CD69 (Coulter, Miami, FL) and FITC-labeled antihuman IFN-γ (Coulter) mAbs. Stained cells were analyzed by FACScan using Lysis II software. Live gating of the CD8+ subset was performed, and 50 000 events were acquired for each analysis.
Tetramer production and staining
MHC-peptide tetramers were produced as described previously.17-21 23 Briefly, BL21(DE3) pLysS (Novagen, Madison, WI) competent cells were transformed with pET11d plasmid (Novagen) encoding HLA-A*2402 heavy chain (a kind gift from Dr H. Takasu, Sumitomo Pharmaceutical, Osaka, Japan) or pET-3a plasmid (Novagen) encoding β2-microglobulin (a kind gift from Dr H. Takizawa, Otsuka Pharmaceutical, Tokushima, Japan) to produce the recombinant proteins. Expression of the HLA heavy chain was limited to the extracellular domain, and C-terminus of their domain was modified by the addition of a substrate sequence for the biotinylating enzyme BirA. Monomeric HLA-peptide complexes were folded in vitro by adding the HLA protein to β2-microglobulin in the presence of the 9-mer, QYDPVAALF. Proteins were dialyzed against water and then concentrated. After purification on gel filtration, the MHC complex was biotinylated by using recombinant BirA enzyme (Avidity, Denver, CO) and then purified by gel filtration. HLA-peptide tetramers were made by mixing the biotinylated MHC with phycoerythrin-labeled streptavidin (Molecular Probes, Eugene, OR) at a molar ratio of 4:1. Tetramers were purified by gel filtration on a Superdex 200 HR 10/30 (Amersham Pharmacia Biotech, Uppsala, Sweden) and concentrated to 3 mg/mL of total proteins and stored at 4°C until use. PBMCs (2 × 106) or CTL clones (2 × 105) were stained with the tetramer at a concentration of 0.1 mg/mL tetramers and Tricolor anti-CD8 mAb (Caltag, Burlingame, CA) at 37°C for 15 minutes. After washing twice, stained cells were fixed with 0.5% paraformaldehyde before analysis with FACScan.
Results
Selection of potential HLA-A24–binding peptides within CMV proteins
To identify potential HLA-A24–binding peptides within amino acid sequences of immediate early (IE), tegument and envelope proteins of CMV were analyzed with a computer program designed to predict HLA-binding peptides, based on estimation of the half-time dissociation of the HLA-peptide complex by access through the World Wide Web site BioInformatics and molecular analysis section (BIMAS) HLA peptide bonding predictions.29 A total of 83 peptides with estimated half-time dissociation scores above 100 were selected (Table1). All the peptides share the HLA-A24–binding motifs as tyrosine at the second residue and phenylalanine or leucine at the ninth or tenth residue.37-39 Next, MHC stabilization assays were performed to test these peptides for HLA-A*2402 binding efficiency using T2-A24 cells. Most peptides increased the HLA-A24 expression on the cells, indicating that these peptides bound and stabilized the HLA complex on the cell surface, but peptides no. 20, 28, 32, 45, 50, and 53 gave negative values for the percent MFI (Table 1) and were excluded from further studies.
Screening of peptides antigenic for an anti-CMV polyclonal CTL line by ELISPOT assay
To identify peptides recognized by CMV-specific CTLs in the context of HLA-A*2402 molecules, the ELISPOT assay was performed using T2-A24 cells as antigen-presenting cells. We chose the assay for the screening because it can test many peptides simultaneously in a few plates using relatively small numbers of CTLs as responders. Moreover, because the sensitive nature of the assay enables 40 spots to be scored as positive, subdominant epitopes could be identified when T cells recognizing such epitopes were present at concentrations no less than 40 in 10 000 cells. As shown in Figure 1, the polyclonal CMV-specific CD8+ T cells established from PBMCs of a CMV-seropositive and HLA-A24+ donor produced significant numbers of IFN-γ spots when incubated with T2-A24 cells pulsed with CMV peptides nos. 60, 80, 81, 82, and 83. Peptide no. 60 was derived from glycoprotein H, and others were from pp65 (open reading frame name UL83). The responders produced negligible spots when incubated with mock-infected autologous fibroblast cells, while more than 1000 spots were detected with CMV-infected autologous fibroblast cells. To ascertain the nature of the peptides recognized by the CMV-specific CD8+ T-cell line, we performed ELISPOT assays with serial dilutions of the 4 peptides derived from pp65. Figure 2shows that peptide no. 81 was still recognized at a concentration of 1 pM. The end points for recognition with the others were 10 pM for peptide no. 83 and 10 nM for peptides no. 80 and 82. The 2 peptides, no. 81 and 83, that were recognized at picomolar levels share the amino acid sequence QYDPVAALF, with an additional C-terminal phenylalanine for no. 83.
Results of ELISPOT assays using serial dilutions of the 4 peptides derived from pp65.
The 2 peptides, no. 81 (●) and 83 (▪), that were recognized at picomolar levels share the amino acid sequence QYDPVAALF, with an additional C-terminal phenylalanine for no. 83. The amino acid sequences of peptides no. 80 (♦) and 82 (▴) are VYALPLKML and QYVKVYLESF, respectively. A total of 1000 polyclonal CMV-specific CD8+ T cells established from PBMCs of a CMV-seropositive and HLA-A24+ donor were cocultured with 50 000 T2-A24 cells in each well in the presence of each peptide at the indicated concentrations.
Results of ELISPOT assays using serial dilutions of the 4 peptides derived from pp65.
The 2 peptides, no. 81 (●) and 83 (▪), that were recognized at picomolar levels share the amino acid sequence QYDPVAALF, with an additional C-terminal phenylalanine for no. 83. The amino acid sequences of peptides no. 80 (♦) and 82 (▴) are VYALPLKML and QYVKVYLESF, respectively. A total of 1000 polyclonal CMV-specific CD8+ T cells established from PBMCs of a CMV-seropositive and HLA-A24+ donor were cocultured with 50 000 T2-A24 cells in each well in the presence of each peptide at the indicated concentrations.
Recognition of the 4 pp65 peptides by single CMV-specific CD8+ T-cell clones
Notably, the sum of the number of T cells that reacted with each of the 5 peptides exceeded that reacting with CMV-infected fibroblast cells (Figure 1). This could be due to either weaker antigen presentation on the CMV-infected fibroblast cells, possibly by the mechanism of class I molecule down-regulation,40-44 or the same T-cell repertoires being reactive with some of the 5 peptides. To test these possibilities, we established T-cell clones from limiting dilution culture of the bulk CTL line stimulated with anti-CD3 mAb, appropriate feeder cells, and IL-2. The results of ELISPOT assays with 2 representative T-cell clones are shown in Figure3. Both clones recognized all 4 peptides at high concentrations. The effect of peptide dilution was the same with bulk CD8 T cells and with the T-cell clones (Figures 2 and 3). Among a total of 54 clones established, 28 clones were reactive for all 4 peptides but not for peptide no. 60—at a concentration of 10 μM—and effects of dilution on recognition were similar for all the clones tested (data not shown). Thus, we conclude that the spots in the wells containing each of the 4 pp65 peptides were produced by the same T-cell repertoires.
Recognition of the 4 peptides derived from pp65 by each CMV-specific CD8+ T-cell clone.
The results of ELISPOT assay with CMV-specific CD8+ T-cell clone no. 1 (A) and no. 3 (B) are shown. A total of 100 CD8+ T cells were cocultured with 50 000 T2-A24 cells in each well in the presence of each peptide at the indicated concentrations. The 2 peptides, no. 81 (●) and 83 (▪), that were recognized at picomolar levels share the amino acid sequence QYDPVAALF, with an additional C-terminal phenylalanine for no. 83. The amino acid sequences of peptides no. 80 (♦) and 82 (▴) are VYALPLKML and QYVKVYLESF, respectively.
Recognition of the 4 peptides derived from pp65 by each CMV-specific CD8+ T-cell clone.
The results of ELISPOT assay with CMV-specific CD8+ T-cell clone no. 1 (A) and no. 3 (B) are shown. A total of 100 CD8+ T cells were cocultured with 50 000 T2-A24 cells in each well in the presence of each peptide at the indicated concentrations. The 2 peptides, no. 81 (●) and 83 (▪), that were recognized at picomolar levels share the amino acid sequence QYDPVAALF, with an additional C-terminal phenylalanine for no. 83. The amino acid sequences of peptides no. 80 (♦) and 82 (▴) are VYALPLKML and QYVKVYLESF, respectively.
Recognition of the nonamer QYDPVAALF at a lower concentration than the octamer or the decamer
Because no. 81 and 83, which were recognized at picomolar levels, share the amino acid sequence QYDPVAALF and amino acid sequences of the peptides no. 80 (VYALPLKML) and 82 (QYVKVYLESF) were irrelevant, QYDPVAALF should be the core sequence of the T-cell epitope. Because class I molecules usually accommodate 8 to 10 amino acids in their antigen-binding sites, the octamer QYDPVAAL was simultaneously tested in CTL assays for its recognition by CTL clone no. 1. As shown in Figure 4, it far less effectively sensitized T2-A24 cells for CTL-mediated cell lysis than the nonamer or the decamer. We did not make and test the octamer YDPVAALF because tyrosine at the second residue is essential for binding to HLA-A24 molecules.37-39 Again, only the nonamer QYDPVAALF (peptide no. 81) could sensitize the target cells for CTL-mediated cell lysis at the concentration of 1 pM. In combination with the results of ELISPOT assays (Figures 2 and 3), this indicates that the nonamer QYDPVAALF is the minimal CTL epitope.
CMV-specific CD8+ CTL clone no. 1 recognition.
Clone no 1. recognizes the nonamer QYDPVAALF (●, peptide no. 81) and the decamer QYDPVAALFF (▪, peptide no. 83) at lower concentrations than octamer QYDPVAAL (▴). ■ indicates TYGPVFMSL derived from EBV latent membrane protein 2A. CTL assays were performed using 51Cr-labeled T2-A24 cells as targets at an effector-target ratio of 5:1 in the presence of each peptide at the indicated concentrations.
CMV-specific CD8+ CTL clone no. 1 recognition.
Clone no 1. recognizes the nonamer QYDPVAALF (●, peptide no. 81) and the decamer QYDPVAALFF (▪, peptide no. 83) at lower concentrations than octamer QYDPVAAL (▴). ■ indicates TYGPVFMSL derived from EBV latent membrane protein 2A. CTL assays were performed using 51Cr-labeled T2-A24 cells as targets at an effector-target ratio of 5:1 in the presence of each peptide at the indicated concentrations.
Recognition by the QYDPVAALF-specific CD8+ T-cell clone of the epitope in the context of HLA-A*2402 molecules
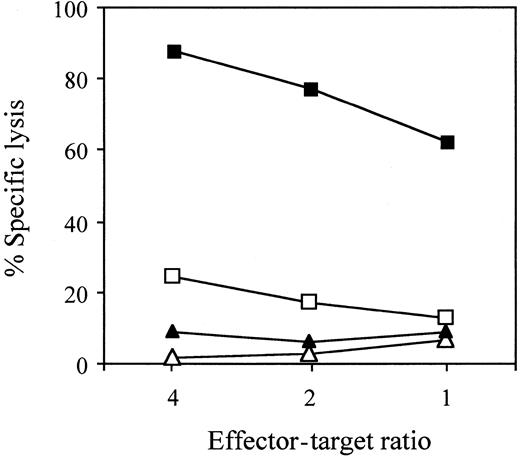
To confirm the HLA restriction, CTL assays were performed using51Cr-labeled T2 or T2-A24 cells pulsed with QYDPVAALF or the control peptide TYGPVFMSL, derived from EBV latent membrane protein 2A (Figure 5). Only T2-A24 cells pulsed with QYDPVAALF were effectively lysed by the CTL clone no. 1. The results indicate that the epitope QYDPVAALF is presented by HLA-A*2402 molecules.
Recognition by the QYDPVAALF-specific CD8+T-cell clone of the epitope in the context of the HLA-A*2402 molecules.
CTL assays were performed using 51Cr-labeled T2-A24 cells pulsed with QYDPVAALF (▪) or control EBV peptide TYGPVFMSL (▴) and using T2 cells pulsed with QYDPVAALF (■) or with the control peptide (▵) as target cells, incubated with CMV-specific CD8+ CTL clone no. 1 at the indicated effector-target ratios in the presence of 100 nM of each peptide.
Recognition by the QYDPVAALF-specific CD8+T-cell clone of the epitope in the context of the HLA-A*2402 molecules.
CTL assays were performed using 51Cr-labeled T2-A24 cells pulsed with QYDPVAALF (▪) or control EBV peptide TYGPVFMSL (▴) and using T2 cells pulsed with QYDPVAALF (■) or with the control peptide (▵) as target cells, incubated with CMV-specific CD8+ CTL clone no. 1 at the indicated effector-target ratios in the presence of 100 nM of each peptide.
Recognition by the QYDPVAALF-specific CD8+ T-cell clone of A*2402+ fibroblast cells infected either CMV or recombinant vaccinia virus expressing pp65
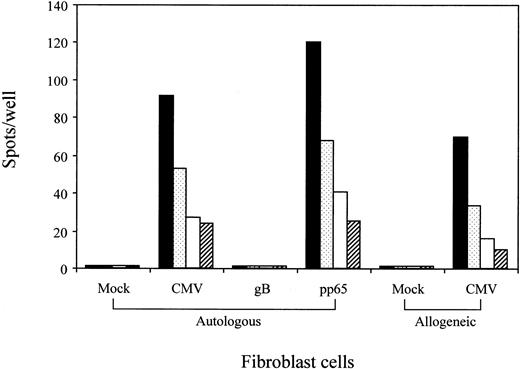
A set of experiments was performed to confirm that the epitope QYDPVAALF is processed within the cells infected with CMV. For that purpose, autologous and allogeneic HLA-A24+ fibroblast cells were mock-infected or CMV-infected and used as antigen-presenting cells for ELISPOT assays using the CTL clone no. 1 as responders. Figure 6 demonstrates that the CMV-infected cells stimulated for IFN-γ production by the T cells. The T cells were cytolytic for the CMV-infected fibroblast cells (data not shown). To confirm that endogenously produced pp65 proteins were processed and presented to the QYDPVAALF-specific CD8+ T cells, autologous fibroblast cells were infected with recombinant vaccinia virus expressing pp65 or glycoprotein B of CMV. The results clearly showed only fibroblast cells infected with recombinant vaccinia virus expressing pp65 to stimulate the T cells for IFN-γ production. The data indicated that pp65 expressed within the cells is endogenously processed to yield the epitope.
Recognition by the QYDPVAALF-specific CD8+ T-cell clone of A24+ fibroblast cells infected either with CMV or recombinant vaccinia virus expressing pp65.
HLA-A24+ autologous and allogeneic fibroblast cells were mock- or CMV-infected and tested for stimulation of QYDPVAALF-specific CD8+ T-cell clone no. 1 in the ELISPOT assay. Autologous fibroblast cells infected with recombinant vaccinia virus expressing pp65 or glycoprotein B (gB) of CMV were also tested for stimulation of the T cells. A total of 200 (▪), 100 ( ), 50 (■), or 25 (▨) CD8+ T cells were cocultured with 10 000 fibroblast cells in each well. Each bar represents the average number of spots in duplicate wells.
), 50 (■), or 25 (▨) CD8+ T cells were cocultured with 10 000 fibroblast cells in each well. Each bar represents the average number of spots in duplicate wells.
Recognition by the QYDPVAALF-specific CD8+ T-cell clone of A24+ fibroblast cells infected either with CMV or recombinant vaccinia virus expressing pp65.
HLA-A24+ autologous and allogeneic fibroblast cells were mock- or CMV-infected and tested for stimulation of QYDPVAALF-specific CD8+ T-cell clone no. 1 in the ELISPOT assay. Autologous fibroblast cells infected with recombinant vaccinia virus expressing pp65 or glycoprotein B (gB) of CMV were also tested for stimulation of the T cells. A total of 200 (▪), 100 ( ), 50 (■), or 25 (▨) CD8+ T cells were cocultured with 10 000 fibroblast cells in each well. Each bar represents the average number of spots in duplicate wells.
), 50 (■), or 25 (▨) CD8+ T cells were cocultured with 10 000 fibroblast cells in each well. Each bar represents the average number of spots in duplicate wells.
Frequencies of QYDPVAALF-specific CD8+ T cells in PBMCs of healthy CMV-seropositive donors
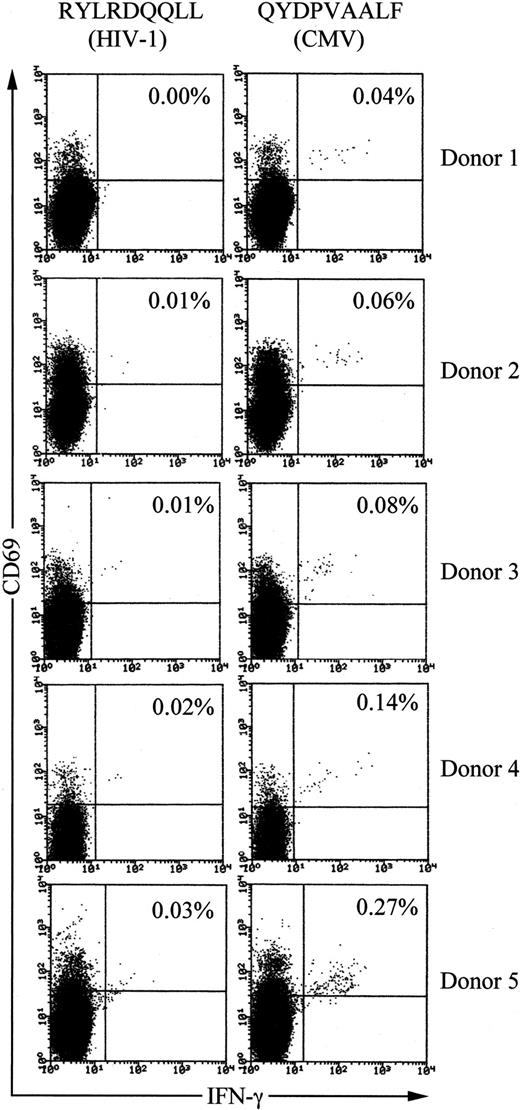
We examined QYDPVAALF-specific CD8+ T-cell frequencies in PBMCs of healthy HLA-A24+, CMV-seropositive, and HIV-seronegative donors using tricolor flow cytometry analysis for detection of antigen-specific IFN-γ–producing T cells.10,13 First, the phycoerythrin-cyanin-5.1–labeled anti-CD8 mAb was used for gating the population. Second, the phycoerythrin-labeled anti-CD69 mAb was employed for enhancement of precise detection of responding T cells. CD69 is up-regulated on activated T cells prior to cytokine production and thus allows more definitive clustering of the true responding fraction.10,13 As a control peptide, we used HLA-A24–binding peptide RYLRDQQLL, derived from the HIV envelope protein.31
When PBMCs from long-term healthy HLA-A24+ and CMV-seropositive individuals (n = 5) were stimulated with peptide QYDPVAALF, 0.04% to 0.27% of total CD8+ T cells produced IFN-γ (Figure 7). The values always exceeded those with control peptide, demonstrating specific reactivity to the CMV peptide. Frequencies of IFN-γ–producing CD8+T cells in PBMCs from CMV-seronegative and HLA-A24+individuals (n = 5) after stimulation with the peptide were below 0.01% (data not shown). These data demonstrate significant populations of CD8+ T cells from long-term healthy HLA-A24+and CMV-seropositive individuals to be specific to the peptide QYDPVAALF.
Frequencies of QYDPVAALF-specific CD8+ T cells in PBMCs of healthy HLA-A24+ and CMV-seropositive donors.
PBMCs were stimulated with either the CMV peptide QYDPVAALF or the control peptide RYLRDQQLL, derived from the HIV envelope protein, for IFN-γ production. After fixation and permeabilization, the cells were stained for CD8, CD69, and IFN-γ. The CD8+ subset was gated and analyzed by flow cytometry. The frequency of IFN-γ–producing cells is shown as the percentage of the total CD8+ T cells. The donors were all seronegative for HIV.
Frequencies of QYDPVAALF-specific CD8+ T cells in PBMCs of healthy HLA-A24+ and CMV-seropositive donors.
PBMCs were stimulated with either the CMV peptide QYDPVAALF or the control peptide RYLRDQQLL, derived from the HIV envelope protein, for IFN-γ production. After fixation and permeabilization, the cells were stained for CD8, CD69, and IFN-γ. The CD8+ subset was gated and analyzed by flow cytometry. The frequency of IFN-γ–producing cells is shown as the percentage of the total CD8+ T cells. The donors were all seronegative for HIV.
Tetramer staining
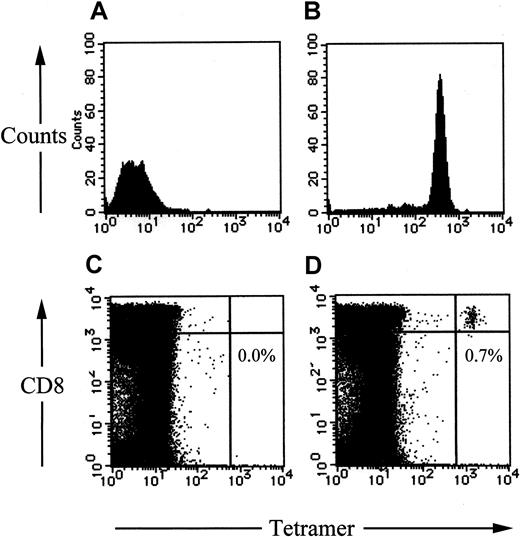
Finally, we made fluorescent-labeled tetrameric MHC-peptide complexes (tetramer) using the nonamer peptide QYDPVAALF. The tetramers specifically bound to the peptide-reactive T-cell clone but not the unreactive clone (Figure 8A,B). QYDPVAALF-specific CD8+ T cells detected by the tetramers were 0.79% in peripheral blood from the donor 5 in Figure 7and 0.0% in that from an HLA-A24+ and CMV-seronegative donor (Figure 8C,D).
HLA-A*2402–QYDPVAALF tetramers specifically bound to the relevant CD8+ T cells.
An HLA-A*2402–restricted EBV-specific CD8+ CTL clone (A), the HLA-A*2402–restricted QYDPVAALF-specific CD8+ CTL clone no. 1 (B), peripheral lymphocytes from a CMV-seronegative and HLA-A*2402+ donor (C), and those from the donor 5 in Figure 7 (D) were stained phycoerythrin-conjugated HLA-A*2402–QYDPVAALF tetrameric complex and an antibody to CD8 conjugated to Tricolor. The cells were analyzed by flow cytometry. The frequency of QYDPVAALF-specific CD8+ T cells in peripheral lymphocytes is shown as the percentage of the total CD8+ T cells (C,D).
HLA-A*2402–QYDPVAALF tetramers specifically bound to the relevant CD8+ T cells.
An HLA-A*2402–restricted EBV-specific CD8+ CTL clone (A), the HLA-A*2402–restricted QYDPVAALF-specific CD8+ CTL clone no. 1 (B), peripheral lymphocytes from a CMV-seronegative and HLA-A*2402+ donor (C), and those from the donor 5 in Figure 7 (D) were stained phycoerythrin-conjugated HLA-A*2402–QYDPVAALF tetrameric complex and an antibody to CD8 conjugated to Tricolor. The cells were analyzed by flow cytometry. The frequency of QYDPVAALF-specific CD8+ T cells in peripheral lymphocytes is shown as the percentage of the total CD8+ T cells (C,D).
Discussion
The identification of new CTL epitopes is generally time-consuming and labor-intensive and consists of (1) establishing virus-specific CTL clones; (2) testing a panel of allogeneic virus-infected target cells that share a part of MHC class I molecules with the CTLs to determine the MHC restriction; (3) transfecting or transducing cells expressing the appropriate MHC class I molecules with each of the segregated virus genes for the target of CTL; (4) making overlapping peptides, eg, 20 amino acid peptides overlapping by 10 amino acids to span a given antigen length for testing the specificity of the T cells; and, finally, (5) mapping the sequences with truncated peptides to determine the minimal epitope. We show in this article a rapid and efficient strategy to find new CTL epitopes, especially presented by MHC molecules of interest. An important step in this process is the use of a computer-based program designed to predict potential HLA-binding peptides within viral proteins.29,33 45 Analysis of amino acid sequences of various proteins of CMV AD169 strain revealed a number of potential HLA-A24–binding peptides, and most of them then functionally stabilized HLA-A*2402 molecules that had been expressed on the peptide transporter-deficient cell line T2. Next and equally important is screening of the peptides by ELISPOT assay for their recognition by bulk CMV-specific CTLs, established from PBMCs of a CMV-immune donor positive for HLA-A24 typing. The ELISPOT assay has many advantages in this regard. First, it allows results with a number of different peptides to be generated simultaneously in a few plates within 3 days. Second, the assay demands relatively small numbers of CTLs, eg, 25 to 100 000 in a well, depending on the concentration of specific T cells. Third, the sensitive and specific nature of the assay enables it to detect minor populations of antigen-specific T cells. For example, 40 spots in 100 000 CD8+ T cells in a well could be scored positive as long as the numbers of background spots were sufficiently low. Fourth, it needs no radioisotopes.
Target antigens of CMV-specific CD8+ T cells have been reported to be restricted to IE proteins,46,47glycoprotein B,34,46 and tegument proteins such as pp65.35,47,48 Thus, we first searched candidate peptides that have the HLA-A24–binding motif by a computer-assisted algorithm among proteins in the IE gene family, glycoproteins, and tegument proteins to find as many epitopes as possible. Among 83 peptides synthesized, 6 were unable to stabilize HLA-A*2402 molecules expressed on T2 cells at all and were excluded from further analysis. The ELISPOT assay using CMV-specific CD8+ T cells maintained by weekly stimulation with CMV-infected autologous fibroblast cells and IL-2 revealed that the T cells recognized almost exclusively peptides derived from pp65 (Figure 1). The results underline the observations of McLaughlin-Taylor et al,35 who showed that pp65 is a target antigen for CMV-specific CTLs in most seropositive donors tested, and the finding of Wills et al47 that 70% to 90% of CMV-specific CTL precursors are pp65-specific. It has been reported that pp65-specific CTL clones recognize CMV-infected target cells in the absence of viral gene expression.34,35 The capacity of CMV to interfere with class I MHC presentation at an early time after infection40-44 might explain why the specificity of the CTL response should be focused on structural proteins, which are relatively abundant immediately after infection, derived from input virus. The pp65 is a major constituent of the virus tegument43 and consequently available to the class I processing pathway immediately after infection. Our data confirm and extend the previous observations in the context of HLA-A*2402 molecules.
Both the bulk CMV-specific CTL line and the CD8+T-cell clones simultaneously recognized peptides no. 80 (VYALPLKML), 81 (QYDPVAALF), 82 (QYVKVYLESF), and 83 (QYDPVAALFF) at concentrations higher than 10 nM. However, only peptides no. 81 and 83, which share a core sequence QYDPVAALF, were able to sensitize T cells at concentrations lower than 100 pM (Figures 2 and 3). Actually, peptides no. 80 (VYALPLKML) and 82 (QYVKVYLESF), which were resynthesized independently, were not able to sensitize the T cells even at a concentration of 10 μM (data not shown). Thus, we reason that the recognition of peptides no. 80 and 82 could be due to cross-contamination between peptides in adjacent wells during synthesis, transfer, storage, or handling and that QYDPVAALF(F) is the T-cell epitope. Because the octamer QYDPVAAL was far less effectively recognized by the T cells (Figure 4), we concluded that the nonamer QYDPVAALF is the minimal epitope. This also indicates that phenylalanine at the C-terminal residue of the nonamer is important for binding to HLA-A24 molecules.38 We did not synthesize octamer YDPVAALF for testing because tyrosine at the second residue is reported to be critical for binding to HLA-A24 molecules.37-39 We do not know yet which is the naturally processed peptide, QYDPVAALF or QYDPVAALFF, or whether they both could be. To answer this question, peptides eluted from HLA-A*2402 molecules of CMV-infected cells should be analyzed for their sequences or molecular weights after fractionating on high-performance liquid chromatography.49 However, the minimal epitope QYDPVAALF can stimulate specific T cells for IFN-γ production (Figure 7) and was durable for making tetrameric MHC complexes (Figure 8). Thus, we did not perform experiments to determine the naturally processed sequence.
The ELISPOT assay demonstrated numbers of CD8+ T cells reacting with the minimal epitope, QYDPVAALF (no. 81), to exceed half of those reacting with autologous CMV-infected fibroblast cells (Figure1), indicating that CMV-specific CD8+ T-cell responses in the donor are highly focused on pp65 and especially the HLA-A*2402–binding peptide, QYDPVAALF. This was also confirmed by the evidence that, among 54 CD8+ T-cell clones established with anti-CD3 mAb stimulation, 28 clones turned out to be specific for the epitope. Thus, as far as the donor is concerned, HLA-A*2402 molecules are dominant presenters of CMV antigens, and QYDPVAALF is a definitely major epitope of CMV-specific CD8+ T cells. To extend this observation, we performed flow cytometric analysis for detecting IFN-γ produced within QYDPVAALF-reactive CD8+ T cells in PBMCs of HLA-A24+ and CMV-seropositive donors. The fact that 0.04% to 0.27% of peripheral CD8+ T cells of the donors specifically produced IFN-γ and the tetramers stained 0.79% of CD8+ T cells of a seropositive individual suggests that the peptide has potential to be a good reagent as a representative CTL antigen to monitor CMV-specific CD8+T-cell responses among HLA-A*2402+ individuals in various clinical settings. Although we tested only limited numbers of donors, the frequencies shown here are relatively lower than those of HLA-A*0201– and HLA-B*0701–restricted pp65-specific CD8+T cells in other populations.17-20 The reason is unclear so far and awaits further study.
Peptide no. 60, NYLDLSALL, derived from glycoprotein H was recognized by bulk CTL at the concentration of 10 μM but not by the pp65-specific CTL clones at the same concentration, indicating that it is a minor epitope presented by HLA-A*2402 molecules; however, this could not be confirmed because CTL clones that specifically recognize NYLDLSALL have not been established so far.
In conclusion, we have identified a possible major CMV-specific CTL epitope from the amino acid sequence of pp65 presented by HLA-A*2402 molecules. Minimal CMV-specific CTL epitopes so far reported are exclusively within the pp65 (Table 2). Our data are in line with the highly focused reaction of CMV-specific CTLs to pp65 in the context of HLA-A*2402 molecules, as is the case with HLA-A*020118,20,21,48 and HLA-B*0702.17The newly identified peptide QYDPVAALF could be used for detection of CMV-specific CD8+ T-cell responses as a representative CTL antigen. In addition, it might be a candidate reagent for peptide vaccines for prevention of CMV disease or expansion of CMV-specific CTLs in vitro for the purpose of adoptive transfer treatment of immunocompromised patients positive for HLA-A*2402.
Minimal CD8+ T-cell epitopes of the human CMV protein pp65
| Sequence . | Start position . | Amino acid length . | HLA . | Reference no. . |
|---|---|---|---|---|
| RPHERNGFTV | 265 | 10 | B7 | 47 |
| TPRVTGGGAM | 417 | 10 | B7 | 47 |
| NLVPMVATV | 495 | 9 | A2 | 22,47 |
| VLGPISGHV | 14 | 9 | A2 | 50 |
| MLNIPSINV | 120 | 9 | A2 | 50 |
| EFFWDANDIY | 511 | 10 | B12(44) | 47 |
| IPSINVHHY | 123 | 9 | B35 | 51 |
| QYDPVAALF | 328 | 9 | A*2402 | This article |
Supported by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (No. 13218152) and the Japan Society for Promotion of Science (No. 12670802 and RFTF 97L00703).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Kiyotaka Kuzushima, Division of Virology, Aichi Cancer Center Research Institute, 1-1 Kanokoden, Chigusa-ku, Nagoya 464-8681, Japan; e-mail:kkuzushi@aichi-cc.pref.aichi.jp.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal