Abstract
The well-established association between TP53 mutations and adverse clinical outcome in a range of human cancers reflects the importance of p53 protein in regulating tumor-cell growth and survival. Although it is theoretically possible for p53 dysfunction to arise through mechanisms that do not involve TP53 mutation, such a phenomenon has not previously been demonstrated in a sporadic tumor. Here, we show that p53 dysfunction in B-cell chronic lymphocytic leukemia (CLL) can occur in the absence of TP53 mutation and that such dysfunction is associated with mutation of the gene encoding ATM, a kinase implicated in p53 activation. Forty-three patients with CLL were examined for p53 dysfunction, as detected by impaired up-regulation of p53 and of the p53-dependent protein p21CIP1/WAF1 after exposure to ionizing radiation (IR). Thirty (70%) patients had normal p53 responses and underwent progressive IR-induced apoptosis. In 13 (30%) patients, p21 up-regulation was markedly impaired, indicating p53 dysfunction. Six (14%) of these patients with p53 dysfunction had increased baseline levels of p53, were found to have TP53 mutations, and were completely resistant to IR-induced apoptosis. In the other 7 (16%) patients with p53 dysfunction, IR-induced p53 up-regulation and apoptosis were markedly impaired, but baseline levels of p53 were not increased, and no TP53 mutations were detected. Each of these patients was found to have at least one ATM mutation, and a variable reduction in ATM protein was detected in all 4 patients examined. This is the first study to provide a direct demonstration that p53 dysfunction can arise in a sporadic tumor by a mechanism that does not involve TP53 mutation.
Introduction
Chronic lymphocytic leukemia (CLL) is characterized by the accumulation of predominantly nondividing clonal mature B cells in the blood, bone marrow, lymph nodes, and spleen and by a highly variable clinical course.1,2 Some of this variability can be attributed to the tumor-suppressor protein p53. Thus,TP53 gene defects in CLL are strongly associated with large-cell transformation,3 resistance to therapy with purine analogues,4-6 and shortened patient survival.4-6
By triggering apoptosis or cell-cycle arrest in response to DNA damage, p53 contributes to the cytotoxic action of many chemotherapeutic agents and protects the genome from mutagenic insult.7-9 In quiescent cells, levels of p53 protein are low owing to its short half-life. After DNA damage, the half-life of p53 becomes prolonged10 and the protein accumulates in the nucleus,11 where it regulates the transcription of a number of genes, including the cyclin-dependent kinase inhibitor p21WAF1/CIP1,12 the proapoptotic protein BAX,13 and the antiapoptotic protein BCL-2.14
TP53 mutations typically prolong the half-life of the protein in the absence of DNA damage and are, therefore, associated with increased basal levels.15 However, even when activated, mutant p53 protein cannot regulate gene expression because of its inability to bind to specific DNA sequences.16
Although TP53 mutations occur in only 10% to 15% of patients with CLL,4-6,17 it is possible that p53 dysfunction occurs in the disease through alternative mechanisms. For example, 15% to 35% of patients have an extra copy of chromosome 12,18,19 which encodes the p53-inhibitory protein MDM2.20 Indeed, MDM2 overexpression has been reported in CLL21,22 and may be associated with trisomy 12.23 However, the effect of such MDM2 overexpression on p53 activation has not been reported and is not necessarily predictable. Thus, the p53-inhibitory function of MDM2 is itself subject to negative regulation,24 and the basal level of this protein might therefore have little impact on p53 activation.
Inactivation of the ATM (ataxia telangiectasia mutated) gene is another potential cause of p53 dysfunction in CLL. ATM is a high-molecular–weight protein kinase encoded at 11q22-23 that has been implicated in p53 activation.25 Thus, ATM can associate with and phosphorylate p53 at serine 1526 and is involved in the dephosphorylation of p53 at serine 37627; both events are associated with p53 activation. Reduced levels of ATM protein have been detected in tumor cells from 30% to 40% of patients with CLL,28,29 and ATM mutations have been demonstrated in a substantial proportion of such cases.29In addition, as many as 20% of CLL patients have a deletion of 11q22-2319 that, in a proportion of cases, is associated with a mutation of the remaining ATMallele.30 31
Although p53 activation in response to ionizing radiation (IR) is impaired in thymocytes from atm-knockout mice32,33 and in lymphoblastoid cells from patients with ataxia-telangiectasia (A-T)34 (most of whom have bi-allelic truncating ATM mutations and absent ATM protein25), it is unknown whether the ATM protein defects that occur in CLL (many of which consist of amino acid substitutions29-31) result in p53 dysfunction. Indeed, given that the mechanisms involved in p53 activation are complex, overlapping and, cell-type dependent,35 36 it is unclear whether ATM is required for p53 activation in CLL cells.
In light of these uncertainties, the aim of the present study was to determine whether p53 dysfunction could occur in CLL in the absence ofTP53 mutations and, if so, to determine the cause. To do this, tumor cells were examined for impaired up-regulation of p53 and of p53-dependent gene products after exposure to IR. Patients with impaired p53 responses were then examined for TP53mutations, and those with wild-type p53, for overexpression of MDM2 and inactivation of ATM. Using this approach, we identifiedATM mutations, but not MDM2 overexpression, as a cause of p53 dysfunction in CLL.
Patients, materials, and methods
CLL patients
Peripheral blood was obtained with informed consent. In all patients, the malignant lymphocytes were morphologically typical and expressed low levels of light chain-restricted surface immunoglobulin, together with CD5 and CD23. Each patient had a lymphocyte count greater than 100 × 109/L.
Cell culture
Mononuclear cells were prepared from whole blood by centrifugation over Lymphoprep (Gibco, Paisley, United Kingdom) and were cultured at 37°C in RPMI and 1% bovine serum albumin (BSA) in the presence of 5% CO2. Culture vessels were precoated in poly(2-hydroxyethyl methacrylate) (Sigma, Poole, Dorset, United Kingdom) to prevent cell adhesion.37 This ensured that cells were not excluded from analysis as a result of sticking to the culture vessel.
Cytotoxic agents
Cells were irradiated at a rate of 3.4 Gy/min using a cesium Cs 137 source. Staurosporine (Calbiochem, Nottingham, United Kingdom) was used at 0.1 μM, a concentration shown in preliminary experiments readily to induce the killing of CLL cells.
Detection of p53, p21, BAX, and BCL-2 by Western blot analysis
Cultured CLL cells (2 × 106) were lysed for 30 minutes at 4°C in modified RIPA buffer containing 25 mM Tris-HCl (pH 7.6), 1% NP40, 5 mM EDTA, 50 mM sodium chloride, 30 mM sodium pyrophosphate, 50 mM sodium fluoride, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, and 1 μg/mL aprotinin. Lysates were centrifuged at 13 000g for 15 minutes, and the supernatants were mixed with sample buffer, incubated for 15 minutes at 95°C, separated by SDS-PAGE, and transferred to a nylon membrane (Immobilon-P; Millipore, Bedford, MA). After blocking in 5% milk, the membranes were probed with rabbit polyclonal antibodies to p21 (C-19; Santa Cruz Biotechnology, Santa Cruz, CA; 0.5 μg/mL) or to BAX (N-20; Santa Cruz Biotechnology; 0.1 μg/mL), with a mouse monoclonal antibody to BCL-2 (clone 100; Santa Cruz; 0.1 μg/mL) or with a mixture of 2 mouse monoclonal antibodies that bind to both wild-type and mutant p53 (pAb 1801; raised against amino acids 46-55) and pAb 421 (raised against amino acids 371-380), both from Oncogene Science (Cambridge, MA) and used at 0.1 μg/mL.38 After washing, the membranes were reacted with a peroxidase-conjugated goat polyclonal antirabbit or antimouse second-layer antibody (Transduction Laboratories, Lexington, KY). After further washing, specific protein bands were visualized using the enhanced chemiluminescence system (Amersham International PLC, Little Chalfont, Buckinghamshire, United Kingdom).
Measurement of ATM by Western blot analysis
CLL cells were lysed by sonication in buffer containing 50 mM Tris-HCl (pH 7.5), 9 M urea, and 150 mM β-mercaptoethanol. This method of protein extraction was used because it minimizes the problem of ATM degradation in vitro. Equal amounts of protein extract were mixed with loading buffer, incubated for 5 minutes at 95°C, separated by SDS-PAGE, and subjected to Western blot analysis as for p21 and BAX. Membranes were cut in 2 pieces. The upper part of the membranes containing high-molecular–weight proteins were probed with a rabbit polyclonal antibody raised to the N-terminal part of the ATM protein, AHP397 (Serotec, Oxford, United Kingdom). Results were confirmed by stripping and reprobing the membranes with a another rabbit polyclonal anti-ATM antibody, FP8r.29 As a control for protein loading, the lower part of each membrane was reprobed with a mouse monoclonal antibody to β-actin (AC-74; Sigma). The major bands corresponding to ATM and β-actin were quantified by densitometry, and the ATM:β-actin ratio of each lane was calculated. To enable comparison between membranes, the ATM: β-actin ratios were normalized with respect to those of the control cells on the same membrane.
Detection of TP53 mutations
Washed CLL cells were lysed in buffer containing 10 mM Tris-HCl (pH 8.0), 0.5 M lithium acetate, 1 mM EDTA, and 2% dodecyl lithium sulfate. Genomic DNA was purified from the lysates by phenol–chloroform extraction and ethanol precipitation.
DNA fragments containing TP53 exons and variable amounts of intron sequence were amplified from approximately 100-ng aliquots of genomic DNA using Taq polymerase and the supplied buffer (Promega, Southampton, United Kingdom), 3 mM MgCl2, 0.4 mM dNTP, and 20 pmol each primer. For exons 5 to 9, a “touchdown” protocol39 was used, with the annealing temperature reduced from 65°C to 55°C over the first 16 of 30 cycles. Exons 4 and 10 were amplified with an annealing temperature of 55°C for 30 cycles. All oligonucleotide primers for polymerase chain reaction (PCR) were designed from the genomic sequence (accession number, X54156), as shown in Table 1. Exons 5 and 6 were coamplified, as were exons 7 and 8, using primers 5/6F with 5/6B and 7/8F with 7/8B, respectively. Exons 4, 9, and 10 were amplified singly using primers 4F with 4B, 9F with 9B, and 10F with 10B, respectively.
TP53 primers used for polymerase chain reaction and sequencing
| Name . | Sequence 5′-3′ . | nt numbers X54156 . |
|---|---|---|
| R1F | AGCGTGCTTTCCACGACGGTG | 910-930 (S) |
| 2SQF | TGGATTGGCAGCCAGACTG | 942-949, 11689-11699 (S) |
| 4F | CAACGTTCTGGTAAGGAC | 11918-11935 (S) |
| 4SQF | GAAAACCTACCAGGGCAG | 12224-12241 (S) |
| 4SQB | GTCACAGACTTGGCTGTC | 12275-12292 (AS) |
| 4SQB2 | AAGCCTAAGGGTGAAGTG | 12388-12405 (AS) |
| 4B | ACAGGAAGCCTAAGGGTGAAGAG | 12388-12410 (AS) |
| 5/6F | TGCTGCCGTGTTCCAGTTGC | 12979-12997 (S) |
| 5SQF | ATCTGTTCACTTGTGCCCTG | 13002-13021 (S) |
| 5SQB | ATCAACCCACAGCTGCAC | 13106-13123 (AS) |
| 6SQF | TGAGCAGCTGGGGCTGGAGAG | 13240-13260 (S) |
| 5/6B | GGAGGGCCACTGACAACCAC | 13474-13493 (AS) |
| 7/8F | GAAGCTTACGAGGCTAAGG | 13599-13617 (S) |
| 7SQF | CTGCTTGCCACAGGTCTCC | 13938-13956 (S) |
| 7SQF2 | CTCACCATCATCACACTG | 14081-14098 (S) |
| 8SQF | TGGGACAGGTAGGACCTG | 14391-14408 (S) |
| 8SQB | GCGGAGATTCTCTTCCTC | 14522-14539 (AS) |
| 7/8B | GCTGGTGTTGTTGGGCAG | 14683-14700 (AS) |
| 9F | CACTAAGCGAGGTAAGCAAGCAG | 14578-14600 (S) |
| 9SQB | GTCTTTGAGGCATCACTG | 14866-14883 (AS) |
| 9B | AGCTACAACCAGGAGCCATTGTC | 14881-14903 (AS) |
| 10F | GCTTTTGATCCGTCATAAAGTC | 17472-17493 (S) |
| 10SQB | AGGATGAGAATGGAATCC | 17740-17757 (AS) |
| 10B | GCTGCCTTTGACCATGAAGGCAG | 17756-17778 (AS) |
| 11SQB | ACGCACACCTATTGCAAG | 18770-18787 (AS) |
| R11B | CTGGGTGCTTCTGACGCACAC | 18780-18800 (AS) |
| Name . | Sequence 5′-3′ . | nt numbers X54156 . |
|---|---|---|
| R1F | AGCGTGCTTTCCACGACGGTG | 910-930 (S) |
| 2SQF | TGGATTGGCAGCCAGACTG | 942-949, 11689-11699 (S) |
| 4F | CAACGTTCTGGTAAGGAC | 11918-11935 (S) |
| 4SQF | GAAAACCTACCAGGGCAG | 12224-12241 (S) |
| 4SQB | GTCACAGACTTGGCTGTC | 12275-12292 (AS) |
| 4SQB2 | AAGCCTAAGGGTGAAGTG | 12388-12405 (AS) |
| 4B | ACAGGAAGCCTAAGGGTGAAGAG | 12388-12410 (AS) |
| 5/6F | TGCTGCCGTGTTCCAGTTGC | 12979-12997 (S) |
| 5SQF | ATCTGTTCACTTGTGCCCTG | 13002-13021 (S) |
| 5SQB | ATCAACCCACAGCTGCAC | 13106-13123 (AS) |
| 6SQF | TGAGCAGCTGGGGCTGGAGAG | 13240-13260 (S) |
| 5/6B | GGAGGGCCACTGACAACCAC | 13474-13493 (AS) |
| 7/8F | GAAGCTTACGAGGCTAAGG | 13599-13617 (S) |
| 7SQF | CTGCTTGCCACAGGTCTCC | 13938-13956 (S) |
| 7SQF2 | CTCACCATCATCACACTG | 14081-14098 (S) |
| 8SQF | TGGGACAGGTAGGACCTG | 14391-14408 (S) |
| 8SQB | GCGGAGATTCTCTTCCTC | 14522-14539 (AS) |
| 7/8B | GCTGGTGTTGTTGGGCAG | 14683-14700 (AS) |
| 9F | CACTAAGCGAGGTAAGCAAGCAG | 14578-14600 (S) |
| 9SQB | GTCTTTGAGGCATCACTG | 14866-14883 (AS) |
| 9B | AGCTACAACCAGGAGCCATTGTC | 14881-14903 (AS) |
| 10F | GCTTTTGATCCGTCATAAAGTC | 17472-17493 (S) |
| 10SQB | AGGATGAGAATGGAATCC | 17740-17757 (AS) |
| 10B | GCTGCCTTTGACCATGAAGGCAG | 17756-17778 (AS) |
| 11SQB | ACGCACACCTATTGCAAG | 18770-18787 (AS) |
| R11B | CTGGGTGCTTCTGACGCACAC | 18780-18800 (AS) |
Nucleotide (nt) numbers correspond to the genomic sequence, accession number X54156. S indicates sense strand; AS, antisense strand. Primer 2SQF is derived from exons 1 and 2.
Total RNA was extracted from washed CLL cells essentially as described previously40 but using TRIzol reagent (Gibco BRL, Paisley, United Kingdom). Approximately 5-μg aliquots were reverse transcribed as described40 using Moloney murine leukemia virus reverse transcriptase (Promega) and an oligo(dT)18 primer. Aliquots (1-5 μL each) of the resultant primary-strand cDNA, diluted 1:10 in H2O, were used to amplify the entire coding region (on a 1339-bp fragment) using primers R1F and R11B (Table 1). PCR conditions and reagents were as described above except that 1.5 mM MgCl2 was used and the annealing temperature was reduced from 60°C to 55°C over the first 10 of 30 cycles.
PCR products from patient samples and negative controls (no added DNA/cDNA) were assessed by agarose gel electrophoresis before sequencing. They were prepared for sequencing either by diafiltration using Nanosep 100 K (Pall Filtron, Lichfield, Staffordshire, United Kingdom) centrifugal concentrators (exons 5-6, 7-8, and 9) or after agarose gel purification and elution using 0.45 μm Nanosep MF (Pall Filtron) or Genelute (Sigma) spin columns (exons 4 and 10 and reverse transcription [RT]-PCR fragments). PCR products amplified from genomic DNA were manually sequenced using a Circumvent kit (New England Biolabs, Hitchin, Hertfordshire, United Kingdom), and primers were end-labeled with γ[32P] adenosine triphosphate (Amersham). The RT-PCR purified fragments were sequenced on an ABI Prism 377 (Applied Biosystems) automated sequencer. For patients 6, 8, 9, 10, 11, 12, 19, and 24, exons 4 to 10 were sequenced from genomic DNA with the following primers (Table 1): exon 4, 4SQB and 4SQB2; exon 5, 5SQF; exon 6, 6SQF; exon 7, 7SQF; exon 8, 8SQF; exon 9, 9SQB; and exon 10, 10SQB. In addition, regions corresponding to coding regions of exons 2, 3, and 11 were sequenced in both directions from cDNA using primers 2SQF, 5SQB, 7SQF2, and 11SQB. Mutations detected in genomic DNA (patients 9-12) were confirmed on cDNA. Patients 35, 36, 39, 40, and 42 were sequenced from cDNA in both orientations using primers 2SQF, 4SQF, 5SQB, 7SQF2, 8SQB, and 11SQB. Thus, in all patients with p53 dysfunction, the entire coding region of TP53 was sequenced in both orientations.
Detection of ATM mutations
CLL patients with p53 defects but without TP53mutations were subjected to ATM mutation analysis using RT-PCR and either restriction endonuclease fingerprinting (REF), a method described by Liu and Sommer,41 or sequencing of the complete coding region of the ATM gene. For REF analysis, each tumor cDNA was subjected to the amplification of 8 overlappingATM fragments using primers listed in Table2. For sequencing of the entire coding region, the amplification of 4 overlapping ATM fragments was initially performed, and then each of the fragments was sequenced with several consecutive primers (Table 2). When feasible, the identified mutation was documented at the tumor DNA level by amplification and sequencing of the corresponding exon from the tumor DNA. In CLL patient 19, ATM mutation analysis was performed from cDNA and from tumor DNA. For this patient, all ATM exons were amplified from the tumor DNA and subsequently fully sequenced using primers previously published.42
ATM primers for reverse transcription-polymerase chain reaction, restriction endonuclease fingerprinting analysis, and cDNA sequencing
| ATM primers for RT-PCR and REF . | ATM primers for cDNA sequencing . |
|---|---|
| Fragment 1104 bp 5′ GTGTTCTGAAATTGTGAACCATGAGTCTAGT 5′ TGGTATCTTCATTAAAAACCTGGTGACAGA Fragment 1629 bp 5′ AGTAGAGGAAAGTATTCTTCAGGATTTCG 5′ CGTTTGCATCAGTAACACTACTATCAG Fragment 1608 bp 5′ GAGGTGGAGGATCAGTCATCCATGAATC 5′ GCGATGGAAAATAGGTGGATTAGGAGCAG Fragment 1121 bp 5′ CAGAGATTGTGGTGGAGTTAGTTATTG 5′ GCATTATGAAGGTCCACTGAAG Fragment 1058 bp 5′ TCTAGAGGCTGTTGGAAGCTGCT 5′ CCATACAAACTATCTGGCTCC Fragment 1233 bp 5′ CTGGAATAAGTTTACAGGATCTTC 5′ GATGATTTCATGTAGTTTTCAATTC Fragment 932 bp 5′ GATGGAGAAAGTAGTGATGAGC 5′ AGTCACCAGATTTCCATATTCTC Fragment 1227 bp 5′ AAGATGTTGTTGTCCCTACTATG 5′ AAGGCTGAATGAAAGGGTAATTC | Fragment 1 (2882 bp) 5′ GTGTTCTGAAATTGTGAACCATGAGTCTAG 5′ TATATGACTTATCTCATTCACTAGCAGATC 5′ TGAATTAGGAGATGAAATTCTTCCCAC 5′ TGACTTTGTCACTGACCACCAGTATAGTTC 5′ CTGGGATTATCAGAACAGCTTCTG 5′ TCCTGCGATTGTTAACATCAAAG 5′AAGCTCCTTTAAAAGCATTAGATACATATG Fragment 2 (2489 bp) 5′ GGCTTTTTCCTGCGATTGTTAACATCAAAGC 5′ AACTACTGCTCAGACCAATACTGTG 5′ GCTGACAATCATCACCAAGTTCG 5′ TTCTTGTAAATATTCTTCCTTATTTTGCCT 5′ ACCTCATTTTCCATCGCATGTGATTAA 5′ GTCCACCATCTGATCTTTATGTAGTTCCAGTTG Fragment 3 (2649 bp) 5′ TGAAAACCTCTATATCACGATTAAGC 5′ TCTAGAGGCTGTTGGAAGCTGCT 5′ GATTCATGATATTTTAGTCCAAGATAC 5′ CTGGAATAAGTTTACAGGATCTTC 5′ GACCATTGCACTTCCGTCAGC 5′ AAACACTCCCAGCTTCTCAAGG 5′ GTTTGTCTGAATTTTATGTTCCCTAAG Fragment 4 (2232 bp) 5′ CAATCCCAGCCTAAAACTTACATACACAGAAT 5′ GTCAATGGCATGATGAAGAGAGACGGA 5′ CAGTGCCAAAAGAAAATGATGGAGGT 5′ TAGCTGGATCCAAGAATTTTTCC 5′ TCTAAAGGCTGAATGAAAGGGTAATTC |
| ATM primers for RT-PCR and REF . | ATM primers for cDNA sequencing . |
|---|---|
| Fragment 1104 bp 5′ GTGTTCTGAAATTGTGAACCATGAGTCTAGT 5′ TGGTATCTTCATTAAAAACCTGGTGACAGA Fragment 1629 bp 5′ AGTAGAGGAAAGTATTCTTCAGGATTTCG 5′ CGTTTGCATCAGTAACACTACTATCAG Fragment 1608 bp 5′ GAGGTGGAGGATCAGTCATCCATGAATC 5′ GCGATGGAAAATAGGTGGATTAGGAGCAG Fragment 1121 bp 5′ CAGAGATTGTGGTGGAGTTAGTTATTG 5′ GCATTATGAAGGTCCACTGAAG Fragment 1058 bp 5′ TCTAGAGGCTGTTGGAAGCTGCT 5′ CCATACAAACTATCTGGCTCC Fragment 1233 bp 5′ CTGGAATAAGTTTACAGGATCTTC 5′ GATGATTTCATGTAGTTTTCAATTC Fragment 932 bp 5′ GATGGAGAAAGTAGTGATGAGC 5′ AGTCACCAGATTTCCATATTCTC Fragment 1227 bp 5′ AAGATGTTGTTGTCCCTACTATG 5′ AAGGCTGAATGAAAGGGTAATTC | Fragment 1 (2882 bp) 5′ GTGTTCTGAAATTGTGAACCATGAGTCTAG 5′ TATATGACTTATCTCATTCACTAGCAGATC 5′ TGAATTAGGAGATGAAATTCTTCCCAC 5′ TGACTTTGTCACTGACCACCAGTATAGTTC 5′ CTGGGATTATCAGAACAGCTTCTG 5′ TCCTGCGATTGTTAACATCAAAG 5′AAGCTCCTTTAAAAGCATTAGATACATATG Fragment 2 (2489 bp) 5′ GGCTTTTTCCTGCGATTGTTAACATCAAAGC 5′ AACTACTGCTCAGACCAATACTGTG 5′ GCTGACAATCATCACCAAGTTCG 5′ TTCTTGTAAATATTCTTCCTTATTTTGCCT 5′ ACCTCATTTTCCATCGCATGTGATTAA 5′ GTCCACCATCTGATCTTTATGTAGTTCCAGTTG Fragment 3 (2649 bp) 5′ TGAAAACCTCTATATCACGATTAAGC 5′ TCTAGAGGCTGTTGGAAGCTGCT 5′ GATTCATGATATTTTAGTCCAAGATAC 5′ CTGGAATAAGTTTACAGGATCTTC 5′ GACCATTGCACTTCCGTCAGC 5′ AAACACTCCCAGCTTCTCAAGG 5′ GTTTGTCTGAATTTTATGTTCCCTAAG Fragment 4 (2232 bp) 5′ CAATCCCAGCCTAAAACTTACATACACAGAAT 5′ GTCAATGGCATGATGAAGAGAGACGGA 5′ CAGTGCCAAAAGAAAATGATGGAGGT 5′ TAGCTGGATCCAAGAATTTTTCC 5′ TCTAAAGGCTGAATGAAAGGGTAATTC |
The left-hand panel shows ATM primers for amplification of 8 overlapping fragments covering the entireATM cDNA from the 5′ to 3′ end, used for REF. The right-hand panel shows primers used for cDNA sequencing. Primers in italics were also used for amplification of 4 overlapping ATM fragments, from which sequencing was carried out.
Measurement of MDM2 by flow cytometry
CLL cells were washed twice in phosphate-buffered saline (PBS), fixed for 60 minutes at room temperature in 1% formaldehyde, washed twice in PBS, permeabilized for 30 minutes at 37°C in 0.5% Tween, washed twice in PBS, and incubated for 30 minutes at room temperature with a mouse monoclonal antibody known to react specifically with MDM2 in formalin-fixed cells (SMP14; Santa Cruz; 5 μg/mL in PBS–BSA–azide) or with a class-specific control antibody. After 2 further washes in PBS, the cells were incubated for 30 minutes at room temperature with a fluorescein isothiocyanate-labeled goat antimouse second-layer antibody (Becton Dickinson; 1:20 dilution in PBS–BSA–azide). After 2 final washes in PBS, staining was quantified by flow cytometry.
Measurement of cell killing
Cultured cells were gently resuspended and added to an equal volume of PBS containing 10 μg/mL propidium iodide (PI; Sigma). After incubation on ice for 10 minutes, cells were analyzed by flow cytometry. Live cells with intact plasma membranes do not stain with PI. In contrast, dead cells that have lost their membrane integrity take up the fluorochrome and, therefore, exhibit bright red fluorescence.43
Detection of apoptosis
The mode of cell death was determined by examining CLL cells for DNA laddering and cleavage of poly(ADP-ribose) polymerase (PARP). Low-molecular–weight DNA was extracted as previously described by Forbes et al.44 Briefly, cultured cells (2.5 × 106) were gently washed in PBS and lysed overnight at 4°C in 500 μL buffer containing 5 mM Tris-HCL (pH 7.5), 5 mM EDTA, and 0.5% NP40. Lysates were centrifuged at 13 000g for 20 minutes, and the supernatants were mixed with 100 μL 5 M sodium chloride and 550 μL propan-2-ol. After overnight incubation at −20°C, the precipitated DNA was pelleted and resuspended in 50 μL buffer containing 10 mM Tris-HCL (pH 7.4) and 1 mM EDTA. Ten-microliter aliquots were mixed with 2.5 μL sample buffer and subjected to agarose gel (1.5%) electrophoresis. During apoptosis, internucleosomal DNA cleavage results in the formation of concatamers of 180 base pairs and a characteristic ladder appearance on electrophoresis.45
To detect PARP-1 cleavage, cultured cells (2.5 × 106) were gently washed in PBS and lysed in 100 μL sample buffer containing 6 M urea. Lysates were sonicated for 15 seconds and heated at 65°C for 15 minutes before they were subjected to SDS-PAGE and Western blot analysis using an anti–PARP-1 mouse monoclonal antibody (clone A6.4.12; Insight Biotechnology, Wembley, Middlesex, United Kingdom). During apoptosis, intact 116-kd PARP-1 is cleaved by caspases into a characteristic 89-kd C-terminal fragment.46
Results
Detection of p53 dysfunction
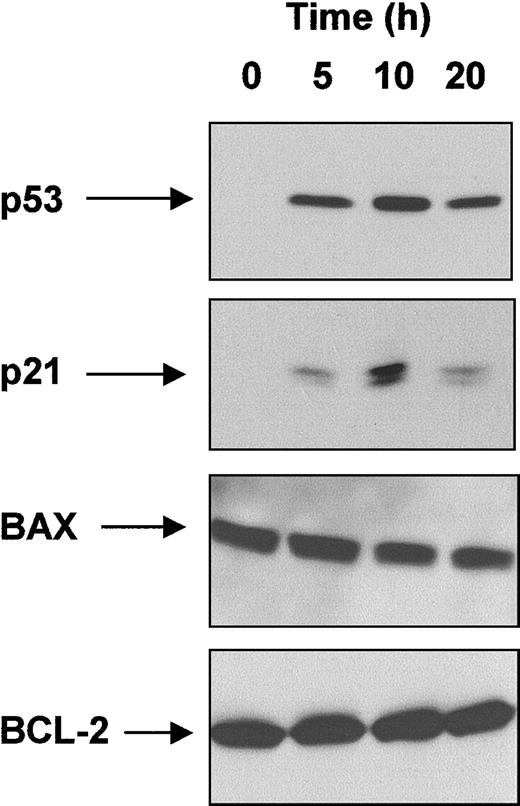
To screen for p53 dysfunction, CLL cells were examined for an impaired p53 response to IR. To do this, we first tested the effect of radiation on the expression of p53 and of several proteins reported in other cell types to be transcriptionally activated (p21, BAX) or repressed (BCL-2) by p53. A time-course was determined using a single CLL patient with wild-type p53. This patient's cells were known to be highly sensitive to IR-induced killing and were predicted to have a normal p53 response to IR (Figure 1).
Effect of IR on p53, p21, BAX, and BCL-2 levels.
Radiosensitive CLL cells from a single patient with wild-type p53 were exposed to γ-radiation (5 Gy) and lysed at the time points shown. Protein extracts were subjected to SDS-PAGE and Western blot analysis, as described in “Patients, materials, and methods.”
Effect of IR on p53, p21, BAX, and BCL-2 levels.
Radiosensitive CLL cells from a single patient with wild-type p53 were exposed to γ-radiation (5 Gy) and lysed at the time points shown. Protein extracts were subjected to SDS-PAGE and Western blot analysis, as described in “Patients, materials, and methods.”
As expected, p53 was undetectable at t = 0. However, after irradiation, p53 levels increased, with more protein being detected at 10 hours than at 5 hours or 20 hours. A similar pattern of expression was observed for p21. In contrast, BAX and BCL-2 were readily detectable at t = 0, and levels of these proteins did not change after irradiation. These findings were consistent with the notion that p53 was regulating the expression of p21 (but not BAX or BCL-2), and they indicated that 10 hours would be an appropriate time point for measuring p53 and p21 accumulation in other patients after radiation.
Tumor cells from 42 additional CLL patients were next examined for p53/p21 expression 10 hours after exposure to IR (Figure2). A normal p53 response (neither p53 nor p21 detectable in nonirradiated cells; both proteins strongly up-regulated by radiation) was observed in 29 of these 42 patients. Impaired up-regulation of p21 was detected in 13 patients. In 6 of them (patients 9, 10, 11, 12, 39, 42), p53 was constitutively increased in nonirradiated cells and became up-regulated further after radiation (type A p53 dysfunction). In the other 7 patients with impaired p21 up-regulation (patients 6, 8, 19, 24, 35, 36, 40), p53 was not detected in nonirradiated cells, and there was marked impairment of radiation-induced p53 accumulation (type B p53 dysfunction).
Detection of p53 dysfunction.
Irradiated and nonirradiated CLL cells were lysed after 10-hour culture. Protein extracts were subjected to SDS-PAGE and Western blot analysis, as described in “Patients, materials, and methods.” After initially probing for p21, membranes were sequentially stripped and reprobed for p53. To take into account any differences in protein loading, membranes were stripped again and reprobed for BCL-2. Dagger and asterisk denote patients with CLL showing types A and B p53 dysfunction, respectively.
Detection of p53 dysfunction.
Irradiated and nonirradiated CLL cells were lysed after 10-hour culture. Protein extracts were subjected to SDS-PAGE and Western blot analysis, as described in “Patients, materials, and methods.” After initially probing for p21, membranes were sequentially stripped and reprobed for p53. To take into account any differences in protein loading, membranes were stripped again and reprobed for BCL-2. Dagger and asterisk denote patients with CLL showing types A and B p53 dysfunction, respectively.
Detection of TP53 mutations
All 13 CLL patients and p53 dysfunction were next examined forTP53 mutations by sequencing the entire coding region of the gene (Table 3). A single TP53mutation was detected in each of the 6 patients with type A p53 dysfunction, and a signal corresponding to the normal sequence was also present in 4 of them (patients 10, 11, 12, 39). In contrast,TP53 mutations were not detected in any of the 7 patients with a type B p53 defect. In each of the 13 patients, the entire coding region could be amplified by RT-PCR. Therefore, the impaired p53 protein expression in the 7 patients with type B dysfunction was unlikely to have resulted from defective transcription caused by mutations in the TP53 promotor region. Taken together, these findings indicate that p53 dysfunction can indeed occur in the absence of TP53 mutation and that this type of dysfunction is associated with normal basal p53 levels and impaired IR-induced p53 protein accumulation and activation.
TP53 status in CLL patients with p53 dysfunction
| Case . | Base change . | Predicted codon change . |
|---|---|---|
| 63-150 | None | None |
| 83-150 | None | None |
| 93-151 | 413 C→T (exon 5) | 138 A→V |
| 103-151 | 430 C→T (exon 5) | 144 Q→stop |
| 113-151 | 584 T→A (exon 6) | 195 I→N |
| 123-151 | 764 T→A (exon 7) | 255 I→N |
| 193-150 | None | None |
| 243-150 | None | None |
| 353-150 | None | None |
| 363-150 | None | None |
| 393-151 | 743 G→A (exon 7) | 248 R→Q |
| 403-150 | None | None |
| 423-151 | 673-681 del 9nt | del 225-227 V,G,S |
| Case . | Base change . | Predicted codon change . |
|---|---|---|
| 63-150 | None | None |
| 83-150 | None | None |
| 93-151 | 413 C→T (exon 5) | 138 A→V |
| 103-151 | 430 C→T (exon 5) | 144 Q→stop |
| 113-151 | 584 T→A (exon 6) | 195 I→N |
| 123-151 | 764 T→A (exon 7) | 255 I→N |
| 193-150 | None | None |
| 243-150 | None | None |
| 353-150 | None | None |
| 363-150 | None | None |
| 393-151 | 743 G→A (exon 7) | 248 R→Q |
| 403-150 | None | None |
| 423-151 | 673-681 del 9nt | del 225-227 V,G,S |
All 13 patients with type B
or A
p53 dysfunction were subjected to TP53 mutation analysis. The entire coding region was sequenced in both orientations from a combination of genomic PCR and RT-PCR products, as detailed in “Patients, materials, and methods.” Nucleotide positions relate to cDNA and are numbered from the ATG codon.
Measurement of MDM2
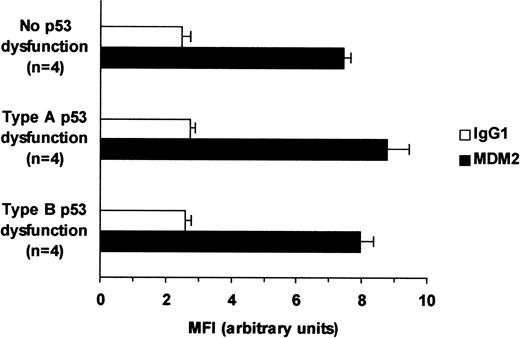
To determine whether the type B p53 dysfunction was caused by MDM2 overexpression, levels of this p53 inhibitory protein were measured by indirect immunofluorescence and flow cytometry (Figure3). Levels of MDM2 were no higher in patients with type B p53 dysfunction than in patients with functionally normal p53 or type A p53 dysfunction. This indicated that type B p53 dysfunction was not caused by MDM2 overexpression.
Relation between MDM2 levels and p53 dysfunction.
MDM2 levels in 4 CLL patients with type B p53 dysfunction and noTP53 gene defects were compared with those in 4 patients with functionally intact p53 and in 4 patients with type A p53 dysfunction and TP53 mutations. Average mean fluorescence intensity values (± SE) obtained by FACS analysis are shown for each group.
Relation between MDM2 levels and p53 dysfunction.
MDM2 levels in 4 CLL patients with type B p53 dysfunction and noTP53 gene defects were compared with those in 4 patients with functionally intact p53 and in 4 patients with type A p53 dysfunction and TP53 mutations. Average mean fluorescence intensity values (± SE) obtained by FACS analysis are shown for each group.
Measurement of ATM
To determine whether the type B p53 dysfunction resulted from ATM inactivation, levels of this protein were measured by Western blot analysis. Because both truncating and missense ATMmutations are associated with a reduction in the amount of full-length ATM protein,29 measurement of the protein can be used to screen for ATM gene defects.
Thirty-one CLL tumor samples were examined for ATM expression by Western blotting using 2 different antibodies. Normal and A-T lymphoblastoid cells were used as positive and negative controls, respectively (Figure 4). Compared with the normal lymphoblastoid cells on each membrane, patients with functionally intact p53 or type A p53 dysfunction (patients 9, 10, 11, 12) had increased amounts of ATM. In contrast, all 4 patients with type B p53 dysfunction (patients 6, 8, 19, 24) had levels of ATM lower than those of the normal lymphoblastoid cells. There was, however, some variation in ATM expression among the 4 patients. Thus, the protein was markedly reduced or undetectable in patients 6, 8, and 24, but in patient 19, ATM levels were only slightly lower than in those with normal lymphoblastoid cells. Nonetheless, these findings confirmed that type B p53 dysfunction was associated with altered levels of ATM protein.
Relation between ATM levels and p53 dysfunction.
ATM and β-actin levels were measured by Western blot analysis in 31 patients with CLL. Normal and A-T lymphoblastoid cell lines (LCL) were used as positive and negative controls, respectively. β-Actin and central ATM bands were quantified by densitometry, and the ATM:β-actin ratio of each lane was calculated. To enable comparisons to be made between individual membranes, the ATM:β-actin ratios were normalized with respect to the control cells on the same membrane. Dagger and asterisk denote patients with types A (patients 9, 10, 11, 12) and B (patients 6, 8, 19, 24) p53 dysfunction, respectively. The Figure shows the results obtained using the FP8r antibody, but similar results were obtained with AHP397.
Relation between ATM levels and p53 dysfunction.
ATM and β-actin levels were measured by Western blot analysis in 31 patients with CLL. Normal and A-T lymphoblastoid cell lines (LCL) were used as positive and negative controls, respectively. β-Actin and central ATM bands were quantified by densitometry, and the ATM:β-actin ratio of each lane was calculated. To enable comparisons to be made between individual membranes, the ATM:β-actin ratios were normalized with respect to the control cells on the same membrane. Dagger and asterisk denote patients with types A (patients 9, 10, 11, 12) and B (patients 6, 8, 19, 24) p53 dysfunction, respectively. The Figure shows the results obtained using the FP8r antibody, but similar results were obtained with AHP397.
Detection of ATM mutations
We next examined all 7 patients with type B p53 dysfunction for the presence of ATM mutations. To do this, the entire coding region of the ATM gene was subjected to REF and sequencing. This was successfully completed in all 7 patients except patient 19, in whom the 5′ end of the transcript could not be amplified despite repeated attempts to do so. Mutations were detected in all 7 patients (Table 4). Two truncating mutations were detected in patient 6, whereas one truncating mutation was detected in patients 24, 35, and 40. In addition, a single silent polymorphism was identified in patients 6 and 24. Two missense mutations were identified in patient 8, whereas one missense mutation was identified in patients 19, 35, and 36. These mutations (except the one in patient 36) all occurred in regions of ATM conserved between mice and humans and were predicted to cause significant changes to amino acids. Furthermore, the sequence alterations in patients 6, 8, 19, and 24 were not present in 40 analyzed meioses. Moreover, none of the alterations in ATM detected in the present study were previously described as polymorphisms in more than 200 A-T patients. Together, these observations strongly suggest that the ATM alterations detected in the present study are true mutations, not polymorphisms.
ATM mutations in CLL patients with type B p53 dysfunction
| Case . | Mutation . | Predicted codon change . | Presence of mutation at DNA level . | Other changes . |
|---|---|---|---|---|
| 6 | 404 delC | Frameshift at 135 | Yes | 6795 C→T (silent F→F) |
| 1426 insA | Frameshift at 475 | Yes | ||
| 8 | 3848 T→C | 1283 L→P | ND | |
| 5945 A→C | 1982 Q→P | ND | ||
| 19 | IVS7-4delT | Not clear4-150 | Yes | |
| IVS11-2insT | Not clear4-150 | Yes | ||
| 7205 A→G | 2402 E→G | Yes | ||
| 24 | 2875 del2nt | Frameshift at 959 | ND | 4473 C→T (silent F→F) |
| 35 | 5188 C→T | 1730 R→stop | ND | |
| 6056 A→G | 2019 Y→C | ND | ||
| 36 | 5134 T→A | 1712 F→I | ND | |
| 40 | 8535 del5nt | 2845 W→stop | Yes |
| Case . | Mutation . | Predicted codon change . | Presence of mutation at DNA level . | Other changes . |
|---|---|---|---|---|
| 6 | 404 delC | Frameshift at 135 | Yes | 6795 C→T (silent F→F) |
| 1426 insA | Frameshift at 475 | Yes | ||
| 8 | 3848 T→C | 1283 L→P | ND | |
| 5945 A→C | 1982 Q→P | ND | ||
| 19 | IVS7-4delT | Not clear4-150 | Yes | |
| IVS11-2insT | Not clear4-150 | Yes | ||
| 7205 A→G | 2402 E→G | Yes | ||
| 24 | 2875 del2nt | Frameshift at 959 | ND | 4473 C→T (silent F→F) |
| 35 | 5188 C→T | 1730 R→stop | ND | |
| 6056 A→G | 2019 Y→C | ND | ||
| 36 | 5134 T→A | 1712 F→I | ND | |
| 40 | 8535 del5nt | 2845 W→stop | Yes |
Tumor cells from the 7 patients with type B p53 dysfunction were subjected to ATM mutation analysis. Eight overlappingATM fragments were obtained for each patient and analyzed by both REF and sequencing of the complete coding region of theATM gene. Mutations were confirmed at the DNA level where indicated. Nucleotide positions relate to cDNA and are numbered from the ATG codon. ND, not done.
Two of the mutations in patient 19 were identified at the DNA level only. Their consequences at the RNA and protein levels cannot, therefore, be entirely predicted. However, both errors are located at the intron/exon boundary, and it is likely that they result in the skipping of exons 8 and 12, respectively.
The difficulty in amplifying the 5′ end of ATM from cDNA in patient 19 was suggestive of major gene disruption. To examine this possibility, the entire coding region of the ATM gene was sequenced exon by exon from genomic DNA. Three mutations were found. One (7205A→G) was previously identified at the RNA level. The other 2 mutations involved the acceptor sites of exons 8 and 12, respectively, and were identified at the DNA level only (Table 4). The consequences of mutations IVS7-4delT and IVS11-2insT cannot be entirely predicted at the RNA and the protein levels. However, it can be assumed that the first splicing error leads to the skipping of exon 8 and to the expression of an unstable truncation mutation and that the error at the acceptor site of exon 12 leads to the skipping of exon 12 and to an in-frame deletion. Large genomic distances did not allow us to use cloning to resolve the distribution of 3 mutations with respect to the 2 ATM alleles in patient 19. Therefore, the possibility remains that apparently full-size ATM protein observed in this patient corresponds either to the protein encoded by the mutant 7205A→G allele or to the protein encoded by the allele carrying an in-frame deletion of exon 12.
Taken together, these findings confirm the presence of ATMmutations in all 7 patients with CLL with type B p53 dysfunction.
Sensitivity to radiation-induced killing
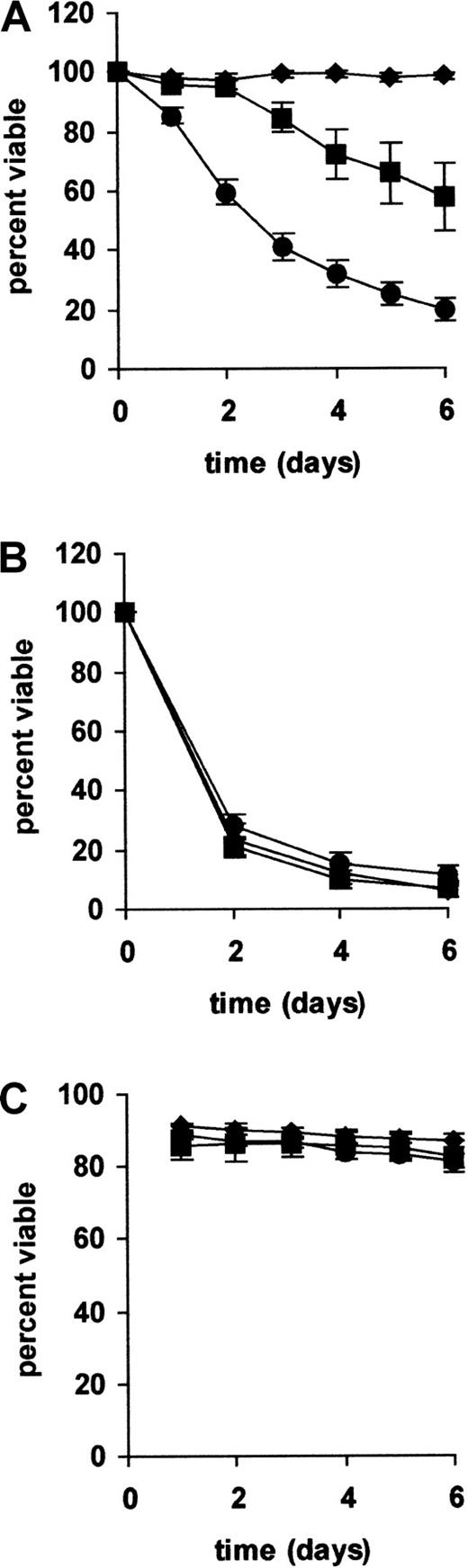
We next sought to establish whether ATM mutations in CLL were associated with impairment of p53-dependent cellular functions. Because CLL cells are predominantly nondividing and refractory to mitogenic stimulation,47 we could not study cell-cycle arrest. Therefore, we examined the other main function of p53—the induction of apoptosis. To do this, CLL cells were tested for their sensitivity to killing by IR, a DNA-damaging agent that induces p53-dependent apoptosis in mouse thymocytes.48 49
CLL cells with no p53 dysfunction underwent progressive cell death after exposure to IR (Figure 5). In 2 patients tested, killing was accompanied by extensive DNA laddering and cleavage of PARP-1 to an 89-kd fragment (data not shown). This confirmed that the cells were dying by apoptosis. In contrast to CLL cells with intact p53 responses, cells from all 6 patients with type A p53 dysfunction and TP53 mutations were completely radioresistant. This confirmed that the IR-induced killing of CLL cells was p53 dependent. The 7 patients with CLL with type B p53 dysfunction and ATM mutations were partially resistant to radiation-induced killing. Thus, little or no cell death was observed within the first 2 to 3 days after irradiation; thereafter, the cells underwent a variable amount of delayed killing.
Relation between p53 dysfunction and radiation-induced killing.
CLL cells from all 43 patients were tested for their sensitivity to killing by (A) γ-radiation (5 Gy) or (B) staurosporine (0.1 μM). Cell viability was measured by PI exclusion and flow cytometry (C). Mean survival values (± SE) are plotted against time. The difference in radiation-induced killing between cells with functionally intact p53 and those with type B p53 dysfunction and ATMmutations was highly significant (P < .001 by Student t test). In contrast, there were no differences among the 3 groups in their sensitivity to staurosporine-induced and spontaneous cell death. ▴ indicates TP53 mutated (n = 6); ▪, ATM mutated (n = 7); and ●, no p53 dysfunction (n = 30).
Relation between p53 dysfunction and radiation-induced killing.
CLL cells from all 43 patients were tested for their sensitivity to killing by (A) γ-radiation (5 Gy) or (B) staurosporine (0.1 μM). Cell viability was measured by PI exclusion and flow cytometry (C). Mean survival values (± SE) are plotted against time. The difference in radiation-induced killing between cells with functionally intact p53 and those with type B p53 dysfunction and ATMmutations was highly significant (P < .001 by Student t test). In contrast, there were no differences among the 3 groups in their sensitivity to staurosporine-induced and spontaneous cell death. ▴ indicates TP53 mutated (n = 6); ▪, ATM mutated (n = 7); and ●, no p53 dysfunction (n = 30).
In contrast to the different cytotoxic responses to IR, all 3 groups of CLL cells showed comparable sensitivity to killing by staurosporine and survived equally well under the culture conditions used (Figure 5). This confirmed that the observed differences in IR-induced killing reflected differences in p53 function rather than differences in susceptibility to apoptosis in general.
Discussion
To our knowledge, the present study provides the first direct demonstration of the notion put forward by Vogelstein and Kinzler in 199250 that p53 dysfunction can occur in sporadic tumors by mechanisms other than TP53 mutation. Although MDM2 overexpression and ATM inactivation have been reported in CLL, the effects of these abnormalities on p53 function in the disease have not previously been studied and were not necessarily predictable. Our demonstration that p53 dysfunction not caused by TP53mutation is associated with ATM inactivation but not with MDM2 overexpression indicates that p53 activation in CLL cells requires ATM but is largely unaffected by variations in basal levels of MDM2. The implication of ATM as a regulator of p53 function in CLL cells is consistent with the clinical observation that CLL patients with defective expression of ATM have, as do those with TP53 gene defects,4-6 a poor prognosis.28
By showing that patients with CLL with TP53 mutations are completely radioresistant, our data indicate that the radiation-induced killing of CLL cells is p53 dependent, as is the case with mouse thymocytes.48,49 Furthermore, our demonstration that CLL cells with ATM mutations are partially resistant to IR-induced killing is consistent with the idea that ATM regulates p53-mediated apoptosis in CLL cells. The fact that some delayed IR-induced killing occurs in all cells with ATM mutations and impaired p53 up-regulation is entirely consistent with the idea that the wild-type p53 in these cells can be activated by alternative pathways. Thus, it is well known that a delayed p53 response can occur in ATM-deficient cells because of redundancy between ATM and other proteins, such as ATR.51 This may explain whyTP53-mutant CLL cells show complete resistance to IR-induced apoptosis, whereas ATM-mutant CLL cells show only partial resistance to such killing. Because p53-mediated apoptosis is thought to underlie the cytotoxic action of many cytotoxic agents,52 our findings implicate ATM mutations as a potential mechanism of drug resistance in CLL and are consistent with the clinical observation that CLL patients with reduced levels of ATM protein respond poorly to therapy.28
Although not every patient with CLL in the present study was examined for TP53 and ATM gene defects, the number of patients with TP53 mutations (14%) was within the expected range of 10% to 15%.4-6,17 Similarly, the frequency ofATM mutations (16%) was close to the 18% found in the only previous study of ATM mutations in which patients were not selected on the basis of having 11q22-23 deletions.29
It is unlikely that the TP53 mutations detected in the present study represent polymorphisms because they have all been previously described in human tumors and because they were all associated with increased baseline levels of p53 protein, impaired radiation-induced up-regulation of p21, and complete resistance to IR-induced killing. Furthermore, the fact that radiation-induced p21 up-regulation and cytotoxicity were completely blocked in patients with p53-mutant CLL that retained a normal copy of the gene is consistent with the idea that mutant p53 can exert a dominant-negative effect over the wild-type protein.53
All 4 CLL patients with ATM mutations who were examined for the ATM gene product had reduced levels of the protein. In agreement with previous reports,29-31 some of these mutations resulted in a frameshift and a premature stop codon, whereas others resulted in an amino acid substitution. The fact that all the alterations within the ATM coding region were associated with reduced ATM protein levels and with impaired radiation-induced up-regulation of p53 and p21 supports the idea that they were true mutations and not polymorphisms.
Because cells from A-T patients (most of whom are compound heterozygotes for truncating ATM mutations) but not from A-T carriers display an impaired p53 response to IR, it is likely that theATM gene defects responsible for impaired p53 activation in CLL cells result from mutation or loss of both alleles. In the 3 CLL patients (patients 6, 8, and 35) in whom more than one ATMmutation was detected, it was not established whether these mutations occurred on the same allele or on different alleles. In one of them (patient 6), however, 2 truncating mutations were associated with complete absence of ATM, and in another (patient 8), 2 missense mutations were associated with marked reduction in the expression of ATM. This is consistent with the mutations being on separate alleles. In each of the other 4 patients with ATM mutations (patients 19, 24, 36, and 40), only one definite ATM mutation was identified within the coding region. In view of the phenotype of these patients with respect to p53 activation, it is likely that a second mutation was present but remained undetected.
It was interesting that in patient 19 the ATM gene was not easily amplified from cDNA, and levels of ATM protein were only slightly reduced. Direct sequencing of the gene from genomic DNA revealed 2 splicing errors in addition to the missense mutation already detected. It is difficult to predict the consequences of these mutations at the RNA and protein levels. However, it can be assumed that the first splicing error would lead to the skipping of exon 8 (166 bp), an event predicted to result in the expression of an unstable truncation mutant. The second splicing error should lead to the skipping of exon 12 (372 bp) and an in-frame deletion. This defect might result in some protein expression, explaining why some ATM protein was detected in this patient. Although the protein should be smaller than wild-type ATM, this size difference (320 kd versus 350 kd) is likely to be difficult to detect by Western blotting. It is, however, unclear how the 3 mutations are distributed with respect to the 2 ATM alleles. It is possible that the 2 splicing errors are on separate alleles. If so, the expressed ATM protein could be the in-frame mutant, with or without the missense mutation. Alternatively, if both splicing errors are on the same allele and the missense mutation is on the other, the expressed ATM protein would be a missense mutant. Because the distances between the 3 mutations (8 kb and 80 kb) preclude the separate cloning of the 2 alleles, it remains open to speculation which of these possibilities is correct.
Examining the p53/p21 response to IR is clearly an effective way of screening CLL patients for TP53 and ATMmutations. Thus, the fact that these defects were found in all cases with p53 dysfunction indicates that the test has a high level of specificity. Furthermore, the frequencies of TP53 andATM mutations detected by this method (14% and 16%, respectively) are similar to their reported frequencies, indicating that the test is likely to have a low rate of false-negative results. Screening for ATM deficiency in CLL patients using this functional approach may be more reliable than measuring ATM protein. For example, some tumors with ATM mutations (as in patient 19) might have near-normal levels of ATM protein. Conversely, it is possible that some CLL patients with no ATM mutations might (genuinely or as a result of in vitro protein degradation) express ATM at levels below those of normal lymphoblastoid cells. Therefore, examining CLL patients for p53 dysfunction highly enriches for an ATM-defective subgroup, though the possibility that some ATM mutations might not result in p53 dysfunction cannot be excluded.
The main disadvantage of using Western blotting to detect p53 dysfunction is that abnormal subclones might, if present, evade detection. In addition, the method is time consuming and difficult to standardize for routine clinical use. To circumvent these problems, we are exploring ways of detecting impaired p53 activation in individual cells using samples of whole blood. If successful, this should greatly facilitate the recognition of CLL patients with TP53 orATM mutations, thereby putting this important, but as yet difficult-to-obtain, prognostic information within easy reach of the clinician.
Supported by grants from the Leukaemia Research Fund (United Kingdom), the North West Cancer Research Fund, the Cancer Research Campaign, and the Kay Kendall Leukaemia Fund.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Andrew R. Pettitt, Department of Haematology, University of Liverpool, Liverpool L69 3GA, United Kingdom; e-mail:andrew.pettitt@rlbuh-tr.nwest.nhs.uk.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal