Abstract
The interaction of von Willebrand factor (vWF) with the platelet receptor glycoprotein Ibα (GPIbα) is important for platelet adhesion at high shear stress. Two functionally important antigenic areas within GPIbα were identified through the characterization of 5 new inhibitory anti-GPIb monoclonal antibodies (mAbs). The binding sites of 3 of these anti-GPIb mAbs, which were intercompeting and potently inhibiting shear stress–induced binding of vWF, were mapped within the N-terminal amino acid (aa) 1-59 area by the use of canine-human chimeras. These antibodies, however, had little or no effect (approximately 40% inhibition) on the binding of vWF induced by either botrocetin or ristocetin. On the other hand, the anti-GPIb mAbs 24G10 and 6B4, which blocked GPIb-vWF binding under all conditions examined, bound to 2 different regions of GPIbα, aa 1-81 and aa 201-268, respectively. The epitope for 6B4 was further narrowed by phage display revealing 2 sets of peptide sequences aligning within aa 259-262 and aa 230-242. In the latter region of GPIbα, the gain-of-function platelet-type von Willebrand disease (PT-vWD) mutations have been identified. Alignment was partially confirmed because the binding of 6B4 to recombinant GPIbα fragments carrying either one of the PT-vWD mutations was considerably impaired but not completely abolished. In contrast, mAb 24G10 bound more strongly to mutant PT-vWD GPIbα. However, although 24G10 competed with 6B4 for binding to platelets, it bound to an epitope within aa 1-81 of GPIbα. In conclusion, 2 functionally important areas within GPIbα were identified: one localized within the leucine-rich repeat N-terminal aa 1-59 area and one composed of residues aa 1-81 in close contact with aa 201-268. Moreover, further support is provided for the existence of an intramolecular interaction between the N-terminal flanking (aa 1-81) and C-terminal flanking (aa 201-268) regions.
Introduction
Adhesion of platelets to sites of vascular injury is critical for hemostasis and thrombosis and is dependent on the binding of von Willebrand factor (vWF) to the glycoprotein Ib (GPIb)/IX/V complex on the platelet surface.1 The GPIb-vWF interaction mediates the initial tethering and subsequent rolling of platelets over collagen in the subendothelium.2 This interaction is of major importance for platelet adhesion at high shear stress and leads to activation of the integrin αIIbβ3, allowing irreversible platelet adhesion, spreading, and thrombus propagation.
There are approximately 25 000 copies of the GPIb/IX/V complex on the membrane of resting platelets. GPIb is a heterodimer consisting of 2 subunits, GPIbα and GPIbβ. The N-terminal domain of GPIbα contains the binding sites for both vWF and thrombin and is characterized by a structural motif consisting of 7 leucine-rich repeats (LRRs), followed by a double disulfide-bonded loop and an anionic sulfated region carboxy terminal to the second loop.3
Several studies in the past 10 years have attempted to identify the crucial interaction sites on both GPIbα and vWF. However, this search is complicated by the fact that GPIbα and vWF only interact with each other in the presence of shear forces. Several systems have been developed that allow the study of flow-mediated platelet adhesion and aggregation, such as perfusion systems4 and viscometers.5 In the absence of shear, the GPIb-vWF interaction can be studied in the presence of nonphysiological modulators such as ristocetin6 or botrocetin.7 The interaction sites on both GPIbα and vWF thus depend on experimental conditions chosen to induce the interaction.
By using synthetic peptides, the vWF-binding site was localized within different areas on the N-terminal domain of GPIbα. Vicente et al8 reported that the peptide S251-Y279 inhibits the GPIb-vWF interaction induced by both ristocetin and botrocetin, whereas the peptide G271-E285 (single-letter amino acid codes) had little effect on ristocetin but was more effective in blocking botrocetin-induced binding. Murata et al9 further confirmed these results by showing that mutating the negatively charged amino acid (aa) between residues 251 and 279 abolished both ristocetin- and botrocetin-induced binding, whereas mutations within the highly charged region that follows (E281-D287) only affected botrocetin-dependent binding. Katagiri et al10 identified the sequence D235-K262 as important for ristocetin-dependent binding and the region F216-T240 as critical for thrombin binding. However, all these synthetic peptides have only a weak inhibitory effect in comparison to the native receptor. In addition, proteolytic fragments of GPIbα revealed that the anionic sulfated tyrosine-rich sequence (Y276-E282) also is important for the botrocetin-dependent binding of vWF and for the binding of thrombin.11
Mutations within the LRR, identified by studies of patients with Bernard-Soulier syndrome, result in either defective or decreased binding of vWF12-14 highlighting the role of the LRR (aa 36-200) in the correct exposure and function of the vWF binding site. This was further demonstrated by Shen et al,15 who used Chinese hamster ovary (CHO) cells expressing canine-human chimeras of GPIbα. The chimeras were constructed at precise boundaries between the different structural regions of GPIbα and were used to map the epitopes of functional blocking anti-GPIb monoclonal antibodies (mAbs) and to further identify the binding sites for vWF. By studying loss-of-function chimeras in which human sequence was replaced by canine and regain-of-function chimeras in which the canine sequence was rehumanized, they identified LRRs 2-4 as important for ristocetin and shear-dependent binding of vWF.15
Finally, within the disulfide loop C209-C248 of GPIbα, 2 gain-of-function platelet-type von Willebrand disease (PT-vWD) mutations (G233V, M239V) have been identified that enhance the interaction of GPIbα with vWF.16 Marchese et al17 demonstrated that beads coated with the mutant (G233V) GPIbα 1-302 fragment translocate on immobilized vWF with a lower velocity compared with beads coated with the nonmutated form. Therefore, it was hypothesized that these PT-vWD mutations regulate vWF binding.
To further investigate the GPIb-vWF interaction, we produced a new panel of murine anti-GPIb mAbs that block the binding of GPIb to vWF. The inhibitory characteristics of these mAbs were studied under shear conditions and under static conditions using the modulators ristocetin and botrocetin. Several approaches were also undertaken to localize the binding site of these mAbs on GPIbα. First, the mAbs were mapped using the canine-human chimeras of GPIbα constructed by Shen et al.15 Second, we used phage-display peptide libraries,18 a method that has proven to be rapid and successful for screening antibodies recognizing both continuous and as discontinuous epitopes.19-22 Finally, the contribution of one particular site to the antibody binding was further confirmed to some extent by mutagenesis studies.
Materials and methods
Production and purification of novel anti-GPIb mAbs and source of others
Human platelet glycocalicin, the extracellular fragment of GPIbα, was isolated from outdated platelet concentrates (Belgian Red Cross, Leuven), as described.23 Anti-GPIb mAbs were prepared by immunization of mice with purified human glycocalicin.24 These anti-GPIb mAbs were subsequently screened for inhibition of ristocetin-induced human platelet aggregation (see below). Inhibitory antibodies were further analyzed. The subtype of the mAbs was determined using an enzyme-linked immunosorbent assay (ELISA) kit (Sigma, St Louis, MO); mAbs were biotinylated using NHS LC biotin (sulfosuccinimidyl 6-[biotinamido] hexanoate) from Pierce (Rockford, IL) according to the manufacturer's instructions. The efficiency of biotinylation was checked by a dot-blot test and was greater than 95% for all the mAbs used. Anti-GPIb mAbs LJ-Ib1 and LJ-Ib10 were a gift from Dr Z. M. Ruggeri (La Jolla, CA), 6D1 was obtained from Dr B. S. Coller (State University of New York, Stony Brook), TM60 was from Dr N. Yamamoto (Tokyo, Japan), AK2 was purchased from Cymbus Biotechnology (Hampshire, United Kingdom), and HIP1 was from Pharmingen (Becton Dickinson, Belgium).
Production and purification of rGPIbα fragment
Shear-independent platelet aggregation
Blood was drawn from healthy volunteers on 3.8% sodium citrate (9:1 vol/vol), and platelet rich plasma (PRP) was prepared by centrifugation at 200g for 10 minutes. Aggregation studies were performed in an Elvi-840 dual-channel aggregometer from Pabish (Brussels, Belgium). Briefly, 200 μL PRP (final concentration, 2 × 108 platelets/mL) was preincubated with serial dilutions of various anti-GPIb mAbs for 3 minutes at room temperature (RT), followed by the addition of ristocetin (1.25 mg/mL) or botrocetin (0.5 μg/mL), and the aggregation response was followed for 5 minutes. Botrocetin was purified from crude Bothrops jararaca venom (Sigma).27 Platelet aggregations in response to α-thrombin (0.2 nmol/L) were performed by Dr M. Jandrot-Perrus (Paris, France).28
Effect of mAbs on shear-induced platelet adhesion
The inhibitory effect of the anti-GPIb mAbs was studied at a shear rate of 2600 seconds−1 in a parallel-plate flow chamber with a slit height of 0.4 mm.29 Human collagen type I (Sigma) was dissolved in 50 mM acetic acid (1 mg/mL), dialyzed extensively for 48 hours against phosphate-buffered saline, and subsequently sprayed onto plastic Thermanox coverslips (Nunc, Rochester, NY). Perfusions were carried out at 37°C with anticoagulated whole blood (low-molecular-weight heparin, 25 U/mL; Clexane, Rhône-Poulenc Rorer, France) obtained from healthy volunteers. Twelve milliliters whole blood was preincubated with various concentrations of the anti-GPIb mAbs for 5 minutes at 37°C, after which it was circulated over the collagen-coated coverslips. After 5-minute perfusion, the coverslips were immersed in methanol and stained with May-Grünwald-Giemsa. Platelet adhesion was evaluated with a light microscope connected to an image analyzer and was expressed as percentage surface coverage with platelets. Before and after every perfusion experiment the platelet count was measured.
Cross-blocking analysis for monoclonal antibody binding to platelets
A binding curve of all mAbs to human platelets was determined by ELISA. Two hundred-microliter aliquots of 2 × 108/mL formaldehyde-fixed platelets were added to microtiter plates precoated with 10 μg/mL poly-L-lysine (100 μL/well; Sigma). The plates were centrifuged at 4°C for 15 minutes at 150g, washed with phosphate-buffered saline, blocked with 3% skimmed milk (2 hours), and incubated with serial dilutions of biotinylated anti-GPIb mAbs (bmAbs) for 1 hour at RT. After another washing step, streptavidin–horseradish peroxidase (HRP; 1/10 000 dilution; Boehringer, Mannheim, Germany) was added, and binding was detected after the addition of ortho-phenylenediamine (OPD; Sigma). The reaction was stopped with 4 M H2SO4, and absorbance was determined at 492 nm.
Competitive inhibition assays between the different anti-GPIb mAbs were performed by preincubating a serial dilution of one unlabeled mAb (50 μL/well) with the platelets for 30 minutes at RT. Then, 50 μL constant concentration of the same or another anti-GPIb bmAb was added to the wells for 1 hour at RT. bmAb was detected after incubation with streptavidin-HRP as described above and was used at a constant concentration experimentally established to saturate 50% of the binding sites.
Inhibition of binding of vWF to a rGPIbα fragment
The GPIb-vWF interaction was studied by ELISA.26The purified rGPIbα fragment (H1-V289) (2 μg/mL) was captured for 2 hours (RT) onto a microtiter plate, precoated with mAb 2D4 (5 μg/mL), and blocked with 3% skimmed milk. mAbs 2D4 and 26D1 (negative control mAb) were nonblocking anti-GPIb mAbs produced in-house. A serial dilution of the inhibitory anti-GPIb mAb was added to the wells, together with purified human vWF, at a concentration of 0.5 μg/mL or 0.15 μg/mL, when ristocetin (300 μg/mL) or botrocetin (0.25 μg/mL) was used as a modulator, respectively. Wells were incubated for 2 hours at RT. Bound vWF was detected after 1-hour incubation with HRP-labeled anti-vWF antibody (DAKO, Glostrup, Denmark) using OPD as described above.
Screening of the antibodies with the canine-human chimeras of rGPIbα
Preparation of the expression vectors and expression of the canine-human chimeras in CHO βIX cells has been described in detail by Shen et al.15 In Figure 3A, an overview of the different chimeras is given. Binding of the anti-GPIb mAbs to the transfected CHO cells was analyzed using fluorescein isothiocyanate-labeled secondary antibody and flow cytometry.15
Screening of antibodies with a phage-display peptide library
A linear 15-mer library (L15; L. Jespers, Leuven, Belgium) and a C-C–linked circular 7-mer library (C7; E. Ruoslahti, La Jolla, CA) were used. Selection for antibody-binding peptides was made using a modified bio-panning technique, with the exception that beads rather than tubes were used for coating mAbs.30 In the first panning round, 2 × 1012 phages from each library (L15 and C7) were added, and bound phages were eluted with 0.1 M glycine-HCl. For the subsequent panning rounds, 1 × 1011eluted phages of the previous round were used as input phages, and a competitive elution was performed by adding 10 μg rGPIbα fragment for 30 minutes at RT.
Phage ELISA
Three types of ELISA were performed to evaluate whether the selected phage bound specifically to the target mAb. In the first ELISA, each pool of phage eluted in the different rounds was screened for binding to the mAb used for selection. In the second ELISA, positive individual colonies from these pools were identified. To this end, a dilution of the eluted phage pool of the last positive panning round was plated on Luria broth (LB) agar (+tetracycline [Tc]) plates. Ninety-six single colonies were picked from these plates and grown overnight in 2 × TY medium (+Tc) in a sterile culture microtiter plate. After centrifugation of the plates, the phage containing supernatant of each individual clone was evaluated in the ELISA. The third type was a competition ELISA—both the monoclonal phage and the rGPIbα fragment competed with each other for binding to the coated mAb. In these competition ELISAs, a serial dilution of the rGPIbα fragment (H1-V 289) was added with a constant concentration of the phages. Phages were used at a concentration sufficient to obtain 50% binding to the respective mAbs.
For all 3 ELISAs, mAbs were coated onto microtiter plates (5 μg/mL in Tris-buffered saline [TBS] overnight at 4°C). Plates were blocked with TBS + 2% skim milk (2 hours, RT). After washing with TBS + 0.1% Tween 20, phage, rGPIbα (in TBS + 2% skim milk), or both were added to the wells. After 1-hour incubation at RT and washing with TBS + 0.1% Tween 20, specifically bound phages were detected using HRP-labeled polyclonal anti-M13 antibody (1/5000 in TBS + 2% skim milk; Pharmacia, Roosendaal, The Netherlands). Final color development was done using OPD, and absorbance was measured at 492 nm.
Immunoblot analyses
Purified phage clones were subjected to 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a nitrocellulose membrane (Schleicher & Schuell, Dassel, Germany), blocked with 3% skim milk in TBS, washed with TBS + 0.05% Tween 80, and incubated with bmAbs (5 μg/mL in TBS + 0.05% Tween 80, 2 hours). After another washing step, the nitrocellulose membranes were incubated with streptavidin-HRP and were developed using the enhanced chemiluminescence detection system from Amersham (Buckinghamshire, United Kingdom).
Sequencing of phage clones
Phages selected for sequencing were amplified overnight in LB (+Tc) medium and were purified twice by precipitation with polyethylene glycol/NaCl. DNA was purified by phenol/chloroform extraction and was sequenced with the Sequenase version 2.0 T7 DNA Polymerase Sequencing Kit (Pharmacia) according to the manufacturer's instructions using32S-dATP (Amersham), 5′-CTCATAGTTAGCGTAACG-3′ for the 15-mer library, and 5′-CCCTCATAGTTAGCGTAACG-3′ for the 7-mer library.
Production of wild-type rGPIbα fragment (H1-R280) and 2 mutants, G233V and M239V
Recombinant GPIbα fragments used in the characterization of antibody binding were produced in insect cells. Wild-type and mutant GpIbα were expressed with a C-terminal histidine-tag and were purified from the harvested secretion medium by metal-affinity chromatography and anion-exchange chromatography (D.F.W. et al, unpublished results, 2001). In ELISA the wild-type fragment does not bind to plasma vWF in the absence of botrocetin and ristocetin, whereas the platelet-type vWD GpIbα fragments bind vWF spontaneously, reflecting the in vivo characteristics of wild-type and platelet-type vWD GpIbα.
Binding of anti-GPIb mAbs to the fragments was determined using plasmon resonance technology (Biacore, Uppsala, Sweden). A standard flow buffer (50 mM Tris pH 7.4, 150 mM NaCl, 0.005% surfactant P-20) was used, and the experiments were performed at RT. Capturing anti-GPIb mAb 2D4 was coupled to a sensor chip–type CM5 using conventional amine-coupling chemistry,31 resulting in a signal of ±1500 resonance units. The wild-type (WT)–rGPIbα fragment or the mutant rGPIbα fragments, at a concentration of 100 nmol/L, were injected at a flow of 5 μL/min for 3 minutes, followed by injection of the anti-GPIb mAbs (50 nmol/L) with a flow rate of 20 μL/min. Association and dissociation phases lasted 5 minutes each. Data were analyzed with the Biaevaluation 3.01 software. Langmuir (1:1) binding kinetics were used to determine the kinetic constants.
Results
Identification of inhibitory anti-GPIb monoclonal antibodies
Out of 2 fusions, a panel of 50 anti-GPIb mAbs was obtained, from which 5 mAbs were identified as inhibitory because they blocked ristocetin-induced human platelet agglutination. To obtain 50% saturation of binding to washed fixed platelets, 0.1, 0.04, 0.08, 0.1, and 0.02 μg/mL 27A10 (IgG1), 12G1 (IgG1), 12E4 (IgG2b), 24G10 (IgG2a), and 6B4 (IgG1), respectively, were required. All mAbs recognized the rGPIbα fragment (H1-V289) by ELISA, but none recognized GPIbα from platelet lysate or the rGPIbα fragment (H1-V289) after SDS denaturation and Western blot analysis.
Binding of vWF to the rGPIbα fragment in the presence of 300 μg/mL ristocetin could be blocked dose dependently by mAbs 6B4 and 24G10 (Figure 1A). On the contrary, mAbs 12G1, 12E4, and 27A10 inhibited the interaction only moderately (a maximal inhibitory effect of only 30% to 40%), even when using a high concentration of the mAbs, whereas the control mAb 26D1 had no effect. Only mAbs 6B4 and 24G10 inhibited botrocetin-induced binding; mAbs 12E4, 12G1, and 27A10 did not, equivalent to the noninhibitory mAb 26D1(Figure 1B). Still, mAb 24G10 was less potent than mAb 6B4, indicating a different mode of action. In addition, we investigated the inhibitory capacities of our antibodies on binding of sodium iodide 125–vWF to fixed, intact platelets and had results similar to those found by ELISA (data not shown).
Inhibitory effects of anti-GPIb mAbs.
Effects on ristocetin-induced (0.3 mg/mL) (A) and botrocetin-induced (0.25 μg/mL) (B) vWF binding to a rGPIbα fragment. mAbs 24G10 (●) and 6B4 (▴) blocked ristocetin-induced binding dose dependently, whereas mAbs 27A10 (▿), 12G1 (■), and 12E4 (⋄) inhibited the binding only moderately. mAbs 27A10, 12G1, and 12E4 did not block the botrocetin-induced binding, in contrast to mAbs 6B4 and 24G10. mAb 26D1 (control, ▪) had no effect when both modulators were used. Binding of vWF was detected with HRP-labeled anti-vWF antibody. Data are the mean of 2 experiments.
Inhibitory effects of anti-GPIb mAbs.
Effects on ristocetin-induced (0.3 mg/mL) (A) and botrocetin-induced (0.25 μg/mL) (B) vWF binding to a rGPIbα fragment. mAbs 24G10 (●) and 6B4 (▴) blocked ristocetin-induced binding dose dependently, whereas mAbs 27A10 (▿), 12G1 (■), and 12E4 (⋄) inhibited the binding only moderately. mAbs 27A10, 12G1, and 12E4 did not block the botrocetin-induced binding, in contrast to mAbs 6B4 and 24G10. mAb 26D1 (control, ▪) had no effect when both modulators were used. Binding of vWF was detected with HRP-labeled anti-vWF antibody. Data are the mean of 2 experiments.
These ELISA results are in agreement with aggregation studies in which mAbs 12E4, 12G1, and 27A10 could not block botrocetin (0.5 μg/mL)-induced human platelet aggregation in PRP, in contrast to mAbs 6B4 and 24G10, which have an IC50 value of 0.8 μg/mL and 2.5 μg/mL, respectively. The IC50 values for inhibition of ristocetin-induced (1.2 mg/mL) platelet aggregation are 2.5 μg/mL, 1.25 μg/mL, 1.3 μg/mL, 0.3 μg/mL, and 0.8 μg/mL for mAbs 27A10, 12G1, 12E4, 6B4, and 24G10, respectively. Only mAb 24G10 showed some inhibitory activity versus platelet aggregation induced by low doses of thrombin: 50 μg/mL 24G10 inhibited 0.2 nmol/L thrombin-induced aggregation by 62% (data not shown). Finally, at a concentration of 5 μg/mL, all the mAbs—12G1, 12E4, 27A10, 24G10, and 6B4—inhibited the adhesion of human platelets from whole blood to human collagen type I in a parallel-plate perfusion chamber at a shear rate of 2600 second−1, whereas mAb 26D1 did not (Table1).
Effect of monoclonal antibodies on shear-induced platelet adhesion to human collagen type I studied in a parallel-plate flow chamber
| Anti-GPIb mAb . | Surface coverage (% of control) . |
|---|---|
| 6B4 | 4.0 ± 1.2 |
| 24G10 | 6.4 ± 2.1 |
| 12G1 | 10.4 ± 4.3 |
| 12E4 | 8.1 ± 5.1 |
| 27A10 | 10.4 ± 3.1 |
| 26D1 | 90 ± 3.5 |
| Anti-GPIb mAb . | Surface coverage (% of control) . |
|---|---|
| 6B4 | 4.0 ± 1.2 |
| 24G10 | 6.4 ± 2.1 |
| 12G1 | 10.4 ± 4.3 |
| 12E4 | 8.1 ± 5.1 |
| 27A10 | 10.4 ± 3.1 |
| 26D1 | 90 ± 3.5 |
At a concentration of 5 μg/mL, all mAbs inhibited shear-induced adhesion, with the exception of the control mAb 26D1. Results are expressed as percentage maximal platelet adhesion (surface coverage) obtained in the absence of mAb (control 32.5% ± 5.2%, set as 100% surface coverage). Mean ± SE, n = 3.
GPIb indicates glycoprotein Ib; mAb, monoclonal antibody.
Cross-blocking analysis for mAb binding to platelets
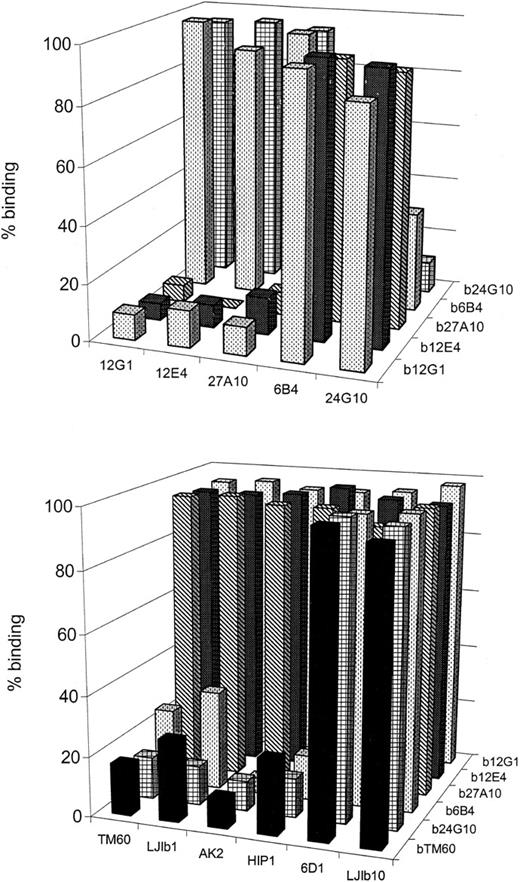
We evaluated whether the 5 anti-GPIb mAbs competed with each other for binding to human platelets (Figure2A). Binding of each biotinylated bmAb could be displaced by its unlabeled counterpart in a dose-dependent manner. Monoclonal antibodies 12G1, 12E4, and 27A10 did compete with each other, nor did they compete with mAbs 6B4 and 24G10, yet mAbs 6B4 and 24G10 cross-blocked the binding of each other. Based on these competition experiments, the mAbs can be divided into 2 groups: 12G1, 12E4, and 27A10 in the first group and 24G10 and 6B4 in the second group. This grouping correlates with their different inhibitory effects on the GPIb-vWF binding. The second group of mAbs (24G10, 6B4) were defined as overall inhibitors, blocking ristocetin-, botrocetin-, and shear-induced GPIb-vWF interaction. The first group of mAbs (12G1, 12E4, 27A10) was clearly less inhibitory with respect to ristocetin-dependent binding of vWF and could not block botrocetin-induced binding but did inhibit the shear-induced binding of vWF.
Competition between several anti-GPIb mAbs for binding to GPIb on human fixed platelets.
Human platelets were incubated with biotinylated mAbs (bmAb) at a concentration resulting in 50% saturation and with 50μg/mL of unlabeled antibody. Bound bmAb was detected by using HRP-labeled streptavidin. Data given are the percentage binding relative to the binding in the absence of mAb. (A) Novel panel of 5 anti-GPIb mAbs versus each other. Two groups of intercompeting mAbs 27A10, 12G1, 12E4 (group 1) and mAbs 6B4 and 24G10 (group 2) were found. (B) This panel of mAbs versus other anti-GPIb mAbs characterized elsewhere. mAbs 27A10, 12G1, and 12E4 did not compete with any of other mAbs, but mAbs 6B4 and 24G10 competed with mAbs LJ-Ib1, AK2, HIP1, and TM60. The binding of TM60 could be equally blocked by mAbs LJ-Ib1, AK2, HIP1, and TM60. Detection was accomplished using HRP-labeled streptavidin. Data given are the percentage binding (relative to control in the absence of mAb).
Competition between several anti-GPIb mAbs for binding to GPIb on human fixed platelets.
Human platelets were incubated with biotinylated mAbs (bmAb) at a concentration resulting in 50% saturation and with 50μg/mL of unlabeled antibody. Bound bmAb was detected by using HRP-labeled streptavidin. Data given are the percentage binding relative to the binding in the absence of mAb. (A) Novel panel of 5 anti-GPIb mAbs versus each other. Two groups of intercompeting mAbs 27A10, 12G1, 12E4 (group 1) and mAbs 6B4 and 24G10 (group 2) were found. (B) This panel of mAbs versus other anti-GPIb mAbs characterized elsewhere. mAbs 27A10, 12G1, and 12E4 did not compete with any of other mAbs, but mAbs 6B4 and 24G10 competed with mAbs LJ-Ib1, AK2, HIP1, and TM60. The binding of TM60 could be equally blocked by mAbs LJ-Ib1, AK2, HIP1, and TM60. Detection was accomplished using HRP-labeled streptavidin. Data given are the percentage binding (relative to control in the absence of mAb).
We also performed competition studies between our 5 mAbs and the mAbs LJ-Ib1, LJ-Ib10, HIP1, 6D1, TM60, and AK2 for binding to platelets (Figure 2B). Binding of mAbs of the first group (12G1, 12E4, 27A10) was not blocked by any of the investigated competing mAbs. However, mAbs 6B4 and 24G10 did compete with mAbs LJ-Ib1, AK2, HIP1, and TM60. The binding of TM60 itself could be equally blocked by LJIb1, AK2, HIP1, and TM60.
Screening the monoclonal antibodies by using canine-human chimeras
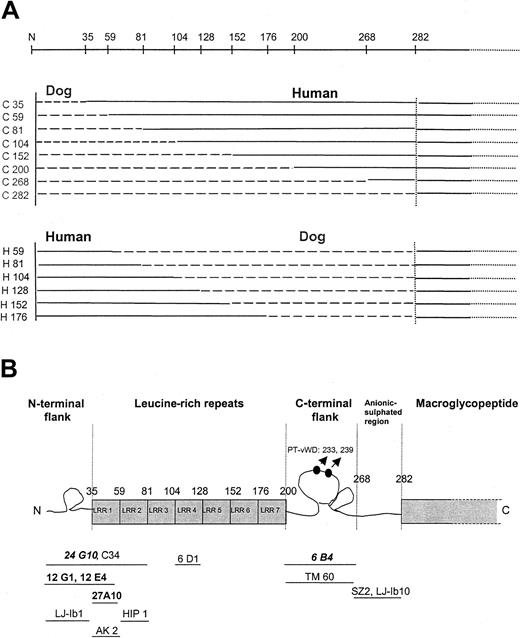
Recently, the binding sites for several other inhibitory anti-GPIb mAbs, characterized by other laboratories, were identified using the same canine-human chimeric system.15 In one set of chimeras, the human part (aa 1-282) was progressively replaced with the canine sequence starting from the N-terminus (Figure3A). Another set of chimeras was constructed such that the canine sequence (aa 1-282) was used as a template and was incrementally replaced with the human sequence. Shen et al15 demonstrated that all chimeras were functionally expressed because they bound to human vWF in the presence of botrocetin, a modulator that does not discriminate between human and canine GPIbα.
Epitope mapping using canine-human chimeras.
(A) Schematic overview of the different canine-human chimeras used for epitope mapping. (B) Scheme of the N-terminal region of GPIbα and localization of the epitopes of several anti-GPIb mAbs. Monoclonal antibodies given are those screened in this article (bold, group 1 mAbs; italic, group 2 mAbs) and those described by Shen et al15 (regular, not bold). Point mutations resulting in PT-vWD gain-of-function mutations (233, 239) are also indicated.
Epitope mapping using canine-human chimeras.
(A) Schematic overview of the different canine-human chimeras used for epitope mapping. (B) Scheme of the N-terminal region of GPIbα and localization of the epitopes of several anti-GPIb mAbs. Monoclonal antibodies given are those screened in this article (bold, group 1 mAbs; italic, group 2 mAbs) and those described by Shen et al15 (regular, not bold). Point mutations resulting in PT-vWD gain-of-function mutations (233, 239) are also indicated.
Our 5 anti–human GPIb mAbs did not bind to canine platelets, indicating that this system can be used for epitope mapping. Binding of the mAbs to the canine-human chimeras was checked by flow cytometry, and the results are summarized in Table 2and Figure 3B. As described by Shen et al,15 the binding domains for each mAb were assigned from the first domain at which the replacement of the human sequence with the canine sequence abolished binding, to the last domain where replacement of the canine sequence by the human sequence resulted in recovered binding.
Binding of anti–glycoprotein Ib monoclonal antibodies to the canine-human and the rehumanized chimeras
| Canine-human chimera . | Group 1 . | Group 2 . | |||
|---|---|---|---|---|---|
| 27A10 . | 12G1 . | 12E4 . | 6B4 . | 24G10 . | |
| C35 | 93 | 2 | 3 | 98 | 2 |
| C59 | 8 | 4 | 4 | 92 | 4 |
| C81 | 2 | 2 | 2 | 98 | 2 |
| C104 | 3 | 3 | 3 | 99 | 3 |
| C152 | 2 | 2 | 2 | 87 | 2 |
| C200 | 2 | 2 | 2 | 97 | 1 |
| C268 | 2 | 2 | 2 | 3 | 3 |
| C282 | 4 | 4 | 4 | 4 | 4 |
| Rehumanized chimera | |||||
| H59 | 84 | 78 | 81 | 8 | 4 |
| H81 | 75 | 73 | 76 | 6 | 73 |
| H104 | 98 | 97 | 99 | 6 | 96 |
| H128 | 72 | 73 | 74 | 5 | 81 |
| H152 | 97 | 98 | 98 | 4 | 96 |
| H176 | 95 | 96 | 97 | 8 | 89 |
| Canine-human chimera . | Group 1 . | Group 2 . | |||
|---|---|---|---|---|---|
| 27A10 . | 12G1 . | 12E4 . | 6B4 . | 24G10 . | |
| C35 | 93 | 2 | 3 | 98 | 2 |
| C59 | 8 | 4 | 4 | 92 | 4 |
| C81 | 2 | 2 | 2 | 98 | 2 |
| C104 | 3 | 3 | 3 | 99 | 3 |
| C152 | 2 | 2 | 2 | 87 | 2 |
| C200 | 2 | 2 | 2 | 97 | 1 |
| C268 | 2 | 2 | 2 | 3 | 3 |
| C282 | 4 | 4 | 4 | 4 | 4 |
| Rehumanized chimera | |||||
| H59 | 84 | 78 | 81 | 8 | 4 |
| H81 | 75 | 73 | 76 | 6 | 73 |
| H104 | 98 | 97 | 99 | 6 | 96 |
| H128 | 72 | 73 | 74 | 5 | 81 |
| H152 | 97 | 98 | 98 | 4 | 96 |
| H176 | 95 | 96 | 97 | 8 | 89 |
Binding was analyzed using fluorescein isothiocyanate–labeled secondary antibody and flow cytometry. Binding is expressed as percentage positive cells.
Binding of mAb 27A10 was lost when the first 59 human aa were changed into canine and binding was re-acquired when the canine sequence was rehumanized to aa 59. Therefore, the epitope is likely to be contained between aa 35 and 59 (first LRR) of GPIbα. Binding of mAbs 12G1 and 12E4 was lost at aa 35 and re-acquired at aa 59, indicating that the epitope is contained within the N-terminal flank of GPIbα and the first LRR (1-59). Binding of mAb 6B4 was lost when aa 201-268 was changed to a canine sequence, and binding was not yet re-acquired at aa 176; therefore, the epitope was likely to be between aa 201 and 282. Binding of mAb 24G10 was lost at aa 35 and re-acquired at aa 81, which implies that the epitope lies within aa 1-81 (N-terminus until the second LRR).
Selection of phage peptides, phage ELISA, sequencing, and sequence alignment
Phages were selected from a linear pentadecamer library (L15) on mAbs 6B4, 4G10, 27A10, 12G1, and 12E4 and, in addition, from the cyclic heptamer library (C7) on mAb 6B4. After 3 rounds of panning, a significant enrichment for phages binding to mAbs 27A10, 12G1, 12E4, and 6B4 was obtained. No phages could be selected for binding to mAb 24G10, despite several selection protocols with varying elution and washing procedures. We do not have a clear explanation for this nonselection. It may be that the epitope of mAb 24G10 is unavailable, though the diversity of the library is high. The same phenomenon has been observed by others.19 20 It might be desirable to screen this particular mAb using a panel of peptide phage libraries or using libraries containing larger peptide inserts.
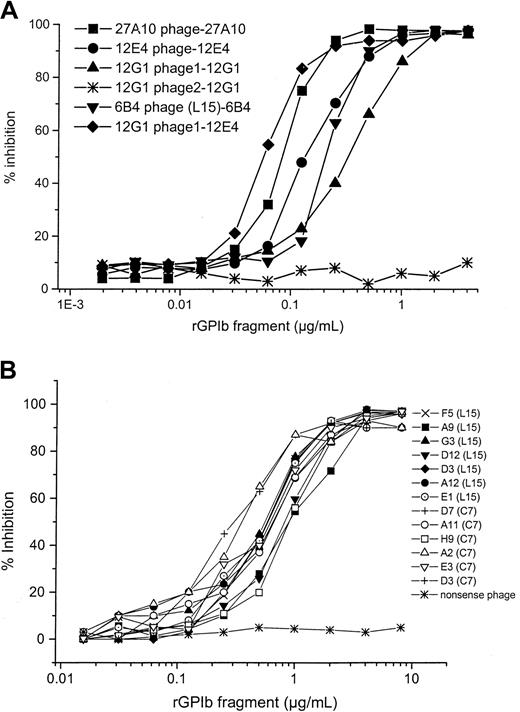
Individual phage clones from the third selection round (using the L15 library) on mAbs 27A10, 12G1, 12E4, and 6B4 were analyzed for binding to the respective mAb on which they where selected (Figure4) and for cross-reactivity with one of the other mAbs. None of the individual phage clones cross-reacted with other mAbs except for the phage (phage 1) selected on mAb 12G1 that also bound to mAb 12E4 (Figure 4). Both mAbs belong to the same group; however, it is unlikely that they are identical because a phage selected for binding to mAb 12E4 did not bind to mAb 12G1 (Figure 4).
Binding of 12E4 phages, 12G1 phages, and 27A10 phages to mAbs 12E4, 12G1, or 27A10.
Phages bound specifically to the mAb on which they were selected. One of the phages, selected on mAb 12G1 (phage 1 (■), cross-reacted with mAb 12E4. Binding of the phages was detected using HRP-labeled anti-M13 antibody, as described in “Materials and methods.” ▪, 12E4 phage and 12E4 mAb; ▾, 12G1 phage 1 and 12G1 mAb; ▿, 12E4 phage and 12G1 mAb; ▵, 27A10 phage and 12G1 mAb; ♦, 27A10 phage and 27A10 mAb; ⋄, 12G1 phage 1 and 27A10 mAb.
Binding of 12E4 phages, 12G1 phages, and 27A10 phages to mAbs 12E4, 12G1, or 27A10.
Phages bound specifically to the mAb on which they were selected. One of the phages, selected on mAb 12G1 (phage 1 (■), cross-reacted with mAb 12E4. Binding of the phages was detected using HRP-labeled anti-M13 antibody, as described in “Materials and methods.” ▪, 12E4 phage and 12E4 mAb; ▾, 12G1 phage 1 and 12G1 mAb; ▿, 12E4 phage and 12G1 mAb; ▵, 27A10 phage and 12G1 mAb; ♦, 27A10 phage and 27A10 mAb; ⋄, 12G1 phage 1 and 27A10 mAb.
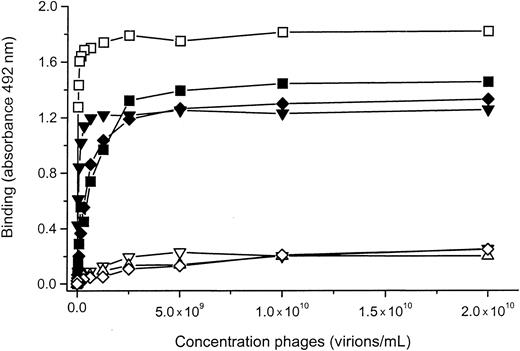
By means of competition ELISA, we could further show that all phages selected from L15 and C7 libraries bound specifically to the antigen-binding site of their respective mAbs because the rGPIbα (H1-V289) fragment was able to compete with the phages for binding to the mAbs (Figure 5A-B). The only exception is mAb 12G1, where only one of the different sequences (phage 1) selected, PVLLFCFLAGRCVSV, was specific (Figure 5A).
Competition between phages and rGPIbα fragment for binding to respective anti-GPIb mAbs.
(A) Competition between the rGPIbα fragment (serial dilution) and phages for binding to the respective mAbs. (B) Competition between the different 6B4 binding phages (selected from the C7 and the L15 libraries) and the rGPIbα fragment for binding to mAb 6B4. In all instances, the rGPIbα fragment was able to compete with the phages for binding to the mAb, with the exception of phage 2 selected on mAb 12G1 in panel A and the nonsense phage in panel B.
Competition between phages and rGPIbα fragment for binding to respective anti-GPIb mAbs.
(A) Competition between the rGPIbα fragment (serial dilution) and phages for binding to the respective mAbs. (B) Competition between the different 6B4 binding phages (selected from the C7 and the L15 libraries) and the rGPIbα fragment for binding to mAb 6B4. In all instances, the rGPIbα fragment was able to compete with the phages for binding to the mAb, with the exception of phage 2 selected on mAb 12G1 in panel A and the nonsense phage in panel B.
Sequence analysis of the phage inserts (from the L15 library) revealed that mAbs 27A10 and 12E4 each recognized a single sequence and that mAb 12G1 recognized 2 sequences (Table 3). Sequence alignment of the different sequences with the GPIbα sequence (aa 1-298) was made with 2 computer programs based on the BLOSUM 62 matrix (BEN best-fit of the GCG package and BioEdit). The alignments always positioned the sequences in the areas previously identified using the canine-human chimeras (not shown). However, to prove that these alignments indeed might identify amino acid residues involved in the binding of the mAbs, further panning experiments under less stringent conditions and with different phage peptide libraries are needed to deduce a consensus sequence. This strategy was followed for mAb 6B4. As selection on this mAb was performed with the L15 and C7 libraries, sequences depicted in Tables 3and 4 were obtained. Two sets of peptide sequences were found that were aligned in the region of GPIbα, identified by the use of the canine-human chimeras. One, comprising the KPGE/D consensus sequence (all C7 and 6 L15 sequences), could be aligned within the region 256-273 (Table 4), a region localized within the second part of the double disulfide loop of GPIbα. Another, comprising a set of peptide 4 sequences from the L15 library, could be aligned within the region 230-242 (Table 4), localized within the first part of the double disulfide loop of GPIbα. The latter part also comprises the PT-vWD mutation at aa 233 (G/V) and 239 (M/V). Sequences D12 and D3, however, aligned weakly within this region. Some of the peptides selected from the L15 library were shorter than expected, possibly because of incomplete ligation efficiency during the construction procedure of the library.32
Sequences of the different L15 phage peptides selected on the anti–glycoprotein Ib monoclonal antibodies
| . | mAb . | Selected peptide sequence . | n . |
|---|---|---|---|
| Group 1 | 6B4 | See Table4 | |
| 24G10 | / | ||
| Group 2 | 12G1-phage 1 | PVLLFCFLAGRCVSV | 7/8 |
| 12G1-phage2 | LRCYSHCTFYFASST3-150 | 1/8 | |
| 12E4 | MCPEMSFVAVHSCAR | 15/15 | |
| 27A10 | GFLSSCNFRFCELVR | 24/24 |
| . | mAb . | Selected peptide sequence . | n . |
|---|---|---|---|
| Group 1 | 6B4 | See Table4 | |
| 24G10 | / | ||
| Group 2 | 12G1-phage 1 | PVLLFCFLAGRCVSV | 7/8 |
| 12G1-phage2 | LRCYSHCTFYFASST3-150 | 1/8 | |
| 12E4 | MCPEMSFVAVHSCAR | 15/15 | |
| 27A10 | GFLSSCNFRFCELVR | 24/24 |
mAb indicates monoclonal antibody; n, number of clones sequenced that carry the displayed sequence; /, no phage peptide selected.
Nonspecific 12G1 binding phage.
Sequences selected on monoclonal antibody 6B4 using both a L15 and a C7 (*) phage peptide library.
| 6B4 clones . | Peptide . |
|---|---|
| A2, E4 | CNKPGERTC* |
| G12, D7, B4 | CDTLKPGEC* |
| A11, D10 | CADQKPGEC* |
| D3 | CYKPGEWAC* |
| H9 | CKPGEVQQC* |
| E3 | CKPVENRAC* |
| D9, F5, B5 | KPGEMR |
| G3 | KPGEMRGAA |
| E8, A9 | KPGDPSALHVVRCWIC |
| GPIb 251-273 | SDKFPVYKYPGKGCPTLGDEGDT |
| E1 | DGRRDVVVRSATFYL |
| D12 | EGLYSPWWPRSLPVL |
| D3 | SSKTRSFGVHLVGPY |
| A12 | LPPGLHVFPLASNRS |
| GPIb 223-245 | NAENVYVWKQG˙VDVKAM˙TSNVAS |
| 6B4 clones . | Peptide . |
|---|---|
| A2, E4 | CNKPGERTC* |
| G12, D7, B4 | CDTLKPGEC* |
| A11, D10 | CADQKPGEC* |
| D3 | CYKPGEWAC* |
| H9 | CKPGEVQQC* |
| E3 | CKPVENRAC* |
| D9, F5, B5 | KPGEMR |
| G3 | KPGEMRGAA |
| E8, A9 | KPGDPSALHVVRCWIC |
| GPIb 251-273 | SDKFPVYKYPGKGCPTLGDEGDT |
| E1 | DGRRDVVVRSATFYL |
| D12 | EGLYSPWWPRSLPVL |
| D3 | SSKTRSFGVHLVGPY |
| A12 | LPPGLHVFPLASNRS |
| GPIb 223-245 | NAENVYVWKQG˙VDVKAM˙TSNVAS |
Amino acids identical with GPIbα residues are italic, peptides of GPIbα studied in Vicente et al8 are underlined, PT-vWD mutations G233V and M239V are marked by ·.
GPIb indicates glycoprotein Ib; PT-vWD, platelet-type von Willebrand disease.
In addition, we tested whether the 2 cysteines present in some L15 phage peptides form a disulfide bridge: purified phages carrying the corresponding peptide sequence were separated on SDS-PAGE under nonreducing and reducing conditions, followed by Western blot analysis, during which phages were detected with the mAb for which they were selected (Figure 6). Although mAbs 27A10, 12E4, and 12G1 recognized their respective phages, mAbs 12G1 and 27A10 no longer did so when reducing conditions were used. For MoAb 12E4, recognition was diminished after reduction, and mAb 12E4 could recognize the cross-reacting 12G1 phage 1 under nonreducing, but not under reducing, conditions (not shown). Results of these Western blot experiments indicate that the cysteines present in the peptides selected on these mAbs (12G1, 27A10, 12E4) indeed form a disulfide bridge, necessary for recognition by their respective mAbs. All the C7 and L15 phages selected on mAb 6B4 could be detected under both nonreducing and reducing conditions, indicating that in this case the disulfide-linked structure is not necessary for recognition by 6B4. As an example, the results with the C7-6B4 phage clone D3 are shown in Figure 6.
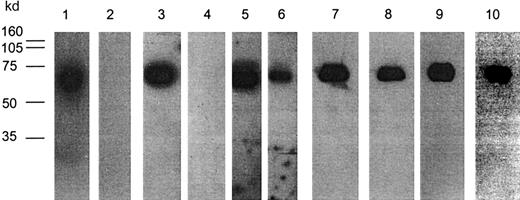
Binding of anti-GPIb mAbs to peptides displayed on phage proteins after SDS-denaturation and Western blotting.
mAb 27A10 (1, 2), mAb 12G1 (3, 4), mAb 12E4 (5, 6), mAb 6B4/clone D3 from the C7 library (7, 8), mAb 6B4/clone E1 from L15 library (9,10). Phages (2 × 1010) were run on SDS-PAGE (10% gel) either under nonreducing (lanes 1, 3, 5, 7, 9) or reducing (lanes 2, 4, 6, 8, 10) conditions, transferred to nitrocellulose membranes, and next probed with respective bmAbs. Detection occurred after incubation with streptavidin-HRP using enhanced chemiluminescence. mAbs 27A10, 12E4, and 12G1 recognized their phages under nonreducing, but not under reducing, conditions except for mAb 12E4. All phages selected on mAb 6B4 could be detected both under nonreducing and reducing conditions. Molecular weight (MW) marker is indicated.
Binding of anti-GPIb mAbs to peptides displayed on phage proteins after SDS-denaturation and Western blotting.
mAb 27A10 (1, 2), mAb 12G1 (3, 4), mAb 12E4 (5, 6), mAb 6B4/clone D3 from the C7 library (7, 8), mAb 6B4/clone E1 from L15 library (9,10). Phages (2 × 1010) were run on SDS-PAGE (10% gel) either under nonreducing (lanes 1, 3, 5, 7, 9) or reducing (lanes 2, 4, 6, 8, 10) conditions, transferred to nitrocellulose membranes, and next probed with respective bmAbs. Detection occurred after incubation with streptavidin-HRP using enhanced chemiluminescence. mAbs 27A10, 12E4, and 12G1 recognized their phages under nonreducing, but not under reducing, conditions except for mAb 12E4. All phages selected on mAb 6B4 could be detected both under nonreducing and reducing conditions. Molecular weight (MW) marker is indicated.
Binding of mAbs to wild-type and mutated rGPIbα fragments
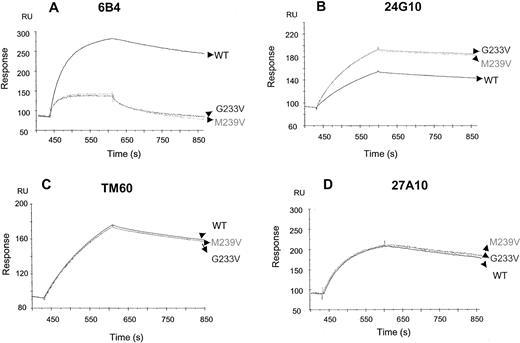
Monoclonal antibodies 24G10, TM60, 6B4, and 27A10 were analyzed for binding to wild-type and rGPIbα fragments (H1-R280) that contain the PT-vWD mutations (G233V and M239V) by Biacore (Figure7). Monoclonal antibodies 6B4, 24G10, and TM60 associated strongly and dissociated slowly, resulting in low apparent dissociation constant values. Monoclonal antibodies 27A10 and TM60 bound equally well to both mutants, G233V and M239V. In contrast, 6B4 had a 5- to 6-fold reduced affinity, whereas 24G10 had a 5-fold enhanced affinity mainly because of an enhanced on-rate.
Binding of anti-GPIb mAbs to recombinant GPIbα fragments.
mAbs 6B4 (A), 24G10 (B), TM60 (C), and 27A10 (D) were bound to wild-type and mutated (G233V and M239V) rGPIbα fragments by Biacore. Wild-type or mutated rGPIbα (H1-R280) fragments (100 nmol/L) were injected for 180 seconds, followed by injection of the anti-GPIb mAbs (50 nmol/L) for 180 seconds, and injection of flow buffer thereafter for another 300 seconds. TM60 and 27A10 bound equally well to both mutants. mAb 6B4 had markedly reduced affinity, whereas mAb 24G10 had enhanced affinity.
Binding of anti-GPIb mAbs to recombinant GPIbα fragments.
mAbs 6B4 (A), 24G10 (B), TM60 (C), and 27A10 (D) were bound to wild-type and mutated (G233V and M239V) rGPIbα fragments by Biacore. Wild-type or mutated rGPIbα (H1-R280) fragments (100 nmol/L) were injected for 180 seconds, followed by injection of the anti-GPIb mAbs (50 nmol/L) for 180 seconds, and injection of flow buffer thereafter for another 300 seconds. TM60 and 27A10 bound equally well to both mutants. mAb 6B4 had markedly reduced affinity, whereas mAb 24G10 had enhanced affinity.
Discussion
The platelet GPIb is an important receptor in the process of platelet adhesion under arterial levels of blood flow. The vWF binding site is located within the N-terminal 300 aa of GPIbα, which contains the 7-tandem LRR (36-200), the conserved N- (1-35) and C- (201-268) terminal flanking sequences, and an anionic sulfated region (269-2823). Several studies using synthetic peptides, proteolytic or mutated fragments of GPIbα, have further defined regions in the N-terminus potentially involved in binding to vWF. We here identified 2 functionally important antigenic areas within GPIbα based on studies with a set of novel anti-GPIb mAbs by analyzing their function, interantibody competition, and binding-site mapping. One such area was defined within the N-terminal region aa 1-59, and a second area was composed of residues aa 1-81 in close contact with the C-terminal region 201-268. Epitope mapping of the mAbs was done using the human-canine chimeras of GPIbα15 and was confirmed, to some extent, by screening using phage peptide libraries.
Studies using the canine-human GPIbα chimeras mapped the epitopes of the first group of 3 intercompeting anti-GPIb mAbs (12G1, 27A10, and 12E4) within the first region aa 1-59 of GPIbα. These 3 mAbs preferentially inhibited shear- and, to some extent, ristocetin-induced vWF binding, but had no effect on botrocetin-induced binding. The functional effects of these mAbs support the suggestion that ristocetin-dependent binding of vWF to GPIb is probably a better reflection of what occurs under shear, in contrast to the botrocetin-induced interaction, as previously suggested,15,33 and recently further disclosed by Dong et al.34 Moreover, the mAbs 12G1, 27A10, and 12E4 did not compete with any other inhibitory mAb tested in this study, further indicative of their unique character and, for another antigenic epitope, important for vWF binding under flow. This demonstration of the importance of the GPIbα N-terminal flanking sequences and adjacent first LRR is in agreement with the findings of Shen et al,15 who demonstrated that the GPIbα canine-human chimeras lacking the N-terminal flank and adjacent first LRR supported vWF binding to a much lesser extent than the wild-type.
The 2 other mAbs (6B4, 24G10) not only competed with each other, but also with a series of inhibitory mAbs from other laboratories (LJ-Ib1, HIP1, AK2, and TM60). The mAbs 6B4 and 24G10 inhibited vWF-GPIb interaction, regardless of whether this was induced by ristocetin, botrocetin, or shear and whether it was in contrast to the mAbs 12G1, 27A10, and 12E4. The mAb 24G10, like TM60, also inhibited thrombin binding to GPIbα. Two disparate binding sites were identified for this group of mAbs—aa 1-81 (N-terminal flank until the second LRR) for mAb 24G10, where LJ-Ib1, AK2, and HIP-1 also bind, and aa 201-282 (C-terminal flank) for mAb 6B4, where TM60 binds within aa 201-268.10 15
To further define the amino acid residues important for binding of mAb 6B4, peptides were selected by phage display using a linear 15-mer and a cyclic 7-mer peptide library. The sequences obtained could be divided within 2 groups and were aligned to the region previously identified by the chimeras (aa 201-282). One group could be aligned to position 230-242, where the gain-of-function platelet-type vWD mutations have been identified. Given that we demonstrated that 6B4 has a discontinuous epitope, it is not surprising that 2 potential binding sites were found. Our alignment results were partially confirmed by demonstrating that mAb 6B4 has a reduced affinity for recombinant GPIbα fragments carrying either of the PT-vWD mutations. However, this finding has to be interpreted with caution, because these mutations are known to have structural implications.35,36Furthermore, binding of 6B4 to these recombinant fragments was not completely abolished, indicating that indeed another binding site is probably involved to some extent. Based on the consensus sequence KPGE/D, which was found within the second group of selected phage sequences, the aligned aa 259-262 might form the second binding site. Indeed, Vincente et al8 demonstrated that a synthetic peptide containing aa 251-265 of GPIbα could block both ristocetin- and botrocetin-induced GPIb-vWF binding. Peptide 231-245 abolished the ristocetin-induced binding, but not the botrocetin-induced binding. On the other hand, peptides 221-235 and 241-255 had no blocking effect at all. Both blocking peptides are within the range of our alignments, indicating that the regions 259-262 and 230-242 of GPIbα indeed contain residues critical for binding of vWF.
Surprisingly, mAb 24G10, also belonging to the same group as mAb 6B4 but interacting with aa 1-81 of GPIbα, instead bound better to the rGPIb PT-vWD mutants, reminiscent of the increased affinity for vWF. This indicates that mutations at positions 233 and 239 may have an influence on the conformation of the aa 1-81 area of GPIbα.
Miller and Lyle37 reported an analogous finding when mapping the epitope of the anti-GPIb mAb C34. This particular mAb binds better to the PT-vWD mutant G233V than the nonmutated GPIbα. By phage- display screening on mAb C34, a mimotope peptide was selected, with a sequence dissimilar to that of GPIbα. A second screening with the mimotope peptide revealed an antimimotope peptide of which the sequence was identical to aa 230-234 of GPIbα. These investigators suggested that an intramolecular interaction occurs between the C34 epitope and the PT-vWD region. Interestingly, when screening mAb C34 with the canine-human chimeras,15 its epitope was found within aa 1-81 of the N-terminal region of GPIbα. These findings are consistent with our hypothesis that an association exists between the GPIbα N-terminal region aa 1-81 and the C-terminal region (a 201-268) comprising the PT-vWD mutations.
Further evidence that the areas aa 201-268 and aa 1-81 are topographically associated comes from antibody cross-blocking studies, because mAbs interacting with the first area prevent binding of mAbs to the second, and vice versa. The anticipated horseshoelike structure of the leucine-rich repeats of GPIbα is also consistent with the possibility that these 2 regions are proximal.38
Other investigators have shown that the region 201-268 of GPIbα plays a role in binding of vWF. For instance, Shen et al15found that the human-canine chimera C268 (in contrast to C200) did not bind to vWF. It is suggested that the region 201-268 of GPIbα contains sequences regulating vWF binding. An antibody that binds to this region might block this regulation and, hence, prevent vWF binding, as seen with our mAb 6B4 and also recently shown by Dong et al.34
Our results are also consistent with the hypothesis that the region aa 230-242 of GPIbα containing the PT-vWD mutations is associated with and has a negative influence on the affinity of aa 1-81 for binding to vWF.16,37 Relief of this negative regulation by gain-of-function mutations might also result in a conformational change in aa 1-81, which we here could directly demonstrate with mAb 24G10 binding to the region aa 1-81. Because ristocetin also binds to GPIbα and may bind to aa 251-279,8 it is possible that ristocetin, apart from its effect on vWF, also helps to relieve the inhibitory action of aa 230-242 on aa 1-81.
On the other hand, it is clear that with the present approaches, we have not fully identified the epitopes of any of our antibodies. Indeed, because none of the antibodies recognized SDS-denatured rGPIbα in Western blot, the antibodies are likely to have a nonlinear epitope. Furthermore, we cannot exclude that loss of binding of the antibodies to the chimeras and mutants to some extent may be due to conformational changes, and alignment of short-peptide sequences has to be looked at with caution. However, because all the data, based on widely different techniques—such as functional effects, antibody cross-competition, studies with chimeras, phage display, and mutants—all confirm one another, the conglomerate evidence indicates that the sequence regions identified within GPIbα are dominant constituents of the epitope of the antibodies.
In conclusion, characterization of a novel panel of anti-GPIb mAbs has provided further insight into the GPIb-vWF interaction. In particular, this study has made the novel finding that 2 binding sites, N-terminal aa 1-59 and aa 1-81 in close contact with aa 201-268, may play a role in vWF binding to GPIbα under high shear stress. Moreover, we have provided additional support for the existence of an intramolecular association between the leucine-rich repeat N-terminal (aa 1-81) and the C-terminal flanking (aa 201-268) domains of GPIbα. Knowledge of the tridimensional structure of GPIbα will probably be the key to a definitive explanation of the mechanisms involved in the regulation of GPIbα receptor activity.
We thank Wim Noppe for his technical assistance with the purification of GPIb and botrocetin. We thank Drs T. J. Kunicki, B. S. Coller, Z. M. Ruggeri, and N. Yamamoto for generously providing the anti-GPIb antibodies; Drs B. Steiner and S. Meyer for the stable CHO cell line that expresses the soluble GPIbα fragment (H1-V289); and Drs L. Jespers and E. Ruoslathi for the phage peptide libraries. We are very grateful to Dr M. Jandrot-Perrus for performing the thrombin-induced platelet aggregation studies.
Supported by European Biomed grant PL 963517 and Levenslijn grant 7.0017.98.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hans Deckmyn, Laboratory for Thrombosis Research-IRC, K U Leuven Campus Kortrijk, E. Sabbelaan 53, B-8500 Kortrijk, Belgium; e-mail: hans.deckmyn@kulak.ac.be.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal