Abstract
Locus control region (LCR) sequences are involved in the establishment of open chromosomal domains. To evaluate the possibility of exploiting the human CD2 LCR to regulate gene expression by Moloney murine leukemia virus (Mo-MLV)–based retroviral vectors in T cells, it was included in vectors carrying the enhanced green fluorescence protein (EGFP) reporter gene; then transduction in vitro of lymphoid and nonlymphoid cell lines was performed. Deletion of the viral enhancer in the Mo-MLV long terminal repeat was necessary to detect LCR activity in the context of these retroviral vectors. It was found that a full-length (2.1 kb), but not a truncated (1.0 kb), CD2 LCR retained the ability to modulate reporter gene expression by Mo-MLV–derived retroviral vectors, leading to a homogeneous, unimodal pattern of EGFP expression that remained unmodified in culture over time, specifically in T-cell lines; on the other hand, viral titer was strongly reduced compared with vectors not carrying the LCR. Lentiviral vectors containing the CD2 LCR could be generated at higher titers and were used to analyze its effects on gene expression in primary T cells. Subcutaneous implantation of genetically modified cells in immunodeficient mice showed that retroviral vectors carrying the CD2 LCR conferred an advantage in terms of transgene expression in vivo, compared with the parental vector, by preventing the down-modulation of EGFP expression. These findings suggest a potential application of this LCR to increase gene expression by retroviral and lentiviral vectors in T lymphocytes.
Introduction
One of the major obstacles to gene transfer in mammalian cells is the gap between gene expression of cellular genes from their genomic loci, which usually occurs at adequate levels, and the relatively poor expression levels obtained when the same gene is expressed from viral vectors, in many cases from heterologous viral promoters. This commonly observed phenomenon clearly constitutes a significant limitation both of gene therapy strategies aimed at treating genetic diseases by transduction of hematopoietic stem cells and of protocols of gene transfer into differentiated primary cells, such as lymphocytes and fibroblasts.1 Differences in gene expression from genomic loci and viral vectors clearly involve multiple levels of complexity, including lack of introns in the transgene, differences between viral and cellular promoters, and absence of some regulatory sequences in viral vectors—such as silencers, insulators, and locus control regions (LCRs)—that regulate the physiological expression of cellular genes. As a consequence, gene expression by viral vectors is often short-term, even though it can transiently occur at higher levels than that of cellular genes.
In some cases, the immune system also may contribute significantly to limiting transgene expression in vivo, specifically when the transgene itself or the viral vector used (ie, adenoviral vectors) is highly immunogenic. Furthermore, the pattern of expression of virally carried therapeutic genes and normal cellular genes is strikingly different: retrovector-encoded genes are expressed heterogeneously in the transduced cells because of their random integration into the host genome, whereas most cellular genes are expressed at homogeneous levels in a defined cell subset. Given these limitations of current gene transfer methods, LCR sequences have recently attracted attention because they might improve the level of gene expression by retroviral vectors (reviewed in2-4). Many studies focused on the β-globin LCR, which could be useful for increasing erythroid-specific synthesis of this protein.5-7 However, it is unclear whether selected LCR elements might be used to improve gene expression in T lymphocytes. A retroviral vector carrying a T-cell–specific LCR could be useful in clinical settings demanding long-term expression of therapeutic genes in lymphoid cells. These might include some primary immunodeficiencies in which T-cell–specific, long-term expression of a relevant gene, such as that coding for ADA, is required to enable the correct development of the immune system and some acquired diseases that might be amenable to treatment with genetically modified lymphocytes that would act as the “host factory” for soluble therapeutic proteins.
To address this issue, we studied an LCR from the humanCD2 gene. This 2.1-kb sequence has been largely studied in transgenic mice, in which it leads to high-level, position-independent gene expression in the T-cell lineage.8-10 Our findings suggested that CD2LCR could be useful for improving gene expression by retroviral and lentiviral vectors in T cells in vitro and in vivo.
Materials and methods
Plasmids
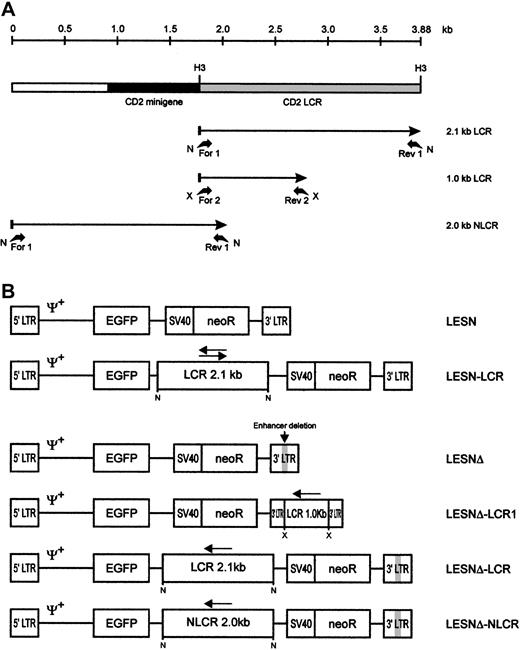
The 2.1-kb CD2 LCR sequence was amplified from plasmid VA hCD2, containing the human CD2 locus,11 by polymerase chain reaction (PCR) using the following primers, carrying a NotI site in the extension (Figure 1A): LCR-for1, 5′-AGAATGCGGCCGC-TGACTAGACCCGTGTCTGCTCAT -3′; and LCR-rev1, 5′-AGAATGCGGCCGC-AAGCTTCTTTACTATGCCATTTCATC-3′. In a set of experiments, an LCR sequence measuring 1 kb and containing a T-cell–specific transcriptional enhancer12 was amplified from VA hCD2 with the following primers, carrying an XbaI site in the extension (Figure 1A): LCR-for2, 5′-GCTCTAGA-TGACTAGACCCGTGTCTGCT-3′; and LCR-rev2, 5′-GCTCTAGA-GGCTGGTCTCAAACTCCT-3′. Finally, a 2.0-kb control sequence, located 5′ to the CD2 LCR and not associated with LCR function, was amplified from the same plasmid with primers (Figure 1A): NLCR-for1, 5′-AGAATGCGGCCGC-TGTGGATGACGCTCATACAACC-3′; and NLCR-rev1, 5′-GTTAACGTTTGTGCATGGAGGC-3′.
Schematic representation of the CD2 LCR fragments and the EGFP transfer vectors.
(A) Map of the CD2 LCR insert from the VA plasmid. Two LCR fragments and the control sequence (NLCR) used for insertion into retroviral vectors are indicated by arrows. Binding sites of the different primers used for amplification and the restriction enzymes used in cloning are indicated. N, NotI; X, XbaI; H3,HindIII. (B) Structure of the retroviral vectors used in this study. An extended Mo-MLV packaging signal (Ψ+) is present in each vector genomic-RNA encoding construct. LCR, 2.1-kb CD2 locus control region; NLCR, 2.0-kb control sequence from theCD2 gene; LCR1, 1-kb CD2 LCR fragment; neoR, neomycin phosphotransferase gene. (gray box) Deletion of the viral enhancer in the LTR; (arrow) orientation of the LCR element in each vector.
Schematic representation of the CD2 LCR fragments and the EGFP transfer vectors.
(A) Map of the CD2 LCR insert from the VA plasmid. Two LCR fragments and the control sequence (NLCR) used for insertion into retroviral vectors are indicated by arrows. Binding sites of the different primers used for amplification and the restriction enzymes used in cloning are indicated. N, NotI; X, XbaI; H3,HindIII. (B) Structure of the retroviral vectors used in this study. An extended Mo-MLV packaging signal (Ψ+) is present in each vector genomic-RNA encoding construct. LCR, 2.1-kb CD2 locus control region; NLCR, 2.0-kb control sequence from theCD2 gene; LCR1, 1-kb CD2 LCR fragment; neoR, neomycin phosphotransferase gene. (gray box) Deletion of the viral enhancer in the LTR; (arrow) orientation of the LCR element in each vector.
PCR was performed in 50 μL standard buffer containing 0.2 μM each primer and 1 U Taq polymerase (PerkinElmer, Foster City, CA) under the following conditions: denaturation for 1 minute at 94°C, annealing for 30 seconds at 65°C, extension for 2 minutes at 72°C, for 30 cycles, with 5 ng plasmid DNA as template. PCR fragments were purified and cloned in the pTA2 vector (Invitrogen, Groningen, The Netherlands). Subsequently, PCR products were excised from this vector by digestion with NotI or XbaI and were sticky-end cloned by standard procedures in the corresponding sites of a retroviral vector termed LESN, which was used as the basic vector, or derivatives of it. The LESN vector (Figure 1B) is a derivative of the LXSN retroviral vector13 and carries the gene for enhanced green fluorescence protein (EGFP) driven by the Mo-MLV long terminal repeat (LTR) and a neomycin resistance (neoR) cassette (Figure 1B). To investigate LCR function in the absence of the viral enhancer, we generated a modified LESN retroviral vector carrying a deletion in the U3 region of the LTR between bases 2976 and 3186 of the parental LXSN genome13 and termed it LESNΔ.
The Mo-MLV Gag-Pol expression constructgag-polgpt harbors the Mo-MLVgag and pol genes under the control of the Mo-MLV LTR and a SV40 polyadenylation signal, but it lacks ψ packaging sequences.14 A plasmid, termed HCMV-G, that expresses the envelope protein G of vesicular stomatitis virus (VSV-G) was used to pseudotype mLV particles.15
Cell culture and transfections
293T human kidney cells were obtained from American Type Culture Collection (Manassas, VA) and were used as a packaging cell line.16 The day before transfection, 1.5 × 106 293T cells were seeded in 25-cm2tissue culture flasks. Cultures were transfected with plasmid DNA using a calcium phosphate precipitation technique.17 293T and murine fibroblastic NIH-3T3 cells were grown in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum (FCS; Life Technologies, Milan, Italy) and 1% L-glutamine. Human Molt-3 and Jurkat cells were grown in RPMI 1640 supplemented with 10% fetal calf serum and 1% L-glutamine.
Transduction of cells with retroviral vectors
Infectious particles were generated by overnight transfection of 293T cells with 3 μg gag-polgpt, 6 μg either LESN, LESN-LCR, LESNΔ, LESNΔ-LCR, LESNΔ-NLCR, or LESNΔ-LCR1 as transducing vectors, and 0.1 μg VSV-G expression construct, as reported.18 Retroviral vector–containing supernatants were collected 48 to 72 hours after transfection, filtered through 0.45-μm filters, and frozen at −80°C until further use. To determine vector titer, serial dilutions of filtered supernatants were layered over NIH-3T3 target cells that had been seeded into 6-well culture plates the day before infection at 0.6 × 105cells per well. Protamine sulfate (8 μg/mL) (Sigma-Aldrich, St Louis, MO) was added to the wells. After 6 to 9 hours at 37°C, 3 mL medium was added; 36 hours later, the cells were split 1:10 in 10-cm diameter Petri dishes, and new medium containing G418 (Life Technologies; 500 μg/mL active compound) was added. After 2-week culture in G418 selective medium, the viral titer was determined as described19 and was expressed as colony-forming units (cfu) per milliliter supernatant.
In experiments with lymphoid cells, because of the low titer of the LCR-carrying vectors, all the relevant supernatants were concentrated 100-fold by ultracentrifugation as described.20Transduction was performed by incubating 1 mL retroviral vector–containing supernatant with 2 × 105 target cells for 6 to 9 hours at 37°C in the presence of protamine sulfate (8 μg/mL). Lymphoid cells were then grown for an additional 4 weeks in G418-containing medium (500 μg/mL) before assessment of EGFP expression.
Proviral DNA analysis
Genomic DNA was obtained from vector-transduced Molt-3 and NIH-3T3 cells using the EasyDNA kit (Invitrogen). For Southern blot analysis, 20 μg DNA was digested to completion with eitherKpnI or XhoI/HindIII (New England Biolabs, Beverly, MA), electrophoresed on 0.8% agarose gels, transferred to nylon membranes (Amersham-Pharmacia Biotech, Little Chalfont, United Kingdom) by Southern blotting as described,21 and hybridized to a 32P-labeled 1.32-kb HindIII/SmaI DNA probe from plasmid pSV2-neo. To determine provirus copy number in the transduced cells, the blot was stripped and rehybridized with a loading control probe (CXCR4).22 After washes under high-stringency conditions, filters were autoradiographed for 48 to 96 hours at −70°C. Band intensities were quantified with an InstantImager (Packard, Meriden, CT) and were used to calculate the relative number of provirus copies per genome in each sample.
Lentiviral vector–mediated gene transfer in primary T cells
All lentiviral vectors were derived from ViGΔBH, which is an simian immunodeficiency virus (SIV)–based lentiviral vector carrying an internal EGFP gene driven by an MLV LTR (details of their construction are available on request).23Vector-containing supernatants were generated by a 3-plasmid vector packaging system based on transient transfections of 293T cells with 6 μg Hgp, a synthetic HIV gag-pol expression plasmid,24 6 μg ViGΔBH or its derivatives, and 0.1 μg VSV-G expression construct, as reported for Mo-MLV–based retroviral vectors. NIH-3T3 cells were used in preliminary experiments for titration of the vectors. Peripheral blood lymphocytes from healthy donors were centrifuged on Ficoll-Hypaque gradients activated with anti-CD3 and were maintained in complete RPMI medium supplemented with 100 U/mL recombinant interleukin-2 (rIL-2; EuroCetus, Milan, Italy). Forty-eight hours after activation, 5 × 105 cells were transduced with 1 mL lentiviral vector–containing supernatants for 6 hours at 37°C; subsequently, peripheral blood lymphocytes were transferred to 24-well plates (Becton Dickinson, Franklin Lakes, NJ) and were cultured for an additional 72 hours before EGFP detection. In a set of experiments, EGFP+ T lymphocytes were sorted by fluorescence-activated cell sorter (FACS) and cultured in 96-well plates in the presence of allogenic irradiated feeder cells (105 cells/well) and IL-2 (100 U/mL) for 6 weeks.
Cytofluorometric analysis
EGFP expression in vector-transduced cells was assessed on an EPICS-Elite cytofluorometer (Coulter, Hialeah, FL) by FACS analysis. At different times after infection, cells were pelleted, washed, fixed, and labeled, where appropriate, with a phycoerythrin-labeled anti–HLA class I monoclonal antibody (mAb) (DAKO, Glostrup, Denmark) or with a PC5-labeled anti–human CD3 mAb (Coulter). Two-color immunofluorescence was carried out as reported25 and was analyzed using the PRISM parameter of the Elite cytofluorometer; the negative control setting for each monoclonal antibody was determined by using labeled immunoglobulin of the corresponding isotype. Mock-transduced cell lines served as the negative control for EGFP-expression analysis. Mean fluorescence intensity (MFI) was calculated by the following formula: MFI = log10(mean × 10) × (1024/4). Coefficient of variation (CV) was calculated by the following formula: CV = [SD/mean] × 100; this parameter describes the homogeneity of the bell-shaped curve of fluorescence expression.
In vivo expression studies
Female severe combined immunodeficiency (SCID) mice (6 to 8 weeks old) were purchased from Charles River (Wilmington, MA). Logarithmically growing human Molt-3 cells transduced with the LESN or the LESNΔ-LCR retroviral vectors were harvested and resuspended in fresh medium at a density of 2 × 108 cells/mL. One hundred microliters of this cell suspension was then injected into the flanks of the mice. Tumor growth was monitored by periodic observation. Mice were killed 1 month later, when the tumors were visible; tumor cells were recovered by microdissection and were analyzed for HLA and EGFP expression. At least 5 animals were used in each group, and the experiment was repeated twice.
Results
Generation of CD2 LCR–carrying retroviral vectors with unmodified LTRs
We first amplified the 2.1-kb CD2 LCR sequence from the VA plasmid11 and cloned it in both orientations downstream of the EGFP cDNA in the LESN retroviral vector (Figure 1). Vectors generated were termed LESN-LCR and LESN-LCRas, and they carried the LCR in the sense or antisense orientation, respectively, relative to theEGFP gene. Retroviral vector–containing supernatants were generated and initially were used to transduce NIH-3T3 cells for determination of the titer of the vector stocks (Table1). Transduction of Molt-3 cells with these vectors, followed by G418 selection, yielded a cell population expressing the EGFP marker at high levels. As shown in Figure2, the pattern of EGFP expression did not differ between cells transduced with the vector carrying the LCR in either orientation compared with the parental LESN retroviral vector; a broad peak of reporter gene expression was detected in either case, as quantitatively indicated by the CV parameter at FACS analysis, which was comparable in the different fluorograms (Figure 2). These findings showed that CD2 LCR did not modulate EGFP expression when this sequence was placed in the context of a retroviral vector carrying full-length LTRs.
Titration of recombinant retroviral vectors carrying CD2 LCR sequences by G418 selection
| Recombinant retroviral construct . | G418-selection (cfu/mL) . |
|---|---|
| LESN | 6.17 × 104 |
| LESN-LCR | 4.60 × 102 |
| LESNΔ | 7.62 × 104 |
| LESNΔ-LCR | 1.18 × 102 |
| LESNΔ-NLCR | 1.02 × 102 |
| LESNΔ-LCR1 | 4.17 × 104 |
| Recombinant retroviral construct . | G418-selection (cfu/mL) . |
|---|---|
| LESN | 6.17 × 104 |
| LESN-LCR | 4.60 × 102 |
| LESNΔ | 7.62 × 104 |
| LESNΔ-LCR | 1.18 × 102 |
| LESNΔ-NLCR | 1.02 × 102 |
| LESNΔ-LCR1 | 4.17 × 104 |
Based on NIH-3T3 cells. Retroviral vectors were produced in 293T cells by cotransfection of gag-polgpt, HCMV-G, and a genomic RNA-encoding vector. The number of neoR cfu/mL was determined after 2-week selection of NIH-3T3 cells in G418-containing medium (500 μg/mL). Average values reported refer to n = 6-8 independent titration experiments.
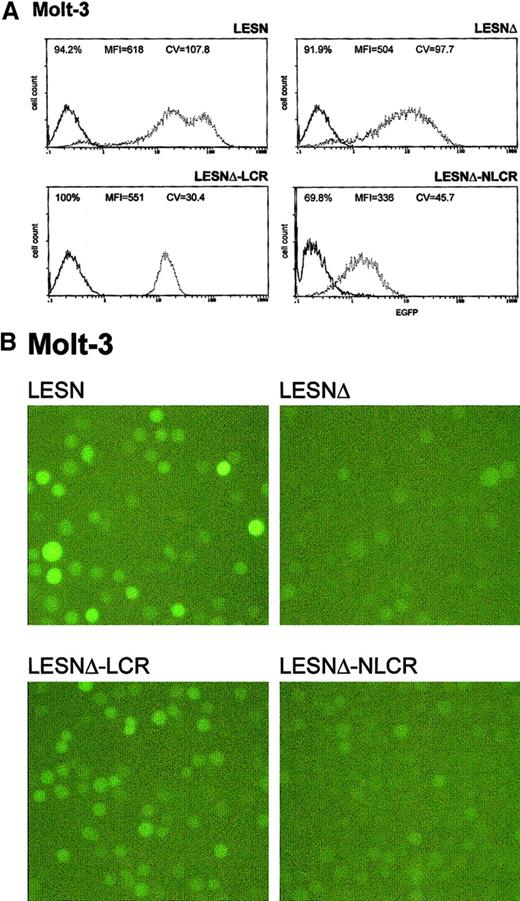
Transduction of the human T-lymphoid cell line Molt-3 with retroviral vectors carrying the CD2 LCR and unmodified LTRs does not change the EGFP expression pattern.
Molt-3 cells were infected with LESN or the LCR-containing retroviral vectors generated by transient transfection of 293T cells and were selected in G418-containing medium for 4 weeks before FACS analysis. LESN-LCR and LESN-LCRas vectors carry the LCR element in the sense or antisense orientation, respectively. Representative patterns of EGFP expression are shown. The percentage of EGFP-positive cells for each construct and MFI are indicated, as is the CV, which describes the profile of the bell-shaped curve. The pattern of the mock-transduced lymphoid cells in each panel is also shown.
Transduction of the human T-lymphoid cell line Molt-3 with retroviral vectors carrying the CD2 LCR and unmodified LTRs does not change the EGFP expression pattern.
Molt-3 cells were infected with LESN or the LCR-containing retroviral vectors generated by transient transfection of 293T cells and were selected in G418-containing medium for 4 weeks before FACS analysis. LESN-LCR and LESN-LCRas vectors carry the LCR element in the sense or antisense orientation, respectively. Representative patterns of EGFP expression are shown. The percentage of EGFP-positive cells for each construct and MFI are indicated, as is the CV, which describes the profile of the bell-shaped curve. The pattern of the mock-transduced lymphoid cells in each panel is also shown.
Generation of CD2 LCR–carrying retroviral vectors with modified LTRs
To investigate LCR function in the absence of the viral enhancer, we generated a modified LESN retroviral vector carrying a deletion in the U3 region of the LTR and termed it LESNΔ (Figure 1B). Preliminary experiments indicated that its EGFP expression in transduced NIH-3T3 cells was strongly attenuated compared with LESN, in terms of MFI (data not shown); this was expected because of the absence of the viral enhancer. We next inserted the 2.1-kb LCR sequence in the LESNΔ vector in the antisense orientation (Figure 1B) and used this vector to transduce lymphoid and nonlymphoid target cells. Furthermore, a 2.0-kb sequence from the human CD2 gene, presumably devoid of LCR activity, was also amplified by PCR from the VA plasmid (Figure 1A), cloned in the LESNΔ vector in the antisense orientation, and used as a control for LCR function in some experiments. The vector carrying this control sequence was termed LESNΔ-NLCR (Figure 1B). Finally, to study whether the full-length LCR was required to exert its activity on gene expression, we constructed a retroviral vector carrying a shorter fragment of the CD2 LCR containing a T-lymphoid–specific enhancer, which was predicted to lack LCR activity, and termed it LESNΔ-LCR1 (Figure 1B).
We first measured the titer of the CD2 LCR–carrying vectors by transduction of NIH-3T3 cells, followed by G418 selection. Deletion of the viral enhancer or the inclusion of a truncated LCR sequence did not reduce viral titer compared with the parental LESN vector; however, all vectors carrying the full-length LCR or the control sequence generated viral titers that were on average 500-fold reduced compared with LESN (Table 1).
Patterns of EGFP expression in lymphoid and nonlymphoid cells
Molt-3 cells transduced by the different recombinant vectors and selected in G418-containing medium were simultaneously analyzed for EGFP expression 4 weeks after gene transfer. The percentage of EGFP+ cells in the different pools ranged from 70% to 100%, with some variability among the experiments and with the recombinant vector used (Table 2); prolonged in vitro culture for an additional 4 weeks did not further increase the fraction of EGFP+ cells, indicating that selection was complete at the time of analysis. We analyzed cells transduced by the LCR-containing vector and observed a unimodal pattern of expression, with MFI at an intermediate level (MFI = 551) between that generated by the LESNΔ vector devoid of LCR (MFI = 504) and the positive control LESN vector (MFI = 618) (Figure3A). The vector carrying the control sequence failed to improve EGFP expression compared with the parental LESNΔ vector; in fact, both percentage of EGFP+ cells and MFI were reduced compared with cells transduced by the parental vector lacking the viral enhancer (LESNΔ), suggesting a negative influence of the NLCR sequence on the expression of the reporter gene (Figure 3A). Changes in the pattern of EGFP expression by LCR-carrying vectors can be quantified by the CV parameter at FACS analysis; this parameter describes the profile of marker gene expression by measuring the width of the bell-shaped curve and, thus, fluorescence homogeneity. The CV value was dramatically reduced in T-lymphoid cells transduced by the LCR-carrying vector (CV = 30.4) compared with LESN-transduced cells (CV = 107.8) (Figure 3A). Similar findings were obtained in 6 independent experiments, which are summarized in Table 2, indicating that the CD2 LCR consistently modulated the EGFP expression pattern in Molt-3 cells. These results were also confirmed by UV microscope observation of the transduced cells; as shown in Figure 3B, the LCR-carrying vector generated a homogeneous fluorescence pattern in transduced Molt-3 cells compared with the highly heterogeneous fluorescence profile generated by the parental LESN vector.
Quantitative analysis of EGFP expression in pools of Molt-3 cells transduced by recombinant retroviral vectors
| Experiment . | LESN . | LESNΔ-LCR . | LESNΔ-NLCR . | LESNΔ . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % . | MFI . | CV . | % . | MFI . | CV . | % . | MFI . | CV . | % . | MFI . | CV . | |
| 1 | 94.2 | 618 | 107.8 | 100 | 551 | 30.4 | 69.8 | 336 | 45.7 | 91.9 | 504 | 97.7 |
| 2 | 96.7 | 678 | 125.2 | 90.9 | 539 | 51.5 | 86.6 | 378 | 54.5 | 94.2 | 501 | 91 |
| 3 | 97.2 | 737 | 114.6 | 98.7 | 500 | 68.2 | 95.5 | 404 | 51.2 | 87.5 | 481 | 99.5 |
| 4 | 95 | 724 | 136.8 | 98.4 | 501 | 72.4 | 97 | 406 | 50.9 | 81.5 | 455 | 91.2 |
| 5 | 98.8 | 882 | 95.3 | 99.1 | 510 | 66.1 | ND | ND | ND | 97.1 | 450 | 78.2 |
| 6 | 90.5 | 651 | 172.8 | 98.4 | 464 | 70.6 | ND | ND | ND | 82.4 | 439 | 86.1 |
| Mean | 95.4 | 715* | 125† | 97.5 | 510* | 59.8† | 87.2 | 381 | 50.5 | 89.1 | 471 | 90.6 |
| SD | 2.9 | 93 | 27.2 | 3.3 | 31 | 16.2 | 12.4 | 32 | 3.6 | 6.3 | 27 | 7.7 |
| Experiment . | LESN . | LESNΔ-LCR . | LESNΔ-NLCR . | LESNΔ . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % . | MFI . | CV . | % . | MFI . | CV . | % . | MFI . | CV . | % . | MFI . | CV . | |
| 1 | 94.2 | 618 | 107.8 | 100 | 551 | 30.4 | 69.8 | 336 | 45.7 | 91.9 | 504 | 97.7 |
| 2 | 96.7 | 678 | 125.2 | 90.9 | 539 | 51.5 | 86.6 | 378 | 54.5 | 94.2 | 501 | 91 |
| 3 | 97.2 | 737 | 114.6 | 98.7 | 500 | 68.2 | 95.5 | 404 | 51.2 | 87.5 | 481 | 99.5 |
| 4 | 95 | 724 | 136.8 | 98.4 | 501 | 72.4 | 97 | 406 | 50.9 | 81.5 | 455 | 91.2 |
| 5 | 98.8 | 882 | 95.3 | 99.1 | 510 | 66.1 | ND | ND | ND | 97.1 | 450 | 78.2 |
| 6 | 90.5 | 651 | 172.8 | 98.4 | 464 | 70.6 | ND | ND | ND | 82.4 | 439 | 86.1 |
| Mean | 95.4 | 715* | 125† | 97.5 | 510* | 59.8† | 87.2 | 381 | 50.5 | 89.1 | 471 | 90.6 |
| SD | 2.9 | 93 | 27.2 | 3.3 | 31 | 16.2 | 12.4 | 32 | 3.6 | 6.3 | 27 | 7.7 |
Retroviral vectors were produced in 293T cells by cotransfection of gag-polgpt, HCMV-G, and a genomic RNA-encoding vector. Molt-3 cells were infected with LESN, LESNΔ, LESNΔ-LCR, and LESNΔ-NLCR retroviral vectors generated by transient transfection of 293T cells and selected in G418-containing medium for 4 weeks before FACS analysis. The percentage of EGFP+ cells for each construct (%), MFI, and CV value are indicated.
ND indicates not determined.
Statistically significant differences (P < .05) according to the Student t test in the MFI values between LESN-transduced and LESNΔ-LCR–transduced cells.
Statistically significant differences (P < .05) according to the Student t test in the CV values between LESN-transduced and LESNΔ-LCR–transduced cells.
Transduction of the human T-lymphoid Molt-3 cells with different retroviral vectors carrying the CD2 LCR and a deletion in the viral enhancer leads to a homogeneous, unimodal pattern of EGFP expression.
(A) Molt-3 cells were infected with LESN, LESNΔ, LESNΔ-LCR, and LESNΔ-NLCR retroviral vectors generated by transient transfection of 293T cells and were selected in G418-containing medium for 4 weeks before FACS analysis. Representative patterns of EGFP expression are shown. The percentage of EGFP+ cells for each construct, MFI, and CV are indicated. The profile of the mock-transduced lymphoid cells in each panel is shown as a continuous line. (B) Representative pattern of EGFP expression in Molt-3 cells transduced with the vectors indicated above each panel. Same cell populations analyzed by FACS and shown in panel A.
Transduction of the human T-lymphoid Molt-3 cells with different retroviral vectors carrying the CD2 LCR and a deletion in the viral enhancer leads to a homogeneous, unimodal pattern of EGFP expression.
(A) Molt-3 cells were infected with LESN, LESNΔ, LESNΔ-LCR, and LESNΔ-NLCR retroviral vectors generated by transient transfection of 293T cells and were selected in G418-containing medium for 4 weeks before FACS analysis. Representative patterns of EGFP expression are shown. The percentage of EGFP+ cells for each construct, MFI, and CV are indicated. The profile of the mock-transduced lymphoid cells in each panel is shown as a continuous line. (B) Representative pattern of EGFP expression in Molt-3 cells transduced with the vectors indicated above each panel. Same cell populations analyzed by FACS and shown in panel A.
To investigate whether these effects on gene expression were specific to Molt-3 cells, we transduced another T-cell line (Jurkat) and NIH-3T3 cells with the different retroviral vectors. As shown in Figure4, infection of Jurkat cells with the LCR-containing vector was associated with a profile of EGFP expression fully comparable to that obtained in Molt-3 cells, and a unimodal pattern of EGFP expression was observed (Figure 4). On the contrary, the CD2 LCR was inactive in non–T cells because the LCR construct generated a similar pattern of EGFP expression in NIH-3T3 cells as the parental vector LESNΔ with a high CV value (Figure 4). Furthermore, the MFI was reduced by one third in NIH-3T3 cells transduced by this vector compared with figures obtained using the LESNΔ vector, thus indicating a possible negative effect of the LCR on gene expression in non–T cells. We also tested the retroviral vectors carrying the LCR or the control element in the opposite orientation with similar findings (data not shown). Therefore, we concluded that LCR activity was conserved in the context of mLV-based retroviral vectors and specific for T lymphoid cells, provided the viral enhancer was removed.
Transduction of human lymphoid Jurkat and murine fibroblastic NIH-3T3 cells with different retroviral vectors carrying the CD2 LCR shows that modulation of EGFP expression is tissue specific.
Cells were infected with LESN, LESNΔ, LESNΔ-LCR, and LESNΔ-NLCR retroviral vectors generated by transient transfection of 293T cells. Four weeks after transduction and selection in G418-containing medium, the percentage of EGFP-expressing cells was quantified by FACS analysis. The percentage of EGFP+ cells for each construct, MFI, and CV are also reported in each panel. Profile of the mock-transduced lymphoid cells is shown as a continuous line.
Transduction of human lymphoid Jurkat and murine fibroblastic NIH-3T3 cells with different retroviral vectors carrying the CD2 LCR shows that modulation of EGFP expression is tissue specific.
Cells were infected with LESN, LESNΔ, LESNΔ-LCR, and LESNΔ-NLCR retroviral vectors generated by transient transfection of 293T cells. Four weeks after transduction and selection in G418-containing medium, the percentage of EGFP-expressing cells was quantified by FACS analysis. The percentage of EGFP+ cells for each construct, MFI, and CV are also reported in each panel. Profile of the mock-transduced lymphoid cells is shown as a continuous line.
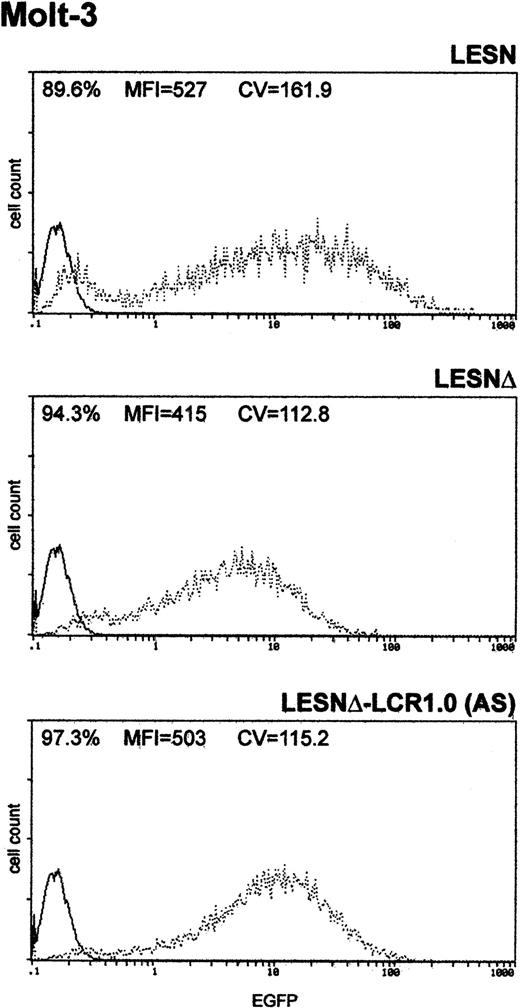
Finally, to determine whether the unimodal pattern of EGFP expression observed required the full-length LCR element, we transduced Molt-3 cells with a retroviral vector carrying a truncated fragment of the LCR sequence, which was expected to lack LCR activity and to retain the enhancer function (Figure 1B). This vector yielded increased EGFP expression compared with the LESNΔ construct, specifically in lymphoid cells (Figure 5); however, a broader pattern of reporter gene expression was observed (CV = 115.2) compared with figures previously generated by cells transduced by the vector carrying the full-length LCR (Figure 3A), thus confirming the lack of LCR activity in cells transduced by this construct.
Transduction of the human T-lymphoid cell line Molt-3 with a retroviral vector carrying a truncated CD2 LCR sequence indicates that LCR activity requires the full-length 2.1-kb element.
Molt-3 cells were infected with LESN, LESNΔ, and LESNΔ-LCR1 retroviral vectors generated by transient transfection of 293T cells. Four weeks after transduction and selection in G418-containing medium, the percentage of EGFP-expressing cells was quantified by FACS analysis. The percentage of EGFP+ cells for each construct, MFI, and CV are indicated. Profile of the mock-transduced lymphoid cells in each panel is shown as a continuous line.
Transduction of the human T-lymphoid cell line Molt-3 with a retroviral vector carrying a truncated CD2 LCR sequence indicates that LCR activity requires the full-length 2.1-kb element.
Molt-3 cells were infected with LESN, LESNΔ, and LESNΔ-LCR1 retroviral vectors generated by transient transfection of 293T cells. Four weeks after transduction and selection in G418-containing medium, the percentage of EGFP-expressing cells was quantified by FACS analysis. The percentage of EGFP+ cells for each construct, MFI, and CV are indicated. Profile of the mock-transduced lymphoid cells in each panel is shown as a continuous line.
Southern blot analysis of transduced cells
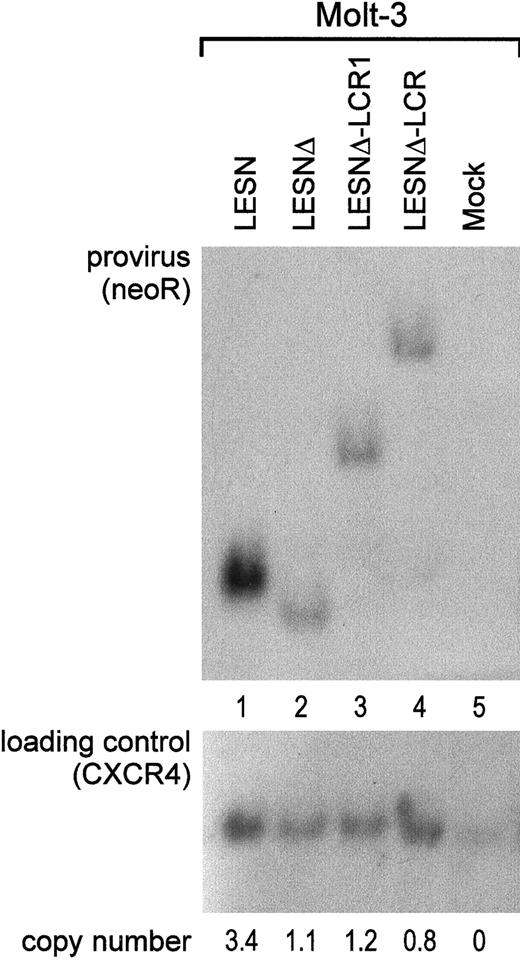
To investigate whether the different patterns of EGFP expression observed in transduced cells might depend on nonspecific provirus rearrangements, genomic DNA samples from pools of transduced cells after G418 selection were analyzed by Southern blotting. We found that the CD2 LCR–carrying vector was not rearranged in Molt-3 transduced cells; digestion with KpnI restriction enzyme yielded a 5.5-kb band (Figure 6A, lane 4) that exactly matched the expected length of an unrearranged provirus. Genomic DNA from LESN-, LESNΔ-, and LESNΔ-LCR1–transduced cells digested with KpnI and hybridized to the same probe yielded bands of 3.6 kb, 3.4 kb, and 4.4 kb, respectively, corresponding to the calculated sizes of the vectors (Figure 6, lanes 1-2). Furthermore, quantitative Southern blot analysis demonstrated only limited variations in the relative number of provirus copies per genome in each sample (Figure 6).
Quantification of gene transfer rates and analysis of proviral DNA structure by Southern blot analysis of transduced Molt-3 cells.
DNA was isolated from bulk cultures of vector-transduced Molt-3 cells in the presence of G418 selection, digested with KpnI that cuts once in each LTR, and analyzed by Southern blotting. The blot was first hybridized with a probe for neoR (upper panel), stripped, and rehybridized with a loading control probe, human CXCR4 (lower panel). Band intensities were quantified with an InstantImager (Packard) and were used to calculate the relative number of provirus copies per genome in each sample, which is indicated. DNA from untransduced Molt-3 cells was used as control (mock).
Quantification of gene transfer rates and analysis of proviral DNA structure by Southern blot analysis of transduced Molt-3 cells.
DNA was isolated from bulk cultures of vector-transduced Molt-3 cells in the presence of G418 selection, digested with KpnI that cuts once in each LTR, and analyzed by Southern blotting. The blot was first hybridized with a probe for neoR (upper panel), stripped, and rehybridized with a loading control probe, human CXCR4 (lower panel). Band intensities were quantified with an InstantImager (Packard) and were used to calculate the relative number of provirus copies per genome in each sample, which is indicated. DNA from untransduced Molt-3 cells was used as control (mock).
Lack of unspecific rearrangements was also confirmed by PCR amplification of the same DNA samples with EGFP-specific primers and primers binding to different sites of the LTR of the vectors (data not shown). Overall, these experiments indicated that retroviral vectors carrying CD2 LCR sequences are genetically stable and do not undergo unwanted rearrangements in transduced cells.
Evaluation of LCR activity by analysis of EGFP expression in single clones
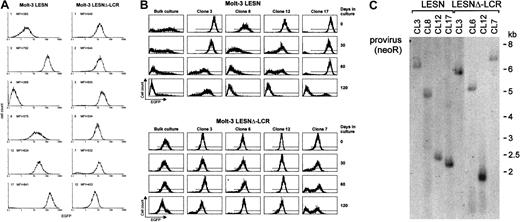
To confirm the changes in EGFP expression profile observed in bulk cultures of T lymphoid cells at the level of single cells, Molt-3 cells transduced by either the LESN or the LESNΔ-LCR retroviral vectors were cloned by limiting dilution; reporter gene expression was analyzed by FACS at different time points and in the absence of G418 selective pressure. As shown in Figure 7A, LESN-transduced clones showed heterogeneous levels of EGFP expression that ranged from very high (as seen in clones 3 and 17) to intermediate (as in clones 8 and 12) and low or very low levels (as seen in clones 1 and 4). On the other hand, 6 of 6 analyzed clones of LESNΔ-LCR–transduced Molt-3 cells expressed EGFP at intermediate levels, with a limited degree of interclonal variation of gene expression (Figure 7A). When the transduced cell lines and the clones derived from them were cultured for 120 days in the absence of G418 and were checked for EGFP expression at regular intervals by FACS (Figure7B), the transgene expression progressively declined over time in LESN-transduced bulk cultures (Figure 7B). In addition, LESN-transduced clones showed down-regulation of EGFP expression with time; in some clones, reporter gene expression became heterogeneous after 30 to 60 days and was reduced to almost background levels by day 120 (Figure7B). On the contrary, LESNΔ-LCR–transduced Molt-3 cells and most clones maintained EGFP expression in vitro over the entire course of the experiment (120 days) (Figure 7B), thus showing that CD2 LCR confers an advantage in terms of duration of gene expression in vitro, in the absence of selective pressure. Southern blot analysis of these clones showed that they carried only one randomly integrated copy of the vector genome (Figure 7C).
Homogeneous and long-term expression of the EGFP reporter gene by independent clones of Molt-3 cells transduced by the retroviral vector carrying the CD2 LCR.
(A) Target cells were transduced with the LESN or the LESNΔ-LCR retroviral vectors generated by transient transfection of 293T cells and selected in G418-containing medium for 4 weeks. After G418 selection, clones of both cultures were obtained by plating at low density (0.3 cells/well) and were analyzed for EGFP expression. MFI for each clone is indicated. (B) Long-term analysis of EGFP expression in vitro in the absence of G418 selective pressure by bulk cultures and representative LESN- or LESNΔ-LCR–transduced Molt-3 clones. (C) Southern blot analysis of proviral DNA shows independent and unique integration sites. Genomic DNA isolated from the clones was digested with XhoI/HindIII, which cut flanking cellular sequences and (once) the proviral DNA, upstream of the neoRgene, and was analyzed by Southern blotting with a probe for neoR.
Homogeneous and long-term expression of the EGFP reporter gene by independent clones of Molt-3 cells transduced by the retroviral vector carrying the CD2 LCR.
(A) Target cells were transduced with the LESN or the LESNΔ-LCR retroviral vectors generated by transient transfection of 293T cells and selected in G418-containing medium for 4 weeks. After G418 selection, clones of both cultures were obtained by plating at low density (0.3 cells/well) and were analyzed for EGFP expression. MFI for each clone is indicated. (B) Long-term analysis of EGFP expression in vitro in the absence of G418 selective pressure by bulk cultures and representative LESN- or LESNΔ-LCR–transduced Molt-3 clones. (C) Southern blot analysis of proviral DNA shows independent and unique integration sites. Genomic DNA isolated from the clones was digested with XhoI/HindIII, which cut flanking cellular sequences and (once) the proviral DNA, upstream of the neoRgene, and was analyzed by Southern blotting with a probe for neoR.
Effects of the LCR on gene expression in vivo
To study expression patterns in vivo, Molt-3 cells were transduced in vitro with either LESN or LESNΔ-LCR retroviral vectors, selected in G418-containing medium for 4 weeks, and implanted subcutaneously into SCID mice. Analysis of EGFP expression in the bulk culture at the time of implantation revealed similar percentages of EGFP+ cells (88.3% vs 91.6%) in LESN- or LESNΔ-LCR–transduced cells (Figure 8); not surprisingly, however, in view of above data (Figures 3, 4), LESN-transduced cells expressed EGFP at higher levels than LESNΔ-LCR–transduced cells, as indicated by the different MFI values generated by the 2 cell populations (MFI = 604 vs MFI = 508), but with a less homogeneous profile, as indicated by the CV value (112.0 vs 80.5). After in vivo growth of Molt-3 cells for 30 days, the animals were killed, and transduced cells were recovered and analyzed by FACS for human HLA class 1 expression and EGFP expression. The HLA antigen, expressed by this lymphoid cell line, was used as a marker of human origin to restrict analysis of EGFP expression to Molt-3 cells and to verify that expression of a cellular gene did not undergo changes after in vivo growth of the cells. EGFP expression was strongly reduced in LESN-transduced cells compared with preimplantation levels; indeed, only 11.2% of the HLA class I+ cells expressed detectable levels of the reporter gene, and the MFI was also greatly reduced compared with in vitro levels (MFI = 419 vs MFI = 604) (Figure 8). Conversely, in the case of LESNΔ-LCR–transduced cells, 72.4% of the Molt-3 cells expressed EGFP at intensity levels comparable to preimplantation figures (MFI = 539 vs MFI = 508) (Figure 8). A more heterogeneous pattern of EGFP expression, as evaluated by the CV parameter, was observed in the samples analyzed after in vivo passage compared with figures generated by Molt-3 cells transduced by the same vector and exclusively cultured in vitro; this may suggest that LCR in vivo might only partially shield the transgene from silencing. Interestingly, the down-modulation of gene expression in vivo, as observed with LESN-transduced cells, was restricted to the transgene, because no changes were detected in the level of expression of the human HLA molecule on recovered Molt-3 cells compared with preimplantation levels (Figure 8). Thus, we concluded that the CD2 LCR could at least partially maintain its functions, also in vivo, by preventing or slowing transcriptional silencing of the transgene.
CD2 LCR partially shields transplanted cells by silencing in vivo.
The human T lymphoid cell line Molt-3 was transduced with the LESN or the LESNΔ-LCR retroviral vectors generated by transient transfection of 293T cells and was selected in G418-containing medium for 4 weeks. At that time, the percentage of HLA class I+and of EGFP-expressing cells was quantified by FACS analysis (in vitro panels). Subsequently, vector-transduced cells were subcutaneously implanted in SCID mice and were grown for 4 weeks before recovery and measurement of HLA-I and EGFP expression (ex vivo panels). The percentage of positive cells for each marker, MFI, and CV of the analyzed cell population are indicated.
CD2 LCR partially shields transplanted cells by silencing in vivo.
The human T lymphoid cell line Molt-3 was transduced with the LESN or the LESNΔ-LCR retroviral vectors generated by transient transfection of 293T cells and was selected in G418-containing medium for 4 weeks. At that time, the percentage of HLA class I+and of EGFP-expressing cells was quantified by FACS analysis (in vitro panels). Subsequently, vector-transduced cells were subcutaneously implanted in SCID mice and were grown for 4 weeks before recovery and measurement of HLA-I and EGFP expression (ex vivo panels). The percentage of positive cells for each marker, MFI, and CV of the analyzed cell population are indicated.
Modulation of EGFP expression by the CD2 LCR in primary T lymphocytes
To evaluate the activity of the LCR in primary T cells, we attempted to transduce them with the different retroviral vectors used in this study. However, because of the poor titer of the LCR-carrying vector, we could not recover EGFP-expressing T cells using the Mo-MLV–based retroviral vectors. Therefore, we switched to lentiviral vectors that have been reported to transduce various types of primary cells, including T lymphocytes, with high efficiency and that might accept LCR elements with reduced loss of titers compared with retroviral vectors.26 Various recombinant EGFP-expressing lentiviral vectors carrying the CD2 LCR upstream of an internal MLV promoter were, therefore, generated (Figure9A) and used to transduce anti-CD3–activated T cells. Preliminary gene transfer experiments on NIH-3T3 cells disclosed that all recombinant vectors generated infectious particles and that high-titer stocks of the LCR-containing vectors could be obtained (Figure 9A). Gene transfer in primary T cells was followed by cytofluorometric analysis of the transduced cells for CD3 and EGFP expression (Figure 9B). All vectors transduced theEGFP gene in T cells, and EGFP expression was detected in 0.1% to 16.7% of cells, depending on the vector used and on the blood donor; results of 5 independent experiments are listed in Table3. The CD2 LCR did not significantly modify the pattern of EGFP expression when placed upstream of the intact MLV LTR within the lentiviral vector, as observed with retroviral vector–transduced lymphoid cells (Figure 9B, panels LTR and LTR-LCR). Furthermore, the lentiviral vector carrying a deletion in the viral enhancer showed greatly reduced EGFP expression in the target cells compared with the parental vector with intact promoter-enhancer sequences (Figure 9B, panels Δ and LTR). Inclusion of the CD2 LCR 5′ upstream of the enhancer-deleted MLV LTR was associated with an increase in EGFP expression, measured by the MFI value, mainly resulting from a reduction in the EGFP+ fraction expressing the marker at arbitrarily defined low levels (Figure 9B, panels Δ and Δ-LCR). This phenomenon was observed in all experiments and was translated into more homogeneous EGFP expression in the transduced T lymphocytes.
Lentiviral vector-mediated gene transfer of LCR-containing vectors in human T lymphocytes.
(A) SIV-based lentiviral vectors carrying EGFP as the reporter gene are schematically shown; details on vector construction are available on request. The 2.1-kb LCR element was cloned in the antisense orientation in lentiviral vectors carrying either the parental MLV LTR (LTR) or an LTR lacking the viral enhancer (Δ) as the internal promoter. The titer of each vector, measured as the transducing unit (TU) per milliliter viral supernatant on NIH-3T3 cells, is indicated. (B) Anti-CD3–activated human T lymphocytes were transduced with the set of lentiviral vectors shown in panel A and were analyzed for CD3 and EGFP expression 72 hours later by FACS. Inclusion of the CD2 LCR 5′-upstream of the enhancer-deleted MLV LTR was associated with a relative reduction of the EGFP+ fraction expressing the marker at arbitrarily defined low levels (left box, quadrant 2) compared with cells expressing EGFP at intermediate levels (right box, quadrant 2) (panels Δ and Δ-LCR). This phenomenon was detected, but less markedly, in cells transduced by the lentiviral vectors carrying the unmodified LTR (panels LTR and LCR-LTR). Percentage values relative to each box and MFI of the entire EGFP+ cell population are indicated. (C) Effects of CD2 LCR on long-term gene expression. Anti-CD3–activated human T lymphocytes were transduced with either the parental (LTR) or the LCR-containing (Δ-LCR) vector and were sorted by FACS. EGFP expression was analyzed by FACS either immediately after sorting (day 0 panels) or after 6-week in vitro culture of the T lymphocytes in the presence of IL-2 (100 U/mL). Down-modulation of EGFP expression was detected in both samples.
Lentiviral vector-mediated gene transfer of LCR-containing vectors in human T lymphocytes.
(A) SIV-based lentiviral vectors carrying EGFP as the reporter gene are schematically shown; details on vector construction are available on request. The 2.1-kb LCR element was cloned in the antisense orientation in lentiviral vectors carrying either the parental MLV LTR (LTR) or an LTR lacking the viral enhancer (Δ) as the internal promoter. The titer of each vector, measured as the transducing unit (TU) per milliliter viral supernatant on NIH-3T3 cells, is indicated. (B) Anti-CD3–activated human T lymphocytes were transduced with the set of lentiviral vectors shown in panel A and were analyzed for CD3 and EGFP expression 72 hours later by FACS. Inclusion of the CD2 LCR 5′-upstream of the enhancer-deleted MLV LTR was associated with a relative reduction of the EGFP+ fraction expressing the marker at arbitrarily defined low levels (left box, quadrant 2) compared with cells expressing EGFP at intermediate levels (right box, quadrant 2) (panels Δ and Δ-LCR). This phenomenon was detected, but less markedly, in cells transduced by the lentiviral vectors carrying the unmodified LTR (panels LTR and LCR-LTR). Percentage values relative to each box and MFI of the entire EGFP+ cell population are indicated. (C) Effects of CD2 LCR on long-term gene expression. Anti-CD3–activated human T lymphocytes were transduced with either the parental (LTR) or the LCR-containing (Δ-LCR) vector and were sorted by FACS. EGFP expression was analyzed by FACS either immediately after sorting (day 0 panels) or after 6-week in vitro culture of the T lymphocytes in the presence of IL-2 (100 U/mL). Down-modulation of EGFP expression was detected in both samples.
Quantitative analysis of EGFP expression in human T lymphocytes transduced by lentiviral vectors
| Experiment . | LTR . | LTR-LCR . | Δ . | Δ-LCR . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % . | MFI . | CV . | % . | MFI . | CV . | % . | MFI . | CV . | % . | MFI . | CV . | |
| 1 | 1.8 | 657 | 168 | 0.3 | 669 | 164 | 1.3 | 512 | 105 | 0.2 | 609 | 119 |
| 2 | 16.7 | 674 | 156 | 0.1 | 689 | 202 | 1.84 | 549 | 100 | 0.2 | 622 | 133 |
| 3 | 2.3 | 663 | 132 | 0.1 | 657 | 135 | 0.7 | 549 | 88 | 0.1 | 589 | 84 |
| 4 | 1.6 | 660 | 176 | 0.3 | 735 | 194 | 2.8 | 564 | 113 | 0.3 | 645 | 135 |
| 5 | 4.5 | 750 | 155 | ND | ND | ND | 1.1 | 600 | 92 | 0.5 | 654 | 87 |
| Mean | 5.38 | 6813-150 | 157 | 0.20 | 688 | 174 | 1.55 | 5553-151 | 100 | 0.26 | 6243-150,3-151 | 112 |
| SD | 6.43 | 39.2 | 16.7 | 0.12 | 34.3 | 30.6 | 0.81 | 31.7 | 10 | 0.15 | 26.4 | 24.6 |
| Experiment . | LTR . | LTR-LCR . | Δ . | Δ-LCR . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % . | MFI . | CV . | % . | MFI . | CV . | % . | MFI . | CV . | % . | MFI . | CV . | |
| 1 | 1.8 | 657 | 168 | 0.3 | 669 | 164 | 1.3 | 512 | 105 | 0.2 | 609 | 119 |
| 2 | 16.7 | 674 | 156 | 0.1 | 689 | 202 | 1.84 | 549 | 100 | 0.2 | 622 | 133 |
| 3 | 2.3 | 663 | 132 | 0.1 | 657 | 135 | 0.7 | 549 | 88 | 0.1 | 589 | 84 |
| 4 | 1.6 | 660 | 176 | 0.3 | 735 | 194 | 2.8 | 564 | 113 | 0.3 | 645 | 135 |
| 5 | 4.5 | 750 | 155 | ND | ND | ND | 1.1 | 600 | 92 | 0.5 | 654 | 87 |
| Mean | 5.38 | 6813-150 | 157 | 0.20 | 688 | 174 | 1.55 | 5553-151 | 100 | 0.26 | 6243-150,3-151 | 112 |
| SD | 6.43 | 39.2 | 16.7 | 0.12 | 34.3 | 30.6 | 0.81 | 31.7 | 10 | 0.15 | 26.4 | 24.6 |
Lentiviral vectors were produced in 293T cells by the triple-transfection protocol. Anti-CD3-activated human T lymphocytes were infected with LTR, LTR-LCR, Δ, and Δ-LCR lentiviral vectors and analyzed by FACS 72 hours after transduction. The percentage of EGFP+ cells for each construct (%), MFI, and CV are indicated.
ND indicates not determined.
Statistically significant differences (P < .05) according to the Student t test in the MFI values between LESN-transduced and LESNΔ-LCR–transduced cells.
Statistically significant differences (P < .05) according to the Student t test in the MFI values between LESNΔ-transduced and LESNΔ-LCR–transduced cells.
To determine the effects of the CD2 LCR on long-term expression of the reporter gene in primary cells, we FACS sorted the EGFP+ T cells transduced by either the LCR-containing vector or the LTR vector and analyzed EGFP expression on the sorted population after 6-week in vitro culture. As shown in Figure 9C, this experiment indicated that both vectors underwent a similar degree of down-modulation of EGFP expression in long-term culture, independent of the presence of the CD2 LCR. Thus, we concluded that the CD2 LCR up-modulated gene expression in primary T cells but could not block the decline in EGFP expression in long-term in vitro cultures.
Discussion
One of the major drawbacks of current gene transfer procedures is that gene expression is often observed only transiently because of transcriptional silencing of the transgene, even though stable gene delivery can be achieved, at least with some viral systems, including retroviral and lentiviral vectors.27 This limitation has long been recognized, and possible solutions to the problem have been suggested, including replacement of viral with cellular enhancers, inclusion in the vector of LTRs from viruses that show a decreased propensity to be silenced in stem cells, such as the murine stem cell virus,28 and insertion of LCRs, matrix attachment sites, and insulators.29-31 The discovery of β-globin LCR sequences greatly improved the outlook for gene therapy of human hemoglobinopathies32 by suggesting that their insertion in retroviral vectors might lead to a dominant chromatin opening and thus shield a linked gene from silencing.33
We selected the human CD2 LCR, a sequence that has been studied in detail8-10 and that is potentially useful for improving gene expression in T-lymphoid cells. Kaptein et al34 also focused on the CD2 LCR and found that it was incapable of generating position-independent expression of the adenosine deaminase transgene delivered by retroviral vectors, but retroviral vectors with unmodified LTRs were used in this study. We observed that a deletion of the viral enhancer might be required to detect LCR activity in this context, and this finding might explain the discrepancies between the 2 studies. Our finding is also strengthened by previous work in transgenic mice indicating that LCR activity might be impaired by retroviral LTRs because of silencer elements.35 36
The main finding of our study is that the CD2 LCR sequence is able to modulate gene expression specifically in T cells, and this translated into a homogeneous, unimodal pattern of EGFP expression in cells transduced by retroviral vectors carrying the LCR compared with controls (Figures 3, 4). A T-cell–specific increase in EGFP expression, in terms of MFI, without significant changes in the pattern of expression was also observed in T cells transduced by a retroviral vector carrying a shortened sequence that maintained the CD2 enhancer but was devoid of LCR activity (Figure 5). However, different figures generated by vectors carrying full-length or shortened LCR sequences might not be directly comparable because of the different design of the 2 vectors. Both the LCR and the enhancer effects were tissue specific—they were found in T-lymphoid cell lines and not in other cell types, such as fibroblastic cells. Despite the use of a selection step that might eliminate silent or unfavorable integration sites, expression among clones varied greatly (Figure 7) and allowed us to study CD2 LCR modulation of transgene expression. However, because of the skewing of G418-selected cell populations for permissive sites, other experiments are needed to evaluate whether the CD2 LCR can contribute to increase the overall rate of transgene expression from unselected integration sites.
We observed that Molt-3 cells transduced by the LCR-containing vector had a lower MFI than cells transduced by the LESN retroviral vector. We attributed this to the fact that EGFP is driven by a cellular enhancer in the former and by a viral one in the latter. This might restrict the exploitation of this vector for gene therapy to pathologic conditions in which even intermediate levels of transgene expression suffice for a therapeutic outcome. In this regard, it is still unknown whether homogeneous expression of the therapeutic gene in transduced cells, albeit at intermediate levels, would be preferable to heterogeneous expression at higher levels.
In vivo experiments, designed to test whether modulation in the pattern of EGFP expression observed in vitro would hold in vivo, disclosed that lymphoid cells transduced by a standard retroviral vector underwent marked reduction in EGFP expression in vivo, as predicted on the basis of previous studies with other cell types, including transduced skin fibroblasts.37 Interestingly, this reduction was specific to the transgene because the endogenous human HLA class I gene was not down-modulated in the same cells. This emphasizes the differences in gene expression between virally transduced and physiologically expressed cellular genes in our system. Strikingly, the CD2 LCR could prevent transgene silencing in most of the implanted lymphoid cells; the percentage of EGFP+ cells and the MFI were similar to those observed in cultured cells. However, EGFP was expressed with a broad range of intensity by ex vivo recovered cells, as opposed to the unimodal pattern of expression observed in cultured cells, thus suggesting that LCR-mediated shielding from silencing in vivo might be incomplete.
A limitation to the exploitation of the retroviral vectors containing LCR sequences is clearly their low titer. The reduction in titer, at least in part, could depend on the increased vector size after the insertion of long sequences (2 kb) from the CD2 locus; indeed, the titer of the control vector, which does not carry any LCR and is approximately the same size as LESNΔ-LCR, is similar to that of the LCR-carrying vector. Furthermore, we cannot rule out that these DNA sequences might encompass regions that are not stable as RNA molecules. A decrease in the titer of viral vectors on inclusion of LCRs was also observed by others.6 7
Inclusion of the CD2 LCR sequence in lentiviral vectors was followed by a less dramatic reduction in vector titer, and it was possible to use lentiviral vectors to transfer the CD2 LCR in primary T cells and to detect its effects on the EGFP expression pattern in the absence of G418 selection. The reason for the difference in performance of retroviral and lentiviral vectors carrying LCR sequences is unknown, but it has been observed by others.26 In our study, titration of the vectors by a real-time PCR assay targeted at theEGFP gene indicated similar amounts of proviral DNA in NIH-3T3 cells transduced by the LCR-containing retroviral and the lentiviral vector (data not shown). This suggests that the 2 vectors might be generated at similar levels by the packaging cells but that they differ in either ability to integrate the proviral DNA or to express the transgene.
Inclusion of the CD2 LCR in lentiviral vectors led to a homogeneous pattern of EGFP expression in the primary T cells, but it did not counteract the down-modulation of reporter gene expression in long-term cultures, as we observed with the clones derived from leukemia cells. This could in principle represent a problem for the exploitation of these vectors in a clinical setting. However, the in vitro assay used might have some limitations in relevance to the in vivo behavior of the genetically modified cells. Thus, the issue of long-term expression in primary cells will likely be definitively addressed only by future studies in a murine transplantation model.
We thank A. Bank for providing gag-polgpt, D. Kioussis for the VA hCD2 plasmid, and K. Uzbela and R. Wagner for ViGΔBH and Hgp plasmids. We also thank P. Gallo for artwork and Ms. P. Segato for help in preparing the manuscript.
Supported in part by Telethon (grant A-126), ISS-AIDS Project, MURST 60% and 40%, Associazione Italiana per la Ricerca sul Cancro, Fondazione Italiana per la Ricerca sul Cancro, Fondazione Città della Speranza, and National Research Council (PF Biotechnology). S.M. is the recipient of a fellowship from Fondazione Italiana per la Ricerca sul Cancro. V.T. is a recipient of an AIRC fellowship.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Stefano Indraccolo, IST-Viral and Molecular Oncology Section and Department of Oncology and Surgical Sciences, University of Padua, Via Gattamelata, 64-35128 Padua, Italy; e-mail: stefano.indraccolo@unipd.it.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal