Abstract
The TEL/PDGFβR gene, which encodes a fusion protein containing the ETS-family member TEL fused to the protein-tyrosine kinase domain of the platelet-derived growth factor receptor-β (PDGFβR), confers interleukin 3 (IL-3)–independent growth on Ba/F3 hematopoietic cells. TEL/PDGFβR mutants have been generated that contain tyrosine-to-phenylalanine (Tyr→Phe) substitutions at phosphorylation sites present in the native PDGFβR to assess the role of these sites in cell transformation by TEL/PDGFβR. Similar to previous findings in a murine bone marrow transplantation model, full transformation of Ba/F3 cells to IL-3–independent survival and proliferation required the TEL/PDGFβR juxtamembrane and carboxy terminal phosphorylation sites. In contrast to previous reports concerning comparable mutants in the native PDGFβR, each of the TEL/PDGFβR mutants is fully active as a protein-tyrosine kinase. Expression of the TEL/PDGFβR fusion protein causes hyperphosphorylation and activation of signal transducer and activator of transcription (STAT5), and this activation of STAT5 requires the juxtamembrane Tyr579 and Tyr581 in the TEL/PDGFβR fusion. Hyperphosphosphorylation of phospholipase Cγ (PLCγ) and the p85 subunit of phosphatidylinositol 3-kinase (PI3K) requires the carboxy terminal tyrosine residues of TEL/PDGFβR. Thus, full transformation of Ba/F3 cells by TEL/PDGFβR requires engagement of PI3K and PLCγ and activation of STAT5. Taken together with the growth properties of cells transformed by the TEL/PDGFβR variants, these findings indicate that a minimal combination of these signaling intermediates contributes to hematopoietic transformation by the wild-type TEL/PDGFβR fusion.
Introduction
Chronic myelomonocytic leukemia (CMML) is characterized by clonal proliferation of myeloid cells and frequent progression to acute leukemia. A recurring cytogenetic abnormality in CMML is the t(5;12)(q33;p13) chromosomal translocation. The resulting gene rearrangement fuses the 5′ region of TEL to the gene encoding the platelet-derived growth factor receptor-β (PDGFβR).1 TEL is a member of the ETS family of transcription factors, and it consists of an amino terminal Pointed domain (PNT) and a carboxy terminal DNA-binding domain that shares sequence homology with the winged helix-turn-helix motif. In the TEL/PDGFβR fusion protein, the amino terminal region of TEL (which contains the PNT domain) is fused to the membrane-spanning segment and the entire cytoplasmic protein-tyrosine kinase domain of PDGFβR. The PNT domain present in the TEL segment mediates homotypic oligomerization of the fusion protein.2,3 Fusion of TEL sequences to PDGFβR results in constitutive oligomerization and activation of protein-tyrosine kinase activity2; numerous examples of receptor tyrosine kinase activation through homotypic oligomerization have been described (reviewed by Lemmon and Schlessinger4).
The Ba/F3 murine hematopoietic cell line, which requires interleukin-3 (IL-3) for survival,5 can be transformed to IL-3 independence by the TEL/PDGFβR fusion.2 TEL/PDGFβR is constitutively phosphorylated on tyrosine residues, and a point mutation corresponding to a kinase-inactivating mutation in the native PDGFβR abrogates TEL/PDGFβR kinase activation and transformation of Ba/F3 cells. Moreover, the PDGFβR tyrosine kinase inhibitor CGP 571486inhibits TEL/PDGFβR kinase activity in vitro, inhibits autophosphorylation in vivo, and abrogates proliferation of Ba/F3 cells transformed by TEL/PDGFβR.7 Thus, hematopoietic transformation by TEL/PDGFβR requires protein-tyrosine kinase activity.
Similar structure-function relationships have been described for the HIP1/PDGFβR fusion8,9 associated with the t(5;7)(q33;q11.2) translocation, and the H4/PDGFβR fusion that is expressed as a consequence of the t(5;10)(q33;q22) translocation.10 Each of these PDGFβR fusions is associated with a phenotype of CMML in humans, but contains a different amino terminal fusion partner and oligomerization motif that serves to activate the PDGFβR kinase activity. These observations suggest that the critical event in the pathogenesis of CMML is the activation of the PDGFβR kinase activity and the subsequent activation of downstream signaling pathways in hematopoietic cells.
Multiple signaling pathways are engaged following ligand activation of the native PDGFβR.11-13 The native PDGFβR autophosphorylates on multiple tyrosine residues that provide docking sites for signaling proteins through their respective SH2 phosphotyrosine-binding domains. Interaction with such intermediates as Src family members, phosphatidylinositol 3-kinase (PI3K), Ras GTPase-activating protein (RasGAP), phospholipase C-γ (PLCγ), the Grb2 small adaptor molecule, and the SHP-2 tyrosine phosphatase have been implicated in signaling by the activated PDGFβR. Moreover, PDGF has been shown previously to induce phosphorylation of multiple STAT proteins and JAK kinases.14 The signal transducers and activators of transcription (STATs) comprise a family of cytoplasmic proteins that are hyperphosphorylated in response to mitogen stimulation; they dimerize, translocate to the nucleus, and serve as transcriptional activators.15 Oncogenic fusion proteins such as P210 and P190BCR/ABL,16TEL/JAK2 fusion variants,17-19 and TEL/PDGFβR20 are each able to cause constitutive STAT signaling.
The TEL/PDGFβR fusion protein might support hematopoietic cell survival and proliferation through pathways similar to those used by the native PDGFβR. We constructed a series of tyrosine-to-phenylalanine (Tyr→Phe) mutants in TEL/PDGFβR at positions corresponding to major autophosphorylation sites in PDGFβR. Although each of the TEL/PDGFβR variants was able to transform Ba/F3 cells to IL-3–independent growth and survival, there was a marked prolongation in disease latency in a murine bone marrow transplant (BMT) assay with increasing Tyr→Phe substitution.21 In striking contrast to our findings using the in vitro Ba/F3 culture system, the ability of TEL/PDGFβR to cause a myeloproliferative disease in the murine BMT assay was markedly attenuated by substitution at the juxtamembrane phosphorylation sites, and bone marrow transduced with these mutants yielded a lymphoproliferative disease rather than myeloid leukemia. We were puzzled by the disparity in these results, and we further characterized the transformation of hematopoietic cells in vitro to better understand the role of individual signaling pathways in each of these model systems. Here we show that phosphorylation of both the juxtamembrane and carboxy terminal tyrosine residues in TEL/PDGFβR is necessary for full transformation in the Ba/F3 assay, and these results are consistent with our previous observations in the murine BMT model.21 Moreover, the juxtamembrane tyrosines Tyr579 and Tyr581 in TEL/PDGFβR are necessary for full activation of STAT5, whereas the carboxy terminal tyrosines are necessary for engagement of PLCγ and PI3K. Taken together, these findings indicate that activation of STAT5 and engagement of PLCγ and PI3K are necessary for full transformation of IL-3–independent Ba/F3 cells.
Materials and methods
Cell culture and retroviral infections
The murine precursor line Ba/F3 was kindly provided by Alan D'Andrea (Dana-Farber Cancer Institute, Boston, MA). The murine myeloid line 32D was a gift from Dong-Er Zhang (Beth Israel-Deaconess Medical Center, Boston, MA). Ba/F3 and 32D cells transformed with MSCV constructs were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum, 1.0 ng/mL of recombinant IL-3 (R & D Systems, Minneapolis, MN), and 1.0 mg/mL G418 in a 5% CO2 incubator at 37°C. Cells were passaged when they reached a density of approximately 0.5 to 1 × 106/mL. For growth curve assays, Ba/F3 cells were infected with each of the MSCV retroviral constructs and were grown in the presence of IL-3 for 2 days. G418 selection was performed for an additional 15 days in the presence of IL-3. Cells were washed and resuspended in medium containing G418 without IL-3. These populations were maintained at a density of 1 × 105 to 1 × 106/mL.
DNA constructs
The TEL/PDGFβR variants were constructed using native PDGFβR F-series mutants described previously.22 The 2.4-kb TEL/PDGFβR complementary DNA (cDNA) was subcloned into the multicloning site of the MSCVneoEB retroviral vector containing a modified murine Maloney leukemia virus long terminal repeat (LTR), and a neomycin-resistance cassette (provided by R. Hawley, Red Cross, Rockville, MD). TEL/PDGFβR Tyr→Phe mutants were generated by subcloning F2, F5, and F7 PDGFβR mutants (provided by A. Kazlauskas, Boston, MA) into the TEL/PDGFβR background.
Protein extracts and immunoprecipitations
The Ba/F3 cells were grown to a density of approximately 1 × 106/mL. Cells were collected by centrifugation and washed in 5 to 10 mL phosphate-buffered saline (PBS) at 4°C. Cells were lysed in 1.0 mL lysis buffer: 20 mM Tris-HCl, pH 7.4, 1% Triton X-100, 0.5 mM EDTA, 150 mM NaCl, 1 mM Na3VO4,25 mM NaF, 10% glycerol, and complete protease inhibitor cocktail (Roche, Indianapolis, IN). Lysates were incubated for 5 minutes at 4°C and were then cleared by centrifugation at 14 000gfor 10 minutes at 4°C. For the assessment of STAT5 phosphorylation in the 32D cell line, cells were washed with PBS supplemented with 0.4 mM Na3VO4, and 20 μM phenylarsine oxide was added to the lysis buffer.
Freshly prepared lysates were used for all immunoprecipitations. Immunoprecipitations were performed by incubating 500 to 1000 μg total cell lysate on a rocker at 4°C for 1 to 2 hours with either polyclonal rabbit anti-βPDGFR tail serum (Pharmingen, San Diego, CA), rabbit anti-PI3K (p85) antiserum (Upstate Biotechnology, Lake Placid, NY), or antibovine PLCγ-1 mixed monoclonal IgG (Upstate Biotechnology). Rabbit polyclonal antibodies against Grb2, RasGAP, STAT5b (which also recognizes STAT5a), or SHP-2 were from Santa Cruz Biotechnology (Santa Cruz, CA). Immunoprecipitates were collected with protein A-Sepharose (Amersham-Pharmacia Biotech, Piscataway, NJ) for rabbit polyclonal antibodies or protein G-Sepharose (Pharmacia Biotech) for mouse monoclonal antibodies. Immunoprecipitates were washed 3 times in lysis buffer and boiled for 5 minutes in sodium dodecyl sulfate (SDS) sample buffer.
Immunoblotting
Samples were separated by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred electrophoretically to either Immobilon-P (Millipore, Cambridge, MA) or to BioBlot-NC (Corning Costar). Samples were blocked with 1% bovine serum albumin (Sigma Fraction V; Sigma Chemicals, St Louis, MO) in wash buffer: 10 mM Tris-HCl, pH 7.4, 0.1% Triton X-100, and 0.9% NaCl. Samples were then incubated for 1 hour with one of the following antibodies: mouse monoclonal 4G10 or horseradish peroxidase (HRP)–conjugated 4G10 antibody against phosphotyrosine (Upstate Biotechnology), polyclonal rabbit anti-βPDGFR tail serum (Pharmingen), rabbit anti-PI3K (p85) antiserum (Upstate Biotechnology), anti–PLCγ-1 monoclonal IgG (Upstate Biotechnology), anti-PTP1D (anti-SHP2) mouse monoclonal IgG (Transduction Laboratories, Lexington, KY), rabbit anti-Grb2 antibody (Santa Cruz Biotechnology), or rabbit anti-STAT5 polyclonal antibody (Santa Cruz Biotechnology). Filters were washed and were incubated with either HRP-conjugated antirabbit IgG or HRP-conjugated antimouse IgG (Amersham). Blots were rinsed and visualized by enhanced chemiluminescence.
In vitro protein kinase assay and phosphoamino acid analysis
Cell lysates were immunoprecipitated with anti-PDGFβR tail antibody as above and washed 3 times with lysis buffer. Immunoprecipitates were washed an additional 2 times with kinase buffer: 20 mM Tris-HCl, pH 7.4, 1 mM Na3VO4, and 10 mM MgCl2. Protein kinase assays were carried out in 30 μL kinase buffer with 10 μCi (0.37 MBq) γ-[32P]-ATP for 10 minutes at 30°C. Kinase assays were washed twice in lysis buffer and loaded onto 7.5% SDS-PAGE. Extraction of radiolabeled proteins from gel slices, acid hydrolysis, and 2-dimensional electrophoresis of phosphoamino acids were performed as described previously.23
For in vitro kinase assays using exogenous substrate, TEL-PDGFβR immune complex kinase assays were performed in the presence of purified bacterial GST-p97 substrate, 20 mM MOPS pH 7.0, 0.2 mM pervanadate, 5 mM MnCl2, 5 μM unlabeled ATP, and 10 μCi (0.37 MBq) γ-[32P]-ATP. Reactions were carried out for 5 minutes and were stopped with SDS-sample buffer containing 50 mM EDTA. Kinase reactions were fractionated by SDS-PAGE and visualized by autoradiography.
Electrophoretic mobility shift assay
Results
Substitution of the major tyrosine phosphorylation sites present in the native PDGFβR impairs cell transformation by the TEL/PDGFβR fusion
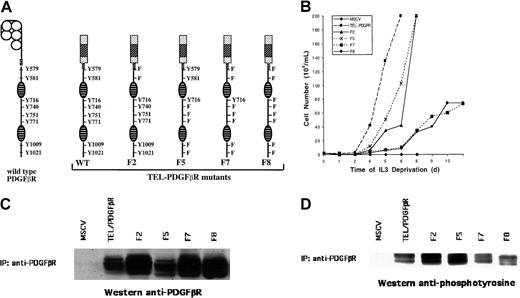
The TEL/PDGFβR fusion was stably expressed by retroviral infection in the Ba/F3 hematopoietic cell line, which requires the continuous presence of IL-3 for survival and proliferation. As shown previously,2 transformation by the TEL/PDGFβR fusion abrogates the requirement of IL-3 for cell survival and proliferation. In contrast, cells transfected with vector alone are not viable in the absence of IL-3 and undergo apoptotic cell death. The major tyrosine phosphorylation sites are known for the native PDGFβR, and these provide critical interaction sites with multiple signaling intermediates that contain SH2 domains. We speculated that similar sites might be necessary for transformation by the TEL/PDGFβR fusion. TEL/PDGFβR variants were constructed that contained Tyr→Phe substitutions at each of the major phosphotyrosine interaction sites present in the native PDGFβR (Figure1A).
Structure, transforming properties, and expression of TEL/PDGFβR mutants.
(A) Schematic diagram of wild-type (WT) PDGFβR and TEL/PDGFβR mutants. At left is depicted the wild-type PDGFβR protein with the major tyrosine phosphorylation sites denoted. Wild-type and mutant TEL/PDGFβR variants are shown that contain Tyr→Phe substitutions at sites corresponding to phosphorylation sites in the native PDGFβR protein. Boxed areas indicate the TEL domain; oval hatched areas denote the split tyrosine kinase domain. Numbering of tyrosine residues corresponds to positions in the wild-type PDGFβR protein. (B) Growth properties of Ba/F3 hematopoietic cells stably transformed with wild-type and mutant TEL/PDGFβR fusion genes. Ba/F3 cells transformed by pMSCV vector with or without wild-type TEL/PDGFβR, F2, F5, F7, or F8 were grown in the absence of IL-3. Cells were maintained at a density of 1 × 105 to 1 × 106/mL. (C) Expression of wild-type and mutant TEL/PDGFβR proteins in stably transformed Ba/F3 cells. Cells were lysed and were immunoprecipitated with antibody recognizing the cytoplasmic domain of the PDGFβR. Immunoprecipitates were separated by SDS-PAGE and immunoblotted with antibody against the PDGFβR cytoplasmic domain. Multiple bands in each lane are the consequence of 2 translational start sites within theTEL gene and of autophosphorylation of TEL/PDGFβR. (D) Tyrosine phosphorylation of wild-type and mutant TEL/PDGFβR proteins. TEL/PDGFβR variants were immunoprecipitated as in panel C and immunoblotted with a monoclonal antibody against protein phosphotyrosine.
Structure, transforming properties, and expression of TEL/PDGFβR mutants.
(A) Schematic diagram of wild-type (WT) PDGFβR and TEL/PDGFβR mutants. At left is depicted the wild-type PDGFβR protein with the major tyrosine phosphorylation sites denoted. Wild-type and mutant TEL/PDGFβR variants are shown that contain Tyr→Phe substitutions at sites corresponding to phosphorylation sites in the native PDGFβR protein. Boxed areas indicate the TEL domain; oval hatched areas denote the split tyrosine kinase domain. Numbering of tyrosine residues corresponds to positions in the wild-type PDGFβR protein. (B) Growth properties of Ba/F3 hematopoietic cells stably transformed with wild-type and mutant TEL/PDGFβR fusion genes. Ba/F3 cells transformed by pMSCV vector with or without wild-type TEL/PDGFβR, F2, F5, F7, or F8 were grown in the absence of IL-3. Cells were maintained at a density of 1 × 105 to 1 × 106/mL. (C) Expression of wild-type and mutant TEL/PDGFβR proteins in stably transformed Ba/F3 cells. Cells were lysed and were immunoprecipitated with antibody recognizing the cytoplasmic domain of the PDGFβR. Immunoprecipitates were separated by SDS-PAGE and immunoblotted with antibody against the PDGFβR cytoplasmic domain. Multiple bands in each lane are the consequence of 2 translational start sites within theTEL gene and of autophosphorylation of TEL/PDGFβR. (D) Tyrosine phosphorylation of wild-type and mutant TEL/PDGFβR proteins. TEL/PDGFβR variants were immunoprecipitated as in panel C and immunoblotted with a monoclonal antibody against protein phosphotyrosine.
In our previous report, each of these TEL/PDGFβR Tyr→Phe substitution mutants was able to transform Ba/F3 cells to IL-3 independence,21 but progressive substitution of these tyrosine residues markedly impaired generation of a myeloproliferative disease in a murine BMT model. To further quantify the transformation of Ba/F3 cells by TEL/PDGFβR, cells were infected with retrovirus expressing each of the TEL/PDGFβR mutants and the neomycin-resistance gene, and cells were then grown for 15 days in the presence of IL-3. IL-3 was then removed from the growth media, and cell proliferation was assayed in the absence of IL-3 (Figure 1B). Substitution of either the juxtamembrane tyrosine residues in the F2 mutant or the carboxy terminal tyrosine residues in the F5 mutant impaired the outgrowth of Ba/F3 cells in the absence of IL-3. Thus, both the juxtamembrane and carboxy terminal phosphorylation sites that are present in the native PDGFβR are necessary for full transformation of Ba/F3 cells by TEL/PDGFβR. Moreover, substitution of both the juxtamembrane and carboxy tyrosine residues in the F7 and F8 variants substantially attenuated the growth of Ba/F3 cells in the absence of IL-3, and this impairment of TEL/PDGFβR transformation was greater than that observed with the individual juxtamembrane or carboxy terminal mutants. No outgrowth of IL-3–independent Ba/F3 cells was observed in cells infected with the empty vector control. Although mutants such as F8 that contain multiple substitutions can ultimately cause IL-3–independent cell outgrowth after long latency,21 the results presented here indicate that both the juxtamembrane and carboxy terminal phosphorylation sites in the TEL/PDGFβR fusion are necessary for full Ba/F3 transformation to IL-3 independence.
To determine the expression of each of the TEL/PDGFβR fusion variants, Ba/F3 cell lysates were immunoblotted with antibody directed against the cytoplasmic tail of the PDGFβR protein. As shown in Figure 1C, the wild-type or variant TEL/PDGFβR protein was expressed in each of the Ba/F3 cell lines, although the level of wild-type and F5 TEL/PDGFβR protein was somewhat reduced in multiple independent Ba/F3 cell lines relative to that in F2, F7, and F8 cell lines.
The TEL/PDGFβR fusion protein has protein-tyrosine kinase activity
The TEL/PDGFβR protein contains the entire protein-tyrosine kinase domain of the native PDGFβR protein. To confirm that the protein was active as a tyrosine kinase, wild-type TEL/PDGFβR protein was immunoprecipitated from Ba/F3 cell lysates and subjected to in vitro autophosphorylation with γ-[32P]-ATP. The wild-type TEL/PDGFβR protein was active in a kinase assay as measured by autophosphorylation (Figure 2A). To determine the amino acid specificity of this kinase, the autophosphorylated TEL/PDGFβR protein was subjected to phosphoamino acid analysis. As shown in Figure 2B, in vitro autophosphorylation occurred entirely on tyrosine residues. Phosphoamino acid analysis of the F8 variant showed that this fusion protein also autophosphorylated on tyrosine residues (data not shown). This result confirmed the activity of the TEL/PDGFβR fusion protein as a protein-tyrosine kinase.
In vitro kinase activity of wild-type TEL/PDGFβR fusion protein in Ba/F3 cells.
(A) Cells were lysed and immunoprecipitated with antibody recognizing the PDGFβR cytoplasmic domain. Immunoprecipitates were washed, subjected to in vitro phosphorylation with γ-[32P]-ATP, and separated by SDS-PAGE. (B) Radiolabeled wild-type TEL/PDGFβR was excised from the gel, acid hydrolyzed to constituent phosphoamino acids, and separated by 2-dimensional electrophoresis. (C) Immune complex kinase assay was performed as above using lysates from 32D cells that express TEL/PDGFβR variants. Phosphorylation of exogenous purified GST-p97 (GST-Gab2) was determined. Expression of TEL/PDGFβR and variants was equivalent in each of the cell lines (data not shown).
In vitro kinase activity of wild-type TEL/PDGFβR fusion protein in Ba/F3 cells.
(A) Cells were lysed and immunoprecipitated with antibody recognizing the PDGFβR cytoplasmic domain. Immunoprecipitates were washed, subjected to in vitro phosphorylation with γ-[32P]-ATP, and separated by SDS-PAGE. (B) Radiolabeled wild-type TEL/PDGFβR was excised from the gel, acid hydrolyzed to constituent phosphoamino acids, and separated by 2-dimensional electrophoresis. (C) Immune complex kinase assay was performed as above using lysates from 32D cells that express TEL/PDGFβR variants. Phosphorylation of exogenous purified GST-p97 (GST-Gab2) was determined. Expression of TEL/PDGFβR and variants was equivalent in each of the cell lines (data not shown).
We next assayed for any attenuation of protein-tyrosine kinase activity that might result from the Tyr→Phe substitution. Although each of the TEL/PDGFβR variants was able to autophosphorylate itself in an immune complex kinase assay (data not shown), the variable expression of the TEL/PDGFβR fusion proteins in Ba/F3 cells complicated quantitative assessment of enzymatic activity. Therefore, we used a series of 32D myeloid cell lines that were stably transformed with wild-type, F2, F7, and F8 TEL/PDGFβR fusions. These 32D cell lines expressed equivalent levels of TEL/PDGFβR fusion protein (Figure 4), and extracts from these cells were therefore used in immune-complex kinase assays. The TEL/PDGFβR fusion proteins were immunoprecipitated with antibody against the carboxy terminal domain of the native PDGFβR, and immune complex kinase assays were performed with γ-[32P]-ATP and a purified GST-p97 (GST-Gab2) fusion protein as an exogenous substrate. As shown in Figure 2C, Tyr→Phe substitution in the TEL/PDGFβR variants produced no attenuation in protein-tyrosine kinase activity directed against GST-Gab2. Similar results were obtained by performing in vitro kinase reactions with unlabeled ATP and then immunoblotting the exogenous substrate with the 4G10 antiphosphotyrosine antibody (data not shown). Therefore, these findings demonstrate that the attenuation of cell transformation by the F7 and F8 TEL/PDGFβR variants was not due to loss of protein-tyrosine kinase activity.
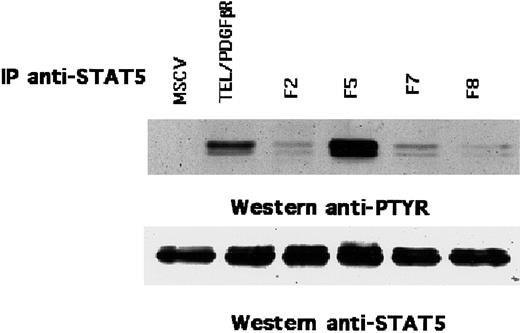
Transformation by TEL/PDGFβR causes hyperphosphorylation of STAT5 on tyrosine residues
The above findings indicated that the juxtamembrane tyrosine residues in TEL/PDGFβR were critical for full transformation of Ba/F3 cells. Comparable tyrosine residues in the native PDGFβR are critical for the activation of the STAT5 transcription factor.24 To assess the potential role of STAT5 phosphorylation on TEL/PDGFβR signaling, Ba/F3 cells that express each of the TEL/PDGFβR variants were immunoprecipitated with antibody against STAT5 and were immunoblotted with the antiphosphotyrosine antibody 4G10. Ba/F3 cells that expressed the native TEL/PDGFβR fusion showed hyperphosphorylation of STAT5 on tyrosine (Figure3). Similarly, the F5 variant also caused STAT5 phosphorylation, and this result indicates that the carboxy terminal phosphorylation sites in TEL/PDGFβR are dispensable for STAT5 activation. In contrast, only minimal phosphorylation of STAT5 was observed in cells that expressed the F2, F7, or F8 variants, and this result indicates that the juxtamembrane Tyr579 and Tyr581 sites are critical for phosphorylation of STAT5 by the TEL/PDGFβR fusion.
Phosphorylation of STAT5 by TEL/PDGFβR in Ba/F3 cells.
Ba/F3 cells that express TEL/PDGFβR were lysed and immunoprecipitated with antibody against STAT5. The immune complex was probed with either 4G10 antiphosphotyrosine (anti-PTYR) antibody (top) or anti-STAT5 antibody (bottom).
Phosphorylation of STAT5 by TEL/PDGFβR in Ba/F3 cells.
Ba/F3 cells that express TEL/PDGFβR were lysed and immunoprecipitated with antibody against STAT5. The immune complex was probed with either 4G10 antiphosphotyrosine (anti-PTYR) antibody (top) or anti-STAT5 antibody (bottom).
STAT5 is hyperphosphorylated in 32D myeloid cells that are transformed with the TEL/PDGFβR fusion protein
Because TEL/PDGFβR expression is associated with myeloid transformation in the murine BMT model21 and in human CMML, we wished to confirm the hyperphosphorylation of STAT5 in the murine 32D myeloid cell line. 32D cells that expressed the wild-type or variant TEL/PDGFβR were immunoprecipitated with antibody to STAT5 and immunoblotted with the 4G10 antiphosphotyrosine antibody. Similar to the results in Ba/F3 cells shown above, TEL/PDGFβR transformation of 32D cells was associated with hyperphosphorylation of STAT5. Moreover, Tyr→Phe substitution at the juxtamembrane sites in the F2, F7, and F8 variants abrogated STAT5 hyperphosphorylation on tyrosine residues (Figure 4). Although F5 TEL/PDGFβR was expressed at a lower level compared to the other variants in 32D cells, this low level of F5 expression was sufficient to cause hyperphosphorylation of STAT5. Similar results were obtained by assessing STAT5 binding to DNA by EMSA (data not shown). Thus, the potential phosphotyrosine sites in TEL/PDGFβR required for STAT5 hyperphosphorylation in 32D myeloid cells are identical to those required for STAT5 phosphorylation in Ba/F3 cells.
STAT5 phosphorylation in 32D cells.
(A) 32D myeloid cells that expressed TEL/PDGFβR variants were lysed and immunoblotted with antibody against TEL/PDGFβR. (B) Cell lysates were also immunoprecipitated with antibody against STAT5 and immunoblotted with antibody against phosphotyrosine or (C) STAT5. Arrow indicates hyperphosphorylated STAT5.
STAT5 phosphorylation in 32D cells.
(A) 32D myeloid cells that expressed TEL/PDGFβR variants were lysed and immunoblotted with antibody against TEL/PDGFβR. (B) Cell lysates were also immunoprecipitated with antibody against STAT5 and immunoblotted with antibody against phosphotyrosine or (C) STAT5. Arrow indicates hyperphosphorylated STAT5.
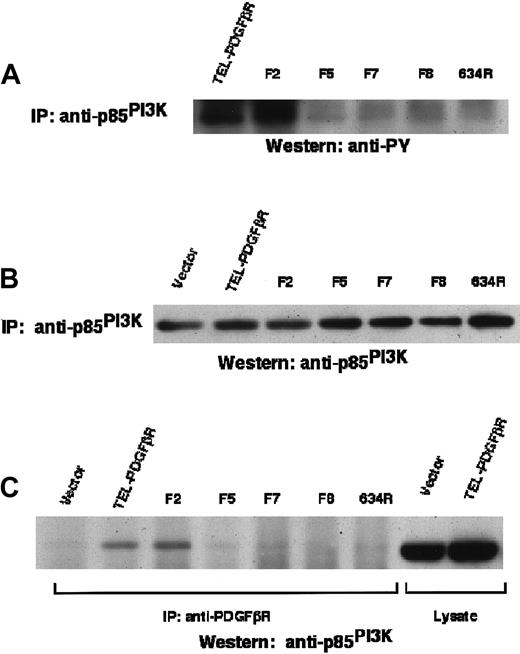
PI3K tyrosine phosphorylation and association with TEL/PDGFβR requires carboxy terminal phosphorylation of TEL/PDGFβR
The findings described above also indicated the critical role of the carboxy terminal tyrosine residues (within the kinase insert domain and carboxy terminal tail) in the transformation of Ba/F3 cells by TEL/PDGFβR. The tyrosine residues 740 and 751 in the kinase insert are necessary for interaction with PI3K by the native PDGFβR.25-27 To assess the role of PI3K in signaling by the TEL/PDGFβR, the level of tyrosine phosphorylation of the 85-kd subunit of PI3K was measured. Ba/F3 cell lysates were immunoprecipitated with antibody against PI3K and were then immunoblotted with antibody against phosphotyrosine (Figure5). Transformation by the wild-type or F2 variant of the TEL/PDGFβR protein was associated with increased p85 PI3K tyrosine phosphorylation. However, transformation by the F5, F7, and F8 variant fusions did not cause increased tyrosine phosphorylation of PI3K. PI3K was expressed at equivalent levels in each of these cell lines.
PI3K phosphorylation by TEL/PDGFβR.
PI3K tyrosine phosphorylation and association with TEL/PDGFβR fusion proteins in Ba/F3 cells. (A) Ba/F3 cell lysates were immunoprecipitated with antibody against the 85-kd subunit of PI3K and immunoblotted with antibody against phosphotyrosine. (B) PI3K immunoprecipitates were immunoblotted with antibody against p85PI3K to assess relative expression and immunoprecipitation. (C) Ba/F3 cells were immunoprecipitated with antibody against the PDGFβR cytoplasmic domain and immunoblotted with antibody against p85PI3K.
PI3K phosphorylation by TEL/PDGFβR.
PI3K tyrosine phosphorylation and association with TEL/PDGFβR fusion proteins in Ba/F3 cells. (A) Ba/F3 cell lysates were immunoprecipitated with antibody against the 85-kd subunit of PI3K and immunoblotted with antibody against phosphotyrosine. (B) PI3K immunoprecipitates were immunoblotted with antibody against p85PI3K to assess relative expression and immunoprecipitation. (C) Ba/F3 cells were immunoprecipitated with antibody against the PDGFβR cytoplasmic domain and immunoblotted with antibody against p85PI3K.
The TEL/PDGFβR and F2 variant also associated with the p85 subunit of PI3K (Figure 5). This interaction was dependent on the intrinsic kinase activity of the fusion protein, because the Arg634 kinase inactive mutant was unable to associate with PI3K. In contrast, the F5, F7, and F8 variants did not associate with PI3K. Collectively, these results show that the presence of the carboxy terminal tyrosine residues in the TEL/PDGFβR fusion protein is necessary for tyrosine phosphorylation of and association with the p85 subunit of PI3K.
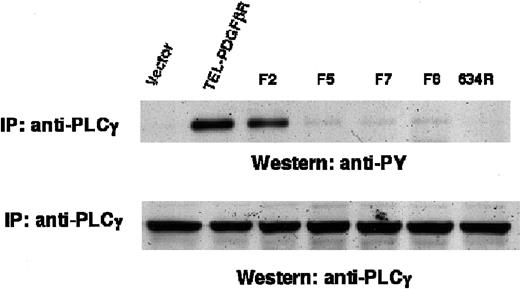
Signaling through PLCγ tyrosine phosphorylation requires carboxy terminal phosphorylation of TEL/PDGFβR
Mitogenic signaling by the native PDGFβR protein occurs through activation of PLCγ, and the engagement of PLCγ is mediated by tyrosine phosphorylation in the carboxy terminal tail. To determine the role of this signaling event in transformation of Ba/F3 cells by the TEL/PDGFβR fusion protein, cell lysates were immunoprecipitated with antibody against PLCγ and immunoblotted with antibody against phosphotyrosine (Figure 6). Transformation of Ba/F3 cells by the wild-type or F2 TEL/PDGFβR fusion was associated with increased tyrosine phosphorylation of PLCγ. This signaling event required TEL/PDGFβR kinase activity because the Arg634 kinase inactive variant did not phosphorylate PLCγ. The transforming variants F5, F7, and F8 also caused no elevation in PLCγ tyrosine phosphorylation. PLCγ was expressed equivalently in each of the Ba/F3 cell lines (Figure 6). Thus, signaling through PLCγ required the presence of the carboxy terminal tyrosine residues in the TEL/PDGFβR fusion protein.
Tyrosine phosphorylation of PLCγ.
Ba/F3 cells were lysed, immunoprecipitated with antibody against PLCγ, and immunoblotted with antibody against phosphotyrosine (top). PLCγ expression was assessed by immunoblotting whole cell lysates with antibody against PLCγ (bottom).
Tyrosine phosphorylation of PLCγ.
Ba/F3 cells were lysed, immunoprecipitated with antibody against PLCγ, and immunoblotted with antibody against phosphotyrosine (top). PLCγ expression was assessed by immunoblotting whole cell lysates with antibody against PLCγ (bottom).
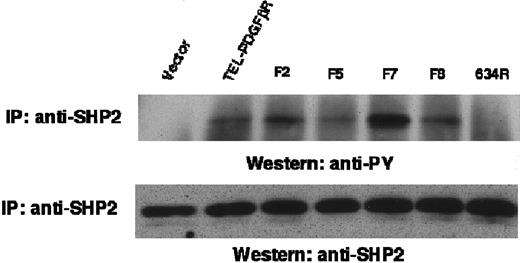
SHP-2 is tyrosine phosphorylated by each of the TEL/PDGFβR variants
To assess the role of SHP-2 in TEL/PDGFβR signaling, Ba/F3 cell lysates were immunoprecipitated with anti–SHP-2 and immunoblotted with antiphosphotyrosine antibody (Figure 7). SHP-2 showed an increased content of phosphotyrosine in Ba/F3 cells transformed by wild-type TEL/PDGFβR compared with vector-control cells or cells transformed with the kinase-inactive Arg634 variant. Moreover, each of the transforming variants (F2, F5, F7, and F8) was associated with elevated SHP-2 tyrosine phosphorylation. There was equivalent expression of SHP-2 in each of these cell lines. Thus, tyrosine phosphorylation of SHP-2 did not absolutely require these 8 major autophophosphorylation sites that are present in the native PDGFβR.
Tyrosine phosphorylation of SHP-2.
Ba/F3 cell lysates were immunoprecipitated with antibody against SHP-2 and immunoblotted with antibody against phosphotyrosine (top) or against SHP-2 (bottom).
Tyrosine phosphorylation of SHP-2.
Ba/F3 cell lysates were immunoprecipitated with antibody against SHP-2 and immunoblotted with antibody against phosphotyrosine (top) or against SHP-2 (bottom).
Each of the TEL/PDGFβR variants is bound to the Grb2 adaptor molecule in vivo
The small adaptor molecule Grb2 forms a stable complex with the Sos nucleotide exchange factor and can mediate signaling from protein-tyrosine kinases through Ras.13 Grb2 can bind to the native PDGFβR directly at the phosphorylated Tyr71628or indirectly through a complex with Shc29,30 or with SHP-2.31 32 Similarly, Grb2 was found in a physical complex with the TEL/PDGFβR fusion protein (Figure8). Surprisingly, each of the TEL/PDGFβR variants (including the F8 variant) was associated with the Grb2 adaptor molecule in an immune complex. Thus, although the major phosphorylation sites present in the native PDGFβR may mediate association of Grb2 with TEL/PDGFβR, Grb2 can also interact directly or indirectly with other potential sites in the TEL/PDGFβR protein.
Association of TEL/PDGFβR with the Grb2 adaptor protein.
Ba/F3 cell lysates were immunoprecipitated with antibody against Grb2 and immunoblotted with antibody against the PDGFβR cytoplasmic domain (top). Expression of Grb2 was determined in whole cell lysates by immunoblotting with antibody against Grb2 (bottom).
Association of TEL/PDGFβR with the Grb2 adaptor protein.
Ba/F3 cell lysates were immunoprecipitated with antibody against Grb2 and immunoblotted with antibody against the PDGFβR cytoplasmic domain (top). Expression of Grb2 was determined in whole cell lysates by immunoblotting with antibody against Grb2 (bottom).
Discussion
The results presented here indicate that multiple phosphorylation sites in the TEL/PDGFβR fusion protein are required for efficient hematopoietic transformation in cell culture, and these data correlate with transformation in a murine BMT assay of leukemogenesis.21 Progressive conservative substitution of tyrosine residues in TEL/PDGFβR at major phosphorylation sites present in the native PDGFβR caused incremental attenuation of IL-3–independent hematopoietic outgrowth. These same substitutions also had profound effects on latency and phenotype in the murine BMT model.
However, one potential explanation for the progressive loss of hematopoietic transformation in the TEL/PDGFβR variant series is that there is progressive disruption of protein tyrosine kinase activity due to the Tyr→Phe substitutions. For example, F2 substitutions in the native PDGFβR have been reported to markedly attenuate kinase activity in response to ligand stimulation,33 although these tyrosines are not necessary for bovine papillomavirus E5-stimulated tyrosine kinase activation of PDGFβR or mitogenic signaling in Ba/F3 cells.34 In addition, Tyr857 in the native PDGFβR, which is preserved in each of the F-series mutants of TEL/PDGFβR, is necessary for the protein-tyrosine kinase activity of the native PDGFβR.35 We used a series of 32D myeloid cell extracts (with uniform levels of TEL/PDGFβR expression) to assay for phosphorylation of an exogenous substrate in an immune-complex kinase assay, and the results presented here indicate that there was no loss of protein-tyrosine kinase activity in the TEL/PDGFβR mutants. Thus, the substitution of major phosphotyrosine interaction sites in the TEL/PDGFβR fusion attenuates activation of signaling intermediates without grossly altering kinase catalytic activity.
We observed in the murine BMT assay that any TEL/PDGFβR mutant that had Tyr→Phe substitutions at positions 579/581 was incapable of causing myeloid disease, and it instead produced long-latency lymphoproliferative disease.21 As noted above, these findings cannot be explained by a decrement in tyrosine kinase activity. These residues are known to mediate STAT5 activation by the native PDGFβR,24 and we tested STAT5 activation by the TEL/PDGFβR to determine whether this signaling event might correlate with phenotypic differences. The findings presented here indicate that the STAT5 transcription factor is hyperphosphorylated in cells that express the wild-type and F5 variant of TEL/PDGFβR but not in the F2, F7, and F8 variants. The tyrosine residues corresponding to Tyr579 and Tyr581 in the native PDGFβR are critical for full phosphorylation of STAT5. Thus, activation of STAT5 in hematopoietic cell culture correlates with transformation of myeloid lineage cells in the murine BMT model.21 These findings are consistent with recent results indicating the critical role of STAT5 in generation of murine myeloproliferative disease by the TEL/Jak2 fusion protein.10 The use of STAT5 knock-out mice in a murine BMT model will be of particular value to further assess the role of STAT5 in myeloid cell transformation by TEL/PDGFβR.
The cytoplasmic tyrosine kinases Src, Yes, and Fyn bind to phosphorylated tyrosine residues at Tyr579 and Tyr581 in the juxtamembrane portion of the native PDGFβR,33 and it is possible that disruption of analogous sites in TEL/PDGFβR attenuates signaling through a Src family member. However, we have not been able to detect TEL/PDGFβR-dependent activation of Lyn or Fyn kinases that are expressed in Ba/F3 cells (data not shown).
Like the PDGFβR protein, TEL/PDGFβR expression in Ba/F3 cells is associated with tyrosine hyperphosphorylation of and association with p85 PI3K and with tyrosine phosphorylation of PLCγ. The phosphorylation of these signaling intermediates is lost in the F5, F7, and F8 variants of TEL/PDGFβR, and this result indicates that the carboxy terminal phosphorylation sites present in the native PDGFβR are necessary for engagement of PLCγ and PI3K in cells that express TEL/PDGFβR. The F5 variant was impaired in its ability to support outgrowth of IL-3–independent Ba/F3 cells, and there was diminished myeloid cell transformation by the F5 variant in the murine BMT model.21 In contrast, the F5 PDGFβR is able to support the survival of Ba/F3 cells in the presence of v-sis or E5 bovine papillomavirus oncoproteins, despite the abrogation of PI3K and PLCγ signaling.34 PLCγ and PI3K signaling may contribute to myeloid transformation by TEL/PDGFβR in the mouse BMT model. Abrogation of these pathways, alone or combined with loss of STAT5 activation in the F7 and F8 mutants, extends the latency of Ba/F3 cell outgrowth.
The F8 TEL/PDGFβR variant was phosphorylated on tyrosine residues in vivo (Figure 1D) and in the immune-complex kinase assay (data not shown), and deletion of the canonical Grb2 interaction site at Tyr716 of the native PDGFβR caused no diminution of Grb2 binding to the TEL/PDGFβR fusion protein. Thus, there are remaining potential sites in F8 for phosphotyrosine-dependent interaction with signaling intermediates such as Grb2. Moreover, a secondary set of novel autophosphorylation sites may have been revealed in F8 TEL/PDGFβR; the compensatory phosphorylation of novel tyrosine residues has been reported for the F5 PDGFβR.36 The TEL domain may also contain phosphorylation sites that mediate interaction with signaling proteins. This conjecture is supported by the finding that tyrosine residues 17 and 27 in TEL/PDGFβR serve as autophosphorylation sites, although mutation of these residues does not impair the ability of TEL/PDGFβR to cause IL-3–independent cell growth.37
Collectively, these data indicate that the activation of a minimal combination of signaling intermediates by TEL/PDGFβR is required for full transforming ability in the Ba/F3 system, as well as in primary hematopoietic cells in the murine BMT assay. For example, the F2 and the F5 mutants had similar attenuation of their ability to confer factor-independent growth of Ba/F3 cells. Moreover, the F7 mutant had even more significant impairment of IL-3–independent cell outgrowth, and these findings indicate that the juxtamembrane and carboxy terminal phosphorylation sites in the TEL/PDGFβR fusion protein are necessary for robust transformation. Correlation of these data with activation of SH2-containing signaling intermediates suggests that activation of either STAT5 or engagement of PI3K and PLCγ yields a similarly attenuated potency for transformation in the Ba/F3 assay, whereas global engagement of these signaling intermediates by the wild-type TEL/PDGFβR mediates the fully transformed phenotype. Moreover, the phosphorylated tyrosine residues in the TEL/PDGFβR fusion might interact with other signaling intermediates other than those that have been characterized here. The overall transformed phenotype may be modulated by additional signaling events because nuclear factor-κB38 and JNK/SAPK39 have been reported to be activated by the TEL/PDGFβR fusion protein. We have assessed Erk1/Erk2 activation in detail by using an in vitro kinase assay and by immunoblotting with a phosphopeptide-specific antibody that recognizes activated Erk1/Erk2 (data not shown), and we have consistently observed an absence of Erk1/Erk2 activation in Ba/F3 and 32D cells that express TEL/PDGFβR.
These findings correlate with recent observations regarding the expression of immediate early genes by an M-CSFR/PDGFβR chimera and related Tyr→Phe mutants.40 Although it had been predicted that each signaling pathway would activate a discrete set of transcriptional targets, Fambrough and coworkers40 demonstrated that the Tyr→Phe chimeric protein mutants activated a nearly identical set of immediate early genes, but there was a quantitative decrease in the level of expression of these genes in the F5 mutant compared with wild-type chimeric protein. The hypothesis that overall signal strength contributes to the physiologic consequences of PDGFβR activation is in consonance with the observations of TEL/PDGFβR mutants both in the context of Ba/F3 cells and in primary murine hematopoietic cells.
In summary, we have shown that multiple signaling pathways involving STAT5, SHP-2, PLCγ, PI3K, and Grb2 are engaged in hematopoietic cells that are transformed by TEL/PDGFβR. STAT5, PLCγ, and PI3K appear to functionally synergize both in the transformation of Ba/F3 cells to IL-3 independence and in myeloid transformation in the murine BMT model.
We thank Ben Neel, James Griffin, and Alan D'Andrea for valuable comments and critical review.
Supported in part by National Institutes of Health (NIH) grant PO1 (DK50654-01), NIH grant K08CA73749-01 (M.C.), and NIH grant K08-CA82261-02 (D.W.S.). D.W.S. is a Scholar of the American Society of Hematology. D.G.G. is an Associate Investigator in the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
D. Gary Gilliland or David W. Sternberg, Howard Hughes Medical Institute, Harvard Medical School, 4 Blackfan Cir, Rm 421, Boston, MA 02115; e-mail: gilliland@calvin.bwh.harvard.edu.

![Fig. 2. In vitro kinase activity of wild-type TEL/PDGFβR fusion protein in Ba/F3 cells. / (A) Cells were lysed and immunoprecipitated with antibody recognizing the PDGFβR cytoplasmic domain. Immunoprecipitates were washed, subjected to in vitro phosphorylation with γ-[32P]-ATP, and separated by SDS-PAGE. (B) Radiolabeled wild-type TEL/PDGFβR was excised from the gel, acid hydrolyzed to constituent phosphoamino acids, and separated by 2-dimensional electrophoresis. (C) Immune complex kinase assay was performed as above using lysates from 32D cells that express TEL/PDGFβR variants. Phosphorylation of exogenous purified GST-p97 (GST-Gab2) was determined. Expression of TEL/PDGFβR and variants was equivalent in each of the cell lines (data not shown).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/98/12/10.1182_blood.v98.12.3390/6/m_h82311781002.jpeg?Expires=1764981059&Signature=s-oI3LZmPNLZjEaQK1yZjJFfiI6G7kaVgc4YBZWeUr-ErNxd742zf8w4y1KFjFZnGsY0qn6hwjVPG649Oc5ASJ0HdF3nyiWIxO-d~9kPfotVSsHknCnQz1QjIXY7RgvSEwkLhvzqonhmYLxVkACetCo67F45zRh5uwxIprm0r1eZOprgXfemafakdc-Mf8r071kN~BLOtEpnyBdQY1diVAmXrI0uH1Uploiv0PxZmV5NPbkRv8XjPPRS4~Q~ei1Kmnycqh-hkoju8Hbvwvstp7x5TMAXaxcEX-YnzsrFdMD2kToTvd81DtekDwTvpqzaIqDZa9tgjcAef9y2CN9mqg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal