Abstract
Rearrangements of the c-myc oncogene have been found in most plasmacytomas induced in mice and human myeloma cell lines (HMCLs) analyzed so far. However, neither induced mouse plasmacytomas nor HMCLs represent relevant models for human multiple myeloma (MM). To evaluate the incidence of c-mycrearrangements in human plasma cell dyscrasias, sets of probes were generated to allow direct assessment of c-myctranslocations on interphase plasma cells by using fluorescence in situ hybridization. After validation of these probes, a large cohort of patients with either newly diagnosed MM (n = 529), relapsed MM (n = 58), primary plasma cell leukemia (PCL; n = 23), monoclonal gammopathy of undetermined significance (n = 65), or smoldering MM (n = 24) were analyzed. C-myc rearrangements were identified in 15% of patients with MM or primary PCL, independently of the stage of the disease (ie, diagnosis or relapse and MM or primary PCL). Analysis of the 2 main translocations observed on karyotyping, ie, t(8;14) and t(8;22), revealed that these specific translocations represented only 25% (23 of 91) of c-mycrearrangements. c-myc rearrangements were then correlated with several other patients' characteristics: illegitimateIgH recombinations, chromosome 13 deletions, and serum β2-microglobulin levels. The only significant correlation was with a high β2-microglobulin level (P = .002), although a trend for association with t(4;14) was observed (P = .08). Thus, c-myc rearrangement analysis in patients with MM revealed a strikingly lower incidence than that in HMCLs and plasmacytomas induced in mice, indicating that data obtained with these models cannot be directly extrapolated to human MM.
Introduction
C-myc rearrangement, especially through chromosomal translocation t(12;15), is a major event in the oncogenesis of plasmacytomas induced in mice.1 This translocation juxtaposes the c-myc proto-oncogene and the gene encoding the immunoglobulin heavy chain (IgH). The main molecular consequence of this translocation is nonregulatedc-myc expression throughout the cell cycle, leading to cellular proliferation. This specific chromosomal rearrangement is observed consistently in several models of induced mouse plasmacytomas. In the model of pristane-induced plasmacytomas, 100% of tumors bear a rearranged form of c-myc.2 In theEμ-IL6 transgenic model, BALB/c mice develop plasmacytoma with t(12;15).3 Similarly, most transgenic mice bearing a Eμ-v-abl transgene will develop plasmacytomas, with 80% of them showing rearrangements ofc-myc.4 Finally, even in theEμ-bcl2 transgenic model, two thirds of the plasmacytomas in the mice bear an Ig–c-mycrearrangement.5
The only mouse model lacking recurrent c-myc activation is the 5T model, which consists of a plasmacytoma occurring spontaneously in 0.5% of C57BL/KW mice.6 This model has been recognized as the most relevant to human multiple myeloma (MM). In MM, cytogenetic studies are much less productive (only 30%-50% of informative cases with use of conventional cytogenetics) and have so far failed to identify any specific rearrangement in 100% of cases. C-mycrearrangements have been observed in some cases of MM. Most of these rearrangements are t(8;14), the human equivalent of the mouse t(12;15). Variant translocations involving the IgLλ locus have also been reported. However, in contrast to results in mouse models, both these translocations occur in less than 10% of patients with MM.7-9
An evaluation of c-myc rearrangements in human myeloma cell lines (HMCLs) highlighted this locus as a hot spot of chromosomal rearrangements, even in human MM. Using an approach based on fluorescence in situ hybridization (FISH), Shou et al10showed that 18 of 20 HMCLs had an 8q24 rearrangement and that another one had a rearrangement involving L-myc. Thus, using specifically dedicated probes, Shou et al10 showed that complex rearrangements involving c-myc, IgH,IgLλ, and other loci were present in most HMCLs. These data prompted us to develop a strategy for analyzing human MM independently of the availability of analyzable clonal metaphases. Using interphase FISH, we found that c-myc rearrangements have been underestimated in human MM, that most of these rearrangements involve nonimmunoglobulin loci, and that their incidence is much lower than that observed in HMCLs and mouse plasmacytomas.
Patients and methods
Patients
A series of 699 patients with either MM (529 with newly diagnosed MM and 58 with relapsed MM), primary plasma cell leukemia (PCL; 23 patients), monoclonal gammopathy of undetermined significance (MGUS; 65 patients), or smoldering MM (SMM; 24 patients) were included in this analysis. These patients were given a diagnosis at different centers of the Intergroupe Francophone du Myélome, and a bone marrow specimen was sent to our laboratory for FISH analysis. Plasma cells from all samples were sorted by using magnetic beads coated with anti-CD138 (Miltenyi Biotec, Paris, France) as reported previously.11
Twenty-two HMCLs available in our laboratory were also analyzed. Eleven were established by us (XG-1,12 XG-2,12XG-5,12 XG-6,12 XG-7,12 SBN-1, MDN, BCN, NAN-1, NAN-2, and NAN-3), and 11 were obtained from other laboratories (LP-1,13 SKMM-1,14OPM-2,15 JJN3,16 Karpas 620,17RPMI 8226,18 L363,19 U266,20AMO-1,21 EJM,22 and ANBL-623). All the 11 HMCLs established in our laboratory and analyzed in this study correspond to early passages of the HMCL. Moreover, results were compared with those obtained in patients from whom the HMCL was derived (SBN-1, MDN, BCN, NAN-1, NAN-2, and NAN-3)
FISH experiments
FISH experiments were done as described previously.11 For FISH analysis of the c-myclocus, we first selected probes enabling detection of 100% of the rearrangements. Because mapping of the 8q24 breakpoints in MM has not been reported, we selected probes for their ability to detectc-myc rearrangements in a series of patients with Burkitt lymphoma, a disorder in which 8q24 translocations are observed in 100% of cases. Most of the translocations are t(8;14), but variant translocations, ie, t(8;22) and t(2;8), have been found in 15% to 20% of patients. Mapping of 8q24 breakpoints in this lymphoma revealed great diversity, with breakpoints located on both sides ofc-myc and dispersed over more than 650 kilobases.
We selected 22 yeast artificial chromosome (YAC) probes according to their location in the 8q24 region in the Centre d'Etude du Polymorphisme Humain database (www.cephb.fr). After labeling with Spectrum Orange–deoxyuridine triphosphate (dUTP; Vysis, Voisins-le-Bretonneux, France) or Spectrum Green–dUTP (Vysis), these probes were tested in 22 patients with Burkitt lymphoma (chimeric YAC probes were discarded). Fifteen patients had t(8;14), 5 had t(8;22), and 2 had t(2;8). Analysis of metaphases allowed classification of the probes as either c-myc centromeric (c-myc-C) probes, ie, those that hybridized on the der(8), or c-myctelomeric (c-myc-T) probes, ie, those that were translocated on one of the partner derivative chromosomes. After this probe selection, we conducted dual-color FISH experiments, combining eachc-myc-C probe (labeled in green) with eachc-myc-T probe (labeled in orange) on peripheral blood mononuclear cells from 5 healthy volunteers and bone marrow cells from 5 healthy donors to select the better set of probes. Criteria for the selection were good signal quality and the smallest gap between the 2 signals. Because of the dispersion of the breakpoints, probes selected for this analysis were necessarily far away from each other and thus may not show a fusion signal in each interphase cell. After this selection, we determined the cut-off level for positivity, ie, the threshold of probe separation enabling assessment of a c-mycrearrangement. Our criterion for fusion (ie, nonrearranged configuration) was a distance between the 2 probes smaller than the size of a signal. The cut-off value was then fixed at the mean ± 3 SDs.
To characterize c-myc rearrangements further, we designed 2 sets of probes enabling direct assessment of t(8;14) and t(8;22) on interphase plasma cells. Translocation t(8;14) was assessed by a fusion between a green-labeled IgH-specific cosmid probe (Ig10, mapping at the centromeric border of the IgHgene11) and an orange-labeled c-myc–specific YAC probe (934E1, encompassing the c-myclocus).24 For translocation t(8;22), we used the same c-myc–specific probe with an orange-labeled P-1 artificial chromosome (PAC) probe specific for IgLλ, which was obtained by screening of the Roswell Park Cancer Institute 1 library (Research Genetics, Inchinnan, Scotland) with a Cλ probe. In the case of t(8;22), a fusion between the 2 probes was observed.
Finally, patients were analyzed for illegitimate IgHrearrangements, especially for the 3 most frequent specific 14q32 translocations—t(11;14), t(4;14), and t(14;16)—as well as for chromosome 13 deletions. Probes for chromosome 4p16 and 13q14 regions were described previously.11 Illegitimate IgHrearrangements were analyzed by using the Ig10 cosmid containing the Cα2 and Cε constant genes (labeled in green) and the yIgH6-9 cosmid (labeled in orange; provided by Michael Kuehl, National Cancer Institute, Bethesda, MD). The latter probe hybridized to the more telomeric VHgenes.10 Illegitimate rearrangements were assessed by a separation of the 2 probes. Because physiologic rearrangements of theIgH gene occur in all cells of the B lineage, the threshold of positivity was evaluated in 2000 nuclei from 5 patients with a myeloid malignant disease (either chronic or acute myeloid leukemia) and fixed at 14.6% (mean ± 3 SDs).
Translocation t(11;14) was identified by using the Ig10 cosmid probe (labeled in green) and the cos 6.22 probe (labeled in orange; provided by Ed Schuuring, Department of Pathology, Leiden, The Netherlands), mapping at 11q13 and containing the CCND1gene.25 To test this probe, 50 patients with MM and t(11;14) previously diagnosed with a commercial probe (Vysis) were reanalyzed with this set of probes. All 50 patients were also positive for t(11;14) on analysis with this probe set. The analysis of 2000 peripheral blood or bone marrow cells from healthy volunteers enabled fixation of the positivity threshold at 10.5% (mean ± 3 SDs).
Translocation t(14;16) was analyzed by using the Ig10 cosmid probe (labeled in green), and a c-maf–specific PAC probe (labeled in orange; provided by Leif Bergsagel, Department of Pathology, Cornell University).26 This probe is localized on the telomeric side of all the 16q23 breakpoints of the t(14;16) reported so far.26 27 Moreover, to test the ability of this set of probes to detect t(14;16), we blindly analyzed 4 HMCLs (JJN3, ANBL-6, BCN, and NAN-2) and 4 patients with cytogenetically proven t(14;16). In all 8 cases, IgH–c-maf fusions were observed in interphase plasma cells. The cut-off value for positivity was fixed at 9.8% (mean ± 3 SDs). For all probes, at least 100 nuclei/patient were counted.
Results
C-myc rearrangements are accurately detected by interphase FISH
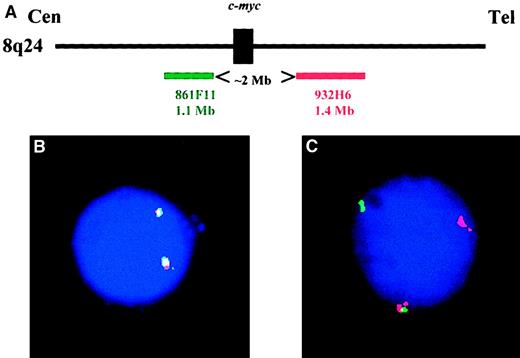
Thirteen of the 22 YAC clones were nonchimeric and localized at 8q24. Six mapped at the centromeric side of 8q24 breakpoints and 7 at the telomeric side. These 13 YAC probes were analyzed (each centromeric clone combined to each telomeric clone) on normal peripheral blood and bone marrow cells to select the set of probes with the lowest false-positive cut-off level. The 861F11 and 932H6 clones yielded the best results and were chosen for the c-myc analysis (Figure1). Two thousand cells were analyzed by 2 observers, and a separation of the 2 probes was observed in 6.7% ± 1.17%. The cut-off value was fixed at 11% (mean ± 3 SDs). We then reanalyzed the 22 specimens from patients with Burkitt lymphoma with this set of probes and showed that they accurately detected thec-myc translocations in all cases.
Configuration of the c-myc probes.
(A) Schematic representation of the 2 YAC probes used in this study (figure is not drawn to scale). (B) A typical plasma cell nucleus lacking any c-myctranslocation: both probes are fused on each allele. (C) Plasma cell nucleus showing a c-myc translocation. One allele is normal (represented by the probe fusion), whereas the second allele is translocated: the green probe remained on the der(8), and the red probe was translocated on the partner chromosome.
Configuration of the c-myc probes.
(A) Schematic representation of the 2 YAC probes used in this study (figure is not drawn to scale). (B) A typical plasma cell nucleus lacking any c-myctranslocation: both probes are fused on each allele. (C) Plasma cell nucleus showing a c-myc translocation. One allele is normal (represented by the probe fusion), whereas the second allele is translocated: the green probe remained on the der(8), and the red probe was translocated on the partner chromosome.
The analysis of sets of probes specific for IgH–c-myc andIgLλ–c-myc rearrangements on normal peripheral blood and bone marrow cells showed that the cut-off values for assessment of these translocations were 9% and 11% (mean ± 3 SDs), respectively. Furthermore, analysis of the Burkitt lymphoma cases with these t(8;14)-specific and t(8;22)-specific probes demonstrated the reliability of these sets of probes for assessment of these 2 specific translocations: there was 100% correlation between cytogenetic and interphase FISH results. To further test the accuracy of thec-myc–specific probes, we analyzed 4 patients with 8q24 abnormalities for whom clonal metaphases had been successfully generated. One patient had a t(8;14)(q24;q32), one a t(8;22)(q24;q11), one a t(8;13)(q24;q14), and one a t(6;8)(q15;q24).
C-myc rearrangements are present in 15% of primary MM tumors and more than half of HMCLs
We first screened the 699 specimens from patients forc-myc rearrangements by using the c-myc-C and c-myc-T set of probes (Figure 1C). A rearrangement was observed in 79 of 529 patients with newly diagnosed MM (15%), 5 of 58 patients with relapsed MM (9%), 3 of 23 patients with primary PCL (13%), 2 of 65 patients with MGUS (3%), and 1 of 24 patients with SMM (4%) (Table 1). The median percentage of plasma cells showing the c-myc rearrangement was 62% (range, 19%-100%). Interestingly, one patient with newly diagnosed MM had a high-level amplification of the c-myc locus, with both probes present in approximately 20 to 40 copies, without rearrangement. The analysis of the 22 HMCLs showed a significantly higher incidence ofc-myc rearrangements, which were observed in 10 of 22 HMCLs (45%; P = .0001 for difference from samples from patients with overt MM and primary PCL). The incidence of c-mycrearrangements was lower in our HMCLs (4 of 11 versus 8 of 11 for others), but the difference was not significant. There was complete concordance between the presence (or absence) of c-mycrearrangements in the original patients and the HMCLs (SBN-1, BCN, MDN, NAN-1, NAN-2, and NAN-3).
C-myc rearrangements in patients
| Diagnosis . | Germline c-myc* . | Classic t(8;14) . | Classic t(8;22) . | Other c-myc partners . | Nonclassic t(8;14)† . | All c-myc rearrangements . |
|---|---|---|---|---|---|---|
| New MM | 449 | 11 | 8 | 61 | 2 | 82/529 (16) |
| Relapsed MM | 53 | 1 | 0 | 4 | 1 | 6/58 (10) |
| Primary PCL | 20 | 0 | 0 | 3 | 0 | 3/23 (13) |
| MGUS | 63 | 0 | 0 | 2 | 0 | 2/65 (3) |
| SMM | 23 | 0 | 0 | 1 | 0 | 1/24 (4) |
| Diagnosis . | Germline c-myc* . | Classic t(8;14) . | Classic t(8;22) . | Other c-myc partners . | Nonclassic t(8;14)† . | All c-myc rearrangements . |
|---|---|---|---|---|---|---|
| New MM | 449 | 11 | 8 | 61 | 2 | 82/529 (16) |
| Relapsed MM | 53 | 1 | 0 | 4 | 1 | 6/58 (10) |
| Primary PCL | 20 | 0 | 0 | 3 | 0 | 3/23 (13) |
| MGUS | 63 | 0 | 0 | 2 | 0 | 2/65 (3) |
| SMM | 23 | 0 | 0 | 1 | 0 | 1/24 (4) |
Values are numbers (%) of patients.
New MM indicates newly diagnosed multiple myeloma (MM); PCL, primary plasma cell leukemia; MGUS, monoclonal gammopathy of undetermined significance; and SMM, smoldering MM.
Germline c-myc indicates that no rearrangement was observed with use of the set of c-myc telomeric and centromeric probes.
Nonclassic t(8;14) refers to patients who hadc-myc–IgH fusion but lacked any c-mycrearrangement on analysis with the c-myc telomeric andc-myc centromeric set of probes.
We then reanalyzed all 699 patients and 22 HMCLs by using the t(8;14)-specific and t(8;22)-specific sets of probes. Fusions betweenIgH and c-myc sequences were observed in 15 patients (13 with newly diagnosed MM and 2 with relapsed MM; Table 1) and in 7 HMCLs (LP-1, SKMM-1, OPM-2, JJN3, Karpas 620, AMO-1, and XG-5), whereas c-myc–IgLλ fusions were found in 8 patients (all with newly diagnosed MM) and one HMCL (RPMI 8226). Interestingly, 3 patients and one HMCL (JJN3) had IgH–c-mycfusions but no rearrangement on analysis with thec-myc–specific set of probes. Similarly, the RPMI 8226 HMCL had c-myc–IgLλ fusion but no c-mycrearrangement. More extensive analysis of the JJN3 HMCL revealed insertion of IgH-specific sequences in the c-myclocus. A similar event was observed previously for the RPMI 8226 HMCL, in which c-myc sequences were inserted in anIgLλ–c-maf rearrangement.10Similar mechanisms might have been involved in the 3 patients withIgH–c-myc fusions and no c-myc rearrangement, since all 3 patients had an extra IgH-specific signal possibly corresponding to the inserted sequences.
Therefore, including these cases with insertions, c-mycrearrangements were observed in 82 of 529 patients with newly diagnosed MM (16%), 6 of 58 patients with relapsed MM (10%), and 12 of 22 HMCLs (55%).
C-myc rearrangements do not correlate with other chromosomal changes but are significantly associated with high serum levels of β2-microglobulin
All 699 patients were analyzed for illegitimate IgHrearrangements, especially the 3 main specific rearrangements (IgH–CCND1, IgH–FGFR3, andIgH–c-maf), and for deletion of the 13q14 region. Furthermore, serum levels of β2-microglobulin were analyzed in all patients with MM or primary PCL at diagnosis to classify patients according to a cut-off value of 3 mg/L. Results are shown in Table2. C-myc rearrangements were observed in 13 of 119 patients without illegitimate IgHrearrangement (11%; not significant [NS]), 14 of 60 patients with t(4;14) (23%; P = .08 for difference from other patients), 17 of 115 patients with t(11;14) (15%; NS), 2 of 14 patients with t(14;16) (14%; NS), and 45 of 302 patients with illegitimate IgH rearrangements involving unknown partners (15%; NS). Analysis of chromosome 13 status (deleted or not) did not reveal any significant correlation with c-mycrearrangements. The only significant correlation was with serum β2-microglobulin levels. C-myc rearrangements were observed significantly more often in patients with β2-microglobulin levels above 3 mg/L (62 of 323 patients versus 29 of 287;P = .002).
Types of c-myc rearrangements and β2-microglobulin (β2-m) levels
| Variable . | Germline c-myc* . | Classic t(8;14) . | Classic t(8;22) . | Other c-myc partners . | Nonclassic t(8;14)† . | All c-myc rearrangements . |
|---|---|---|---|---|---|---|
| No 14q32 rearrangement | 106 | 0 | 2 | 11 | 0 | 13/119 (11) |
| t(4;14)(p16;q32) | 46 | 1 | 1 | 12 | 0 | 14/60 (23) |
| t(11;14)(q13;q32) | 98 | 2 | 1 | 14 | 0 | 17/115 (15) |
| t(14;16)(q23;q32) | 12 | 1 | 0 | 1 | 0 | 2/14 (14) |
| 14q32 rearrangement with other partners | 260 | 8 | 4 | 30 | 3 | 45/302 (14) |
| No chromosome 13 deletion | 273 | 7 | 2 | 35 | 3 | 47/317 (15) |
| Chromosome 13 deletion | 249 | 5 | 6 | 33 | 0 | 44/293 (15) |
| β2-m less than or equal to 3 mg/L | 260 | 4 | 3 | 20 | 2 | 29/287 (10) |
| β2-m greater than 3 mg/L | 262 | 8 | 5 | 48 | 1 | 62/323 (19) |
| Variable . | Germline c-myc* . | Classic t(8;14) . | Classic t(8;22) . | Other c-myc partners . | Nonclassic t(8;14)† . | All c-myc rearrangements . |
|---|---|---|---|---|---|---|
| No 14q32 rearrangement | 106 | 0 | 2 | 11 | 0 | 13/119 (11) |
| t(4;14)(p16;q32) | 46 | 1 | 1 | 12 | 0 | 14/60 (23) |
| t(11;14)(q13;q32) | 98 | 2 | 1 | 14 | 0 | 17/115 (15) |
| t(14;16)(q23;q32) | 12 | 1 | 0 | 1 | 0 | 2/14 (14) |
| 14q32 rearrangement with other partners | 260 | 8 | 4 | 30 | 3 | 45/302 (14) |
| No chromosome 13 deletion | 273 | 7 | 2 | 35 | 3 | 47/317 (15) |
| Chromosome 13 deletion | 249 | 5 | 6 | 33 | 0 | 44/293 (15) |
| β2-m less than or equal to 3 mg/L | 260 | 4 | 3 | 20 | 2 | 29/287 (10) |
| β2-m greater than 3 mg/L | 262 | 8 | 5 | 48 | 1 | 62/323 (19) |
Values are numbers (%) of patients.
Germline c-myc indicates that no rearrangement was observed with use of the set of c-myc telomeric and centromeric probes.
Nonclassic t(8;14) refers to patients who hadc-myc–IgH fusion but lacked any c-mycrearrangement on analysis with the c-myc telomeric andc-myc centromeric set of probes.
Discussion
Rearrangements of the c-myc locus have been observed in most plasmacytomas induced in mice, regardless of the model of generation (transgenic models with several oncogenes or pristane-induced plasmacytomas1), but not in the spontaneous 5T model in C57Bl mice. Shou et al10 reported a high incidence (90%) of such rearrangements in HMCLs. Comparisons between the nature of c-myc rearrangements in the 2 models reveal important similarities. In animal models, c-mycrearrangements involved the IgH locus, through t(12;15) translocations. In HMCLs, most (15 of 18) rearrangements involved theIgH locus, through a large number of different molecular recombinations. Classic t(8;14) was observed in 5 cases, whereas other rearrangements involved insertions of sequences specific for one or the other gene within the other locus. In 2 cases, the insertional events involved c-myc and IgLλ. Finally, in 3 HMCLs,c-myc was rearranged with other unknown partners. Thus, these data in mouse plasmacytomas and HMCLs highlightedc-myc deregulation as a possible major molecular event in the oncogenesis of MM.
However, some data reported by Shou et al10 suggest that these rearrangements might appear late in the natural history of human MM and are secondary rather than primary oncogenetic events. These data prompted us to analyze primary human MM tumors to determine the place of c-myc in MM oncogenesis. The first step was to generate a set of probes enabling detection of all the c-mycrearrangements on interphase plasma cells. Because some of thec-myc rearrangements occurred through insertional events (as shown by Shou et al10), our set of probes would miss these rearrangements. Moreover, some rearrangements occurred through insertion of IgLλ sequences within the c-myclocus or insertion of c-myc sequences within theIgLλ locus. To identify these types of recombinations, we also generated 2 sets of probes enabling detection ofc-myc–IgH and c-myc–IgLλ recombinations on interphase plasma cells. A large cohort of patients (almost 700), including patients with newly diagnosed MM, patients with relapsed MM, and patients with primary PCL, as well as patients with nonmalignant plasma cell dyscrasia (MGUS and SMM) were analyzed by using the 3 sets of probes.
Our results were different from those obtained in either plasmacytomas induced in mice or HMCLs. First, the incidence of c-mycrearrangements in patients with plasma cell malignant disease was somewhat lower (15%) than that observed in the 14 patients analyzed by Shou et al (7 of 14).10 Second, whereas mostc-myc rearrangements described in the models involvedIgH or IgLλ sequences, rearrangements in patients occurred through translocations with other, unknown partner regions. Third, we found a lower incidence of c-mycrearrangements in HMCLs (55%). These differences do not appear to be related to a lack of sensitivity of our sets of probes, since we accurately detected all the rearrangements observed in the 22 specimens from patients with Burkitt lymphoma and because our results with 8 HMCLs that were also analyzed by Shou et al10 were in complete agreement with the findings of those researchers.
Analysis of our large cohort of patients showed thatc-myc rearrangements are likely to occur as soon as diagnosis, since the incidence was not higher in patients analyzed at relapse of MM than in those with newly diagnosed MM (16% versus 10%). Moreover, we observed c-myc rearrangements in 2 of 65 and 1 of 24 patients with MGUS and SMM, respectively. However, a more detailed analysis revealed a wide patient-to-patient range in the percentage of plasma cells with the rearrangement (19%-100%). This finding favors the hypothesis that c-myc rearrangements are secondary events, thereby supporting the results of Shou et al,10 who detected c-myc rearrangements only in subsets of clonal metaphase plasma cells.
Furthermore, in our current study, most c-myc rearrangements occurred through translocation involving nonimmunoglobulin genes (68 of 91 [75%]). Classic translocations, ie, t(8;14) and t(8;22), were observed in 15 and 8 cases, respectively. Because cytogenetic analyses were not routinely performed in our series, the nature of the chromosomal partner in the other 68 patients is largely unknown. Only 2 patients (both with primary PCL) underwent successful cytogenetic analysis, one with a t(6;8)(q15;q24) and one with a t(8;13)(q24;q14).28 In both cases, metaphase and interphase FISH accurately detected the abnormality, thereby showing involvement of the c-myc locus. We also found a novel recurrent translocation, ie, t(6;8)(q15;q24), in 2 patients with primary PCL.28 In an update of their multicolor FISH analysis, Sawyer et al29 described several translocations involving 8q24 and another partner, such as 16q22, 1p13, and 6q21. Thus, many partners seem to be involved in c-mycderegulation, even though t(8;14) and t(8;22) appear to occur most frequently in cytogenetic analyses.
The discrepancy between the incidence of c-myc in our series and the series reported by Shou et al10 has no clear explanation. A possible reason for the disparity is the difference between the probes used in the 2 studies. However, we did detect c-myc rearrangements in the 8 HMCLs that were also analyzed by Shou et al.10 Another possible explanation is that c-myc rearrangements are acquired during HMCL evolution. Most of the HMCLs established in our laboratory have been established only recently or, in the case of the XG HMCL, correspond to early passages. Moreover, we analyzed 6 primary patient/derived HMCL pairs and found a complete agreement in results (2 with and 4 withoutc-myc rearrangement). In contrast, most of the HMCLs obtained from other laboratories were established several years ago and may have acquired their c-myc rearrangements.
We compared the c-myc configuration with other characteristics in patients with either MM or primary PCL, including illegitimate IgH rearrangements, chromosome 13 deletion, and serum β2-microglobulin levels. A random distribution ofc-myc rearrangements within each 14q32 subgroup was observed. There was only one weak association, with t(4;14) (P = .08). Similarly, no correlation was found with chromosome 13 deletion, a prognostic variable identified by several groups, including ours.30-32 In contrast, we found a strong association between c-myc rearrangement and a high serum β2-microglobulin level, a well-known indicator of poor prognosis. If the correlation between c-myc rearrangement and a high labeling index is confirmed, and according to the correlation with a high β2-microglobulin serum level, patients with these indicators should have a poor outcome. No prognostic analysis has yet been done in our series (because the follow-up time is still too short), but this will constitute a major challenge in the future.
In conclusion, our data do not confirm those obtained in HMCLs and plasmacytomas induced in mice. However, most of the murine models are not good reproductions of the human disease, and many biologic findings in mouse plasmacytomas are not relevant for human MM. The most relevant model of human MM is the spontaneous 5T model, in whichc-myc rearrangements are rarely reported. The data presented here provide additional support for use of this model in future animal studies.
We thank Axelle Daviet, Stéphanie Saulnier, and Hélène Masson for excellent technical assistance.
Supported by grants from the Association de Recherche contre le Cancer and the Fondation de France.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hervé Avet-Loiseau or Régis Bataille, Laboratoire d'Hématologie, Institut de Biologie, 9 quai Moncousu, 44093 Nantes Cedex 01, France; e-mail:havetloiseau@chu-nantes.fr or frb@sante.univ-nantes.fr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal