Abstract
p15INK4b and p16INK4a proteins are cell cycle regulators involved in the inhibition of G1 phase progression. High frequency of methylation of both genes has been reported in multiple myeloma (MM), but it remains to be determined how and when these alterations contribute to tumorigenesis. Monoclonal gammopathy of undetermined significance (MGUS) represents an early disease stage in a fraction of MMs. Plasma cells from 33 patients with MGUS and 33 patients with MM were isolated and analyzed forp15INK4b and p16INK4amethylation by methylation-specific polymerase chain reaction. Selective methylation was found in 19% forp16INK4a, 36% forp15INK4b, and 6.5% for both genes in MGUS, and frequencies were similar in MM suggesting that methylation of these genes is an early event, not associated with transition from MGUS to MM. p15INK4b andp16INK4a gene methylation might contribute to immortalization of plasma cells rather than malignant transformation in the natural history of MM.
Introduction
Multiple myeloma (MM) has been related to monoclonal gammopathy of undetermined significance (MGUS). Patients with MGUS show a significant risk of progression to MM with an annual actuarial risk of malignant transformation of 0.8%, and up to 33% of newly diagnosed MM patients had a previous history of MGUS.1,2 Genes involved in the occurrence of MGUS and those promoting malignant transformation from MGUS to MM have not been identified.3 One of the most frequent gene alterations in MM is methylation of the p15INK4b andp16INK4a genes in the 5′ upstream region.4-10 p15INK4b and p16INK4aproteins are cell cycle regulators involved in the inhibition of G1 phase progression. Both proteins associate with cyclin-dependent kinases 4 and 6 (CDK4, CDK6) and cyclin D-CDK4/6 complexes, and inhibit their kinase activities. The p16INK4a andp15INK4b genes can be inactivated by homozygous deletion, point mutation, or methylation in various tumor types. Frequencies of p16INK4a orp15INK4b gene methylation up to 75% have been reported in MM and in myeloma-derived cell lines.4,10However, it is not known if p16INK4a,p15INK4b, or both genes are already methylated in some MGUS or if methylation occurs during malignant transformation. The number of clonal plasma cells in the bone marrow from MGUS appears to be very low and Zandecki et al showed that, in contrast to myeloma cells, which are usually really monoclonal, several cytogenetic subclones may coexist within MGUS, suggesting that detection ofp16INK4a and p15INK4bgene methylation would need a sensitive method of analysis.11,12 In order to investigate if methylation of the p16INK4a and p15INK4bgenes occurs early in MGUS or later in MM, we selected plasma cells from patients with MGUS and patients with MM and analyzedp16INK4a and p15INK4bgene methylation using the methylation-specific polymerase chain reaction (MS-PCR).13
Study design
Bone marrow mononuclear cells from patients with MGUS and patients with MM were isolated by Ficoll Hypaque sedimentation and plasma cells were purified using the anti-CD138 plasma cell isolation system (Miltenyi-Biotec, Bergisch Gladbach, Germany) according to the manufacturer's recommendations. Purities of positive and negative fractions were analyzed in 6 MGUS samples and 6 MM samples by flow cytometry analysis using a phycoerythrin (PE)–conjugated mouse antihuman CD138 monoclonal antibody (Beckman Coulter, Miami, FL) or control isotype added simultaneously to cell separation. Morphologic evaluation of the positive fraction was also performed on stained cytocentrifuge slides. Purified plasma cells were frozen and stored in liquid nitrogen before use.
DNA extracted from CD138-selected cells was modified for MS-PCR by bisulfite using the CpGenome DNA Modification Kit (Intergen, Purchase, NY) and p16INK4a andp15INK4b gene-promoter regions were amplified with DNA-methylated and -unmethylated specific primers using CpG WIZ amplification kits (Intergen) according to the manufacturer's recommendations. Reactions were hot-started using AmpliTaq Gold (Perkin Elmer, Foster City, CA). A first step of denaturation at 95°C for 10 minutes was followed by 35 cycles of amplification (30 seconds at 95°C, 30 seconds at 60°C, 30 seconds at 72°C), and by a final 10-minute extension at 72°C. Controls without DNA were performed for each set of PCR reactions. PCR products (10 μL) were loaded on 2% agarose gels stained with GelStar nucleic acid gel stain (BioWhittaker, Walkersville, MD) and visualized under ultraviolet (UV) illumination directly and with a gel scan software analysis system (Bio-Print, Marne la Vallée, France). DNA from the HeLa cell line, which has been reported to be unmethylated forp15INK4b and p16INK4a, was used as negative control. DNA from the Raji cell line, previously reported to have p15INK4b andp16INK4a extensive methylation, and from the RPMI-8226 MM cell line, which is methylated for thep16INK4a but not thep15INK4b gene, were used as positive controls.4 14
Deletion of chromosome 13 was also searched in 21 MGUS samples and 17 MM samples by fluorescence in situ hybridization (FISH) using the D13S319-probe (Vysis, Downers Grove, IL) mapping at 13q14 as previously reported.15
Results and discussion
Thirty-three samples of plasma cells isolated from patients with MGUS and 33 samples from patients with MM were analyzed by MS-PCR. Purity of the positive fraction was more than 91% in the 6 MGUS samples and 6 MM samples. Examinations of cytocentrifuge slides showed plasma cell morphology in all cells. CD138+ cells in all negative fractions were less than 1%. Patient characteristics are presented in Table 1.
Patient characteristics and results of methylation-specific polymerase chain reaction
| . | MGUS . | . | Multiple myeloma . | . |
|---|---|---|---|---|
| Number of patients | 33 | 33 | ||
| Sex ratio | 1.06 | 2 | ||
| Median age (years) | 59 (43-80) | 64 (39-83) | ||
| Median time from diagnosis to MS-PCR (months) | 0 (0-246) | 8 (0-156) | ||
| Median percent of plasma cell in bone marrow | 1.5 (1-8) | 19 (1-86) | ||
| Monoclonal component isotype | ||||
| IgG | 26 | 22 | ||
| IgA | 6 | 7 | ||
| Other | 1 | 4 | ||
| Light chain of the monoclonal protein | ||||
| Kappa | 22 | 27 | ||
| Lambda | 11 | 6 | ||
| Deletion of chromosome 13 | 2/21 | 7/17 |
| . | MGUS . | . | Multiple myeloma . | . |
|---|---|---|---|---|
| Number of patients | 33 | 33 | ||
| Sex ratio | 1.06 | 2 | ||
| Median age (years) | 59 (43-80) | 64 (39-83) | ||
| Median time from diagnosis to MS-PCR (months) | 0 (0-246) | 8 (0-156) | ||
| Median percent of plasma cell in bone marrow | 1.5 (1-8) | 19 (1-86) | ||
| Monoclonal component isotype | ||||
| IgG | 26 | 22 | ||
| IgA | 6 | 7 | ||
| Other | 1 | 4 | ||
| Light chain of the monoclonal protein | ||||
| Kappa | 22 | 27 | ||
| Lambda | 11 | 6 | ||
| Deletion of chromosome 13 | 2/21 | 7/17 |
| . | . | Stage . | ||
|---|---|---|---|---|
| I/II . | III . | Relapsed or refractory . | ||
| Methylation of: | ||||
| p16INK4a | 6/31 (19%) | 1/6 (16.5%) | 3/19 (16%) | 2/5 (40%) |
| p15INK4b | 12/33 (36%) | 2/6 (33%) | 4/18 (22%) | 1/5 (20%) |
| p16INK4a andp15INK4b | 2/31 (6.5%) | 0/6 (0%) | 2/18 (11%) | 1/5 (25%) |
| . | . | Stage . | ||
|---|---|---|---|---|
| I/II . | III . | Relapsed or refractory . | ||
| Methylation of: | ||||
| p16INK4a | 6/31 (19%) | 1/6 (16.5%) | 3/19 (16%) | 2/5 (40%) |
| p15INK4b | 12/33 (36%) | 2/6 (33%) | 4/18 (22%) | 1/5 (20%) |
| p16INK4a andp15INK4b | 2/31 (6.5%) | 0/6 (0%) | 2/18 (11%) | 1/5 (25%) |
MGUS indicates monoclonal gammopathy of undetermined significance; MS-PCR, methylation-specific polymerase chain reaction; IgG, immunoglobulin G.
In agreement with previous reports, both the RPMI-8226 and Raji cell lines were methylated for the p16INK4agene.4 Results of MS-PCR in MGUS and MM samples are summarized in Table 1. Selective methylation was found in 19% forp16INK4a, 36% forp15INK4b, and 6.5% for both genes in MGUS. No correlation could be made between methylation and gender, age, isotype, or level of M-component. MS-PCR showed in all MGUS and MM samples the 154-bp and 162-bp bands corresponding to respective amplifications of unmethylated p16INK4a andp15INK4b genes (Figure1). It has been previously shown thatp16INK4a and p15INK4bgene methylation in acute leukemia is heterogeneous and that, even in heavily methylated cell lines, unmethylatedp16INK4a and p15INK4bgene DNA could be detected.16 As several subclones may coexist in MGUS, it is also possible that thep16INK4a and p15INK4bgenes were methylated in only a fraction of clonal cells.11 12 Although we cannot rule out that, especially in MGUS, MS-PCR detected unmethylated DNA in non-clonal cells. Analysis of the CD138− fraction of 16 MGUS samples and 9 MM samples showed that among 8 MGUS samples and 4 MM samples methylated for thep15INK4b gene, and 5 MGUS samples and 3 MM samples methylated for the p16INK4a gene in CD138-selected cells, one MGUS sample and one MM sample were also methylated for p16INK4a, and one MM sample was methylated for p15INK4b in the CD138− fraction. Flow cytometry analysis of this MGUS sample showed that 0.3% of cells remained CD138+ after separation in the negative fraction. No methylation of either thep16INK4a or p15INK4b gene was detected in the CD138− fraction of MGUS and MM samples unmethylated in CD138+-selected cells. These data suggest that methylation occurs preferentially in CD138+ cells. However, given the fact that few CD138 cells persisted in negative fractions, we cannot rule out that other bone marrow cells were methylated in few patients.
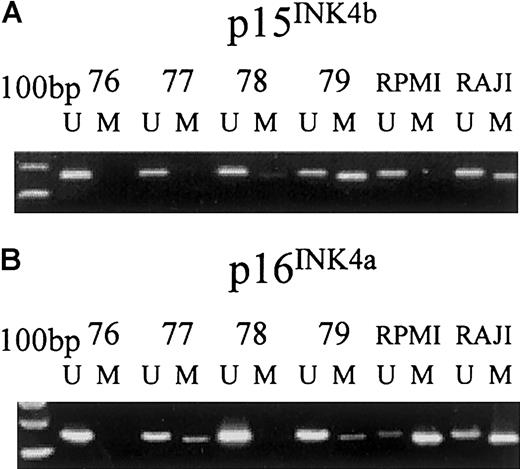
Methylation-specific PCR of thep15INK4b and p16INK4agenes in selected plasma cells from patients with MGUS.
MS-PCR was performed with specific primers for unmethylated (U) and methylated (M) p15INK4b and p16INK4a alleles in Raji and RPMI-8226 cell lines as control and in CD138-selected MGUS plasma cells (samples 76, 77, 78, and 79). PCR product of appropriate molecular weight indicates the presence of unmethylated and/or methylated (A) p15INK4b (162 bp and 154 bp for U and M, respectively) and (B) p16INK4a alleles (154 bp and 145 bp for U and M, respectively) in that sample.
Methylation-specific PCR of thep15INK4b and p16INK4agenes in selected plasma cells from patients with MGUS.
MS-PCR was performed with specific primers for unmethylated (U) and methylated (M) p15INK4b and p16INK4a alleles in Raji and RPMI-8226 cell lines as control and in CD138-selected MGUS plasma cells (samples 76, 77, 78, and 79). PCR product of appropriate molecular weight indicates the presence of unmethylated and/or methylated (A) p15INK4b (162 bp and 154 bp for U and M, respectively) and (B) p16INK4a alleles (154 bp and 145 bp for U and M, respectively) in that sample.
Frequencies of methylation of the p15INK4b andp16INK4a genes were not significantly different among MGUS samples and MM samples (P = .44 and .88, respectively; chi square test), and also between stage I/II and stage III/relapsed or refractory patients with MM (P = .9 and .637, respectively; Fisher exact test). Results were obtained on purified CD138 cells, thus limiting possible variations of results related to variable plasma cell marrow infiltration. Among 21 patients with MGUS analyzed by FISH in our experiments, 2 of them showed deletion of chromosome 13 in plasma cells and both were methylated for the p15INK4b gene without methylation ofp16INK4a. Deletion of chromosome 13 was found in 7 of 17 patients with MM; 2 of them showed exclusive methylation ofp15INK4b gene and one showed exclusive methylation of p16INK4a. Monosomy 13 has been associated with the transition from MGUS to MM and to poor prognosis of MM.15,17 These findings suggest thatp16INK4a and p15INK4bgene methylations and acquisition of monosomy 13 are distinct events in the evolution of MGUS. Ng et al reported similar incidence ofp15INK4b and p16INK4agene methylation in pretreated and posttreated patients with MM.10 Inactivation of p16INK4a gene has been reported in benign tumors like adenomas and in premalignant lesions.18-20 Loss of p16INK4a gene expression by methylation occurs early in the establishment of cell lines from primary culture. These data suggest thatp15INK4b and p16INK4agene methylations are not associated with malignant transformation from MGUS to MM but rather might contribute to immortalization of plasma cells.
Fourteen MGUS samples (12 of them without methylation ofp16INK4a) had p15INK4bgene methylation. Frequent methylation of thep15INK4b gene has only been described in MM, Burkitt lymphoma, acute leukemia, and myelodysplastic syndromes (MDS).14,21 p15INK4b protein is one of the effectors of regulatory effect of transforming growth factor-β (TGF-β). TGF-β can antagonize in vitro the effect of interleukin 6 (IL-6) in normal B cells, but in contrast can also trigger IL-6 secretion by malignant plasma cells and does not alter pRb phosphorylation in these cells.22 Mutation of the TGF-β receptor and alteration of TGF-β signal transduction have been described in various human and murine tumors, and TGF-β is a key regulator of bone marrow stem cells.23 Methylation of thep15INK4b gene might be a mechanism for plasma cells in MGUS to escape to TGF-β inhibitory effect.
Ploidy, immunophenotype, and cytokine expression profiles have been proposed as possible specific characteristics of high-risk MGUS. However, because it takes decades to evaluate malignant transformation rates, the only known risk factors of transformation of MGUS toward hematologic malignancies remain the level and kinetics of the increase of the M-component. Given the low number of patients analyzed and the short follow-up, we are not currently able to know whether methylation of the p15INK4b andp16INK4a genes might define a subset of patients with MGUS who are likely to develop MM. Moreover, we do not know yet extensively which other molecular events occur during the transformation of MGUS toward MM. Such information is needed to design experiments comparing properties of methylated and unmethylated clones isolated from MGUS.
In conclusion, we showed that p15INK4b andp16INK4a gene methylations are present at similar incidences in patients with MGUS and patients with MM, supporting the idea that alteration of the regulation of G1 phase of the cycle is a very early event in the history of MM.
Supported by the Ligue Contre le Cancer (Comité du Nord and Comité du Pas de Calais).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Bruno Quesnel, Service des Maladies du Sang, CHU Lille, 1 Place de Verdun, 59037 Lille, France; e-mail:bquesnel@nordnet.fr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal