Abstract
The development of an immune response to infused factor VIII is a complication affecting many patients with hemophilia A. Inhibitor antibodies bind to antigenic determinants on the factor VIII molecule and block its procoagulant activity. A patient-derived inhibitory immunoglobulin G4κ antibody (BO2C11) produced by an immortalized memory B-lymphocyte cell line interferes with the binding of factor VIII to phospholipid surfaces and to von Willebrand factor. The structure of a Fab fragment derived from this antibody complexed with the factor VIII C2 domain was determined at 2.0 Å resolution. The Fab interacts with solvent-exposed basic and hydrophobic side chains that form a membrane-association surface of factor VIII. This atomic resolution structure suggests a variety of amino acid substitutions in the C2 domain of factor VIII that might prevent the binding of anti-C2 inhibitor antibodies without significantly compromising the procoagulant functions of factor VIII.
Introduction
Factor VIII is a large, 2332-residue plasma glycoprotein that acts as a regulatory cofactor in the process of blood coagulation.1-3 It binds to activated factor IX (factor IXa) in the presence of calcium and negatively charged phospholipids that are presented at the surface of activated platelets to form a membrane-associated, proteolytically active complex. Upon complex formation, the Vmax (maximum velocity) of factor IXa is increased by approximately 200 000-fold, promoting the rapid activation of its substrate, the serine protease factor X. The proteolytic conversion of factor X to its active form, Xa, is a central control point in the coagulation cascade, leading to activation of thrombin, formation of a fibrin mesh, and establishment of a stable blood clot. The binding of factor VIIIa and other activated proteins to these membrane surfaces allows for localization of procoagulant processes to sites of vascular damage.
The factor VIII sequence contains 6 sequential domains arranged in the order A1-A2-B-A3-C1-C2.4-6 The A domains are homologous to one another and display sequence similarity to the copper-binding protein ceruloplasmin. They are flanked by short spacer sequences that are highly acidic. The C domains are also homologous to each other and have a weak homology to the discoidin protein fold family (eg, the lipid-binding domain of galactose oxidase).7,8 The circulating form of the factor VIII protein is a metal bridged heterodimer consisting of a heavy chain (A1-A2-B) and a light chain (A3-C1-C2). This form of factor VIII is bound tightly to von Willebrand factor (vWF). Factor VIII is processed further by specific thrombin cleavages into a heterotrimeric form. This active form, factor VIIIa, dissociates from vWF and binds to negatively charged phospholipids on activated platelet surfaces. The carboxyl terminal C2 domain of factor VIII contains binding sites for vWF and for negatively charged phospholipids. The binding of factor VIIIa to membranes involves stereoselection for O-phospho-L-serine, the negatively charged head group of phosphatidylserine (PS).9 The binding of factor VIII or VIIIa to vWF or PS is mutually exclusive, even though the activation of factor VIII involves cleavages outside the C2 domain. This implies that the binding sites for vWF and PS overlap on the C2-domain surface.
Hemophilia A is a congenital bleeding disorder that is due to deleterious factor VIII gene mutations. These mutations may block factor VIII expression or secretion and may involve premature truncations, sequence rearrangements, or single-residue substitutions. To date, a total of 28 missense mutations at 21 different amino acids within the C2 domain have been associated with hemophilia A.10 The crystal structure of the factor VIII C2 domain was reported recently.11 A mechanism for the interaction of factor VIII with negatively charged phospholipid surfaces, as well as explanations for the molecular basis of hemophilia A point mutations localized to this domain, were proposed on the basis of this study.10 11
Current therapy for patients with hemophilia A involves therapeutic infusions of factor VIII. Inhibitory antibodies against factor VIII, which may occur transiently or may persist as a serious long-term complication, are generated in up to 35% of patients with severe hemophilia A.12-16 These patients generally have very low or undetectable factor VIII antigen. Patients who have mild or moderate hemophilia A, which is usually associated with missense mutations, can also develop inhibitory antibodies. This is less common and occurs in 3% to 13% of patients.17,18 Antibody inhibitor development has been associated with the mutations R593C and W2229C (single-letter amino acid code), with about a 40% inhibitor incidence for each mutation. Autoantibodies to factor VIII can also develop in postpartum women or in individuals with various underlying disease states, but this is very rare, as it occurs in one per million individuals.19 20
The primary antigenic epitopes on factor VIII have been localized to the A2 and C2 domains.21-26 The A3 and C1 domains and the acidic region between A1 and A2 have also been implicated, although epitopes in these regions occur less frequently.27,28Inhibitory antibodies against the A2 domain generally allow factor VIII to form complexes with vWF or factor IXa, but the proteolytic activation of factor X is blocked.13 In contrast, inhibitory antibodies specific to the C2 domain have been shown to prevent the binding of factor VIII to vWF and to membrane surfaces that expose PS. Two studies have localized a C2-domain epitope to the regions between residues 2181 to 224322 and 2248 to 2312.22 29 Blocking the interaction between factor VIII and vWF greatly reduces the half-life of factor VIII in the circulation, while antibodies that prevent the binding of factor VIIIa to membrane surfaces abolish its procoagulant cofactor function.
To gain further insight into the interaction of autoantibodies and alloantibodies against factor VIII at the molecular level, a factor VIII–specific human IgG4κ antibody (BO2C11) was produced by a cell line derived from the memory B-cell repertoire of a patient with hemophilia A with a strong inhibitor response.30 Biosensor measurements indicated that although the association rate of factor VIII with BO2C11 is slower than that measured for factor VIII binding to vWF, the dissociation rate of the factor VIII:BO2C11 complex is 100-fold slower than that of the factor VIII:vWF complex.30 Thus, the current working model for the inactivation of factor VIII by BO2C11 is that the antibody forms a tight, stable complex with factor VIII (or factor VIIIa) as it dissociates from vWF. The antibody blocks the C2-domain membrane-binding site, thus effectively sequestering factor VIII and neutralizing its procoagulant effects. The antibody complex may also lead to accelerated clearance of the bound factor VIII, but this has not been measured directly.
Materials and methods
The recombinant C2 domain was expressed and purified as described previously.11 The factor VIII C2-domain–specific human IgG4κ monoclonal antibody (BO2C11) was digested with papain, and the Fab fragment was purified using standard methods. Approximately equimolar amounts of Fab and C2 protein were combined in 10 mM HEPES, 0.1 M NaCl, pH 7.5, at approximately 10 mg/mL total protein concentration. Crystals were grown at room temperature in 16% polyethylene glycol (PEG) 8000, 0.1 M HEPES (pH 7.0), 0.2 M NaCl. The crystals were flash frozen in the same solution, plus a final concentration of 25% vol/vol PEG 400 as cryoprotectant. Data were collected to 2.0 Å resolution at the Advanced Light Source (Berkeley, CA) beamline 5.0.2 and processed using Denzo/Scalepack program suite.31 The space group was P212121 with unit cell parameters a = 64.5 Å, b = 73.7 Å, andc = 112.4 Å. The structure was solved by molecular replacement with the use of EPMR version 2.1.32 The models used for molecular replacement during independent, iterative searches were the recombinant factor VIII C2 domain,11 the constant domain of a human IgG4κ Fab (pdb acc.: 1BBJ), and a molecular model of the variable domain that used optimal CDR loop length and sequence alignment (pdb: 1GC1 and 1AD9). The constant-domain search model was found by searching the Protein Data Bank for a structure of a Fab of homologous classification. The heavy and light variable-chain sequences were used independently to locate optimal search models, and the program Blast (Basic local alignment search tool) was used to search the Protein Data Bank (the VL and VH sequences are in the European Molecular Biology Laboratory (EMBL) Nucleotide Sequence Database under the accession numbers AJ224084 and AJ224083, respectively). As each phase solution was determined, it was fixed in orientation as subsequent rounds were performed. Model building of all deleted loops and side chains was performed with XFIT33 version 3.7, and the structure was refined using CNS34 after removing 10% of the data for cross-validation purposes. The final model consists of 564 amino acids and 477 water molecules. The Rcryst and Rfree both decreased at every step of the refinement, and the final Rcryst was 20.4% and Rfree was 25.5%. The stereochemical validity of the final model was verified using Procheck.35 Data and refinement statistics are shown in Table 1.
Data and refinement statistics
| Type of data . | Category . | Result . |
|---|---|---|
| Diffraction data | ||
| Resolution | 2.0 Å | |
| Source | P212121 | |
| Unit cell (a) | 64.5 Å | |
| (b) | 73.7 Å | |
| (c) | 112.4 Å | |
| Wavelength | 1.07 Å | |
| Unique reflections | 36 867 | |
| Redundancy | 7.7 | |
| Completeness* | 99.6 (98.8) | |
| Rmerge | 6.1 | |
| Refinement | ||
| Resolution range | 50-2.0 Å | |
| Rcryst | 20.4% | |
| Rfree(10%) | 25.5% | |
| No. of protein atoms | 4335 | |
| No. of solvent atoms | 477 | |
| Ramachandran dist. | 87.8, 11.4 | |
| (% core, allowed, generously allowed, disallowed) | 0.4, 0.4† | |
| rms bonds, angles | 0.0054, 1.33 | |
| Average 〈B〉1-153 | 24.25 | |
| Molecular replacement‡ | ||
| Model 1 (variable domain) | Correlation coefficient 0.416 R value 0.733 | |
| Model 2 (C2 domain) | Correlation coefficient 0.381 R value 0.753 | |
| Model 3 (constant domain) | Correlation coefficient 0.564 R value 0.651 |
| Type of data . | Category . | Result . |
|---|---|---|
| Diffraction data | ||
| Resolution | 2.0 Å | |
| Source | P212121 | |
| Unit cell (a) | 64.5 Å | |
| (b) | 73.7 Å | |
| (c) | 112.4 Å | |
| Wavelength | 1.07 Å | |
| Unique reflections | 36 867 | |
| Redundancy | 7.7 | |
| Completeness* | 99.6 (98.8) | |
| Rmerge | 6.1 | |
| Refinement | ||
| Resolution range | 50-2.0 Å | |
| Rcryst | 20.4% | |
| Rfree(10%) | 25.5% | |
| No. of protein atoms | 4335 | |
| No. of solvent atoms | 477 | |
| Ramachandran dist. | 87.8, 11.4 | |
| (% core, allowed, generously allowed, disallowed) | 0.4, 0.4† | |
| rms bonds, angles | 0.0054, 1.33 | |
| Average 〈B〉1-153 | 24.25 | |
| Molecular replacement‡ | ||
| Model 1 (variable domain) | Correlation coefficient 0.416 R value 0.733 | |
| Model 2 (C2 domain) | Correlation coefficient 0.381 R value 0.753 | |
| Model 3 (constant domain) | Correlation coefficient 0.564 R value 0.651 |
Rcryst indicates refinement R-factor; Rfree, cross-validation R-factor on 5% of randomly selected reflections.
Completeness is reported for all reflections and for the highest resolution shell.
Two residues (A52 and H2315) are in the disallowed region of the Ramachandran plot. Both residues have good electron density to indicate proper conformation.
Values reported by EPMR version 2.1.
Average temperature factor of crystallographic structure.
Results
The structure of the C2-domain–BO2C11 Fab complex was solved to 2.0 Å resolution (Table 1). The factor VIII C2-domain construct consists of residues 2171 to 2332. The heavy and light chains of the Fab fragment consist of residues 1 to 211 and 1 to 212, respectively. Electron density of the entire complex was of excellent quality except for 3 residues at the C2-domain amino and carboxyl termini, 4 residues in the light chain, and 13 residues in the heavy chain. None of these residues was involved in the binding interface. The crystallographic model contains a total of 477 water molecules, 41 of which reside in the binding region of the complex. The core structure of the C2 domain is completely conserved, displaying an 8-stranded β–sandwich fold. The amino and carboxyl terminal regions of the C2 domain are linked by an internal disulfide bond joining C2174 to C2326. Two exposed hydrophobic β turns and an underlying pair of basic residues (R2215 and R2220), all previously hypothesized to participate in the C2-membrane–binding site, formed critical interactions at the antibody interface (Figures1 and 2; Table 2). An area of approximately 1200 Å2 on the surface of each molecule is buried in the protein interface. The C2-epitope surface is basic in character, whereas the Fab surface is quite acidic at the interface, indicating a favorable overall charge complementarity (Figure3).
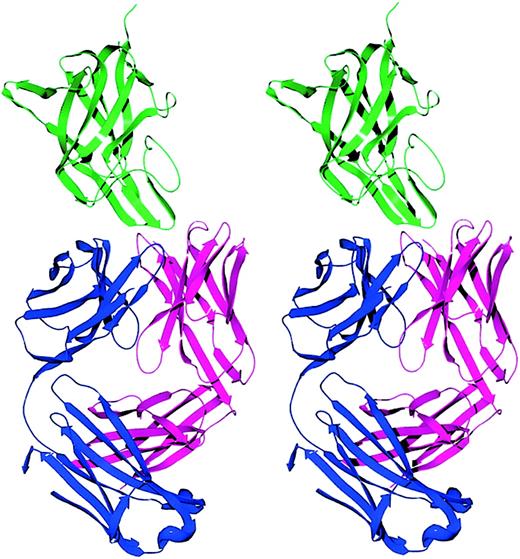
Stereo ribbon diagram of the factor VIII C2-domain/BO2C11 Fab complex.
The light chain, heavy chain, and C2 domain are displayed in blue, magenta, and green, respectively. The BO2C11 Fab displays a typical immunoglobulin fold with 18 β strands in the light chain and 19 β strands in the heavy chain. Two β hairpins from the C2 domain project into the CDR loops of the Fab fragment.
Stereo ribbon diagram of the factor VIII C2-domain/BO2C11 Fab complex.
The light chain, heavy chain, and C2 domain are displayed in blue, magenta, and green, respectively. The BO2C11 Fab displays a typical immunoglobulin fold with 18 β strands in the light chain and 19 β strands in the heavy chain. Two β hairpins from the C2 domain project into the CDR loops of the Fab fragment.
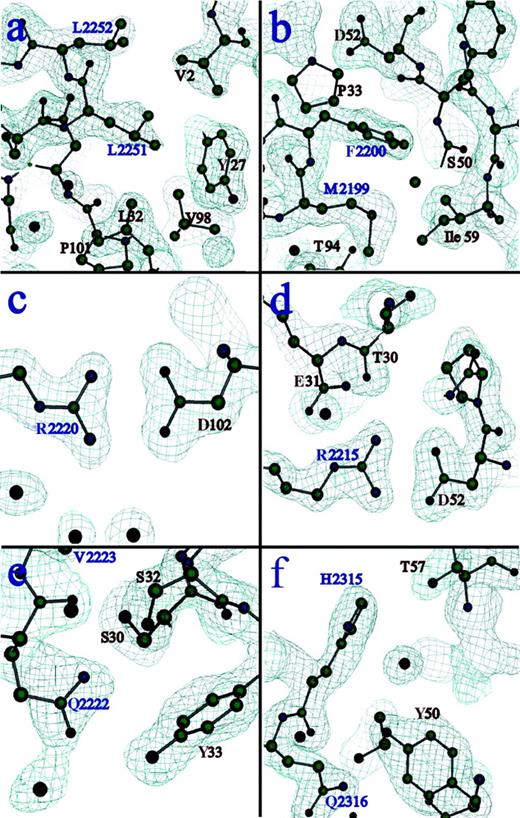
Electron density at residues involved in critical interactions of the binding interface.
The first antigenic peptide of C2 (residues 2250 to 2253), corresponding to an exposed hydrophobic β turn containing L2251 and 2252, forms primarily hydrophobic interactions with residues from CDR-H1, CDR-H3, and the amino terminus of the heavy chain (A). The second antigenic region from C2 (residues 2197 to 2203), corresponding to a second exposed hydrophobic β turn containing M2199 and F2200, exhibits more extensive van der Waals contact with the Fab surface, and there are more polar interactions than in the first epitope (B). Two R residues from the C2 domain, both of which are proposed to interact with anionic lipid head groups when factor VIII binds to platelet membranes, form salt links with D residues on the Fab surface. R2220 lies within a cleft between the 2 hydrophobic hairpin turns (C) and interacts with D102 of the heavy chain. R2215 resides on the third loop at this end of the C2 molecule and interacts with D52 of the heavy chain (D). An adjacent loop contains residues Q2222 and V2223, which contact the Fab surface directly (E). V2223 is another hydrophobic residue that was solvent-exposed in the free C2 structure, and it was proposed that it may make additional contacts with membrane surfaces. Finally, residues H2315 and Q2316 participate in polar interactions with specific residues and buried water molecules at the C2-Fab interface (F). See Table 2 for a list of all contacts in this interface.
Electron density at residues involved in critical interactions of the binding interface.
The first antigenic peptide of C2 (residues 2250 to 2253), corresponding to an exposed hydrophobic β turn containing L2251 and 2252, forms primarily hydrophobic interactions with residues from CDR-H1, CDR-H3, and the amino terminus of the heavy chain (A). The second antigenic region from C2 (residues 2197 to 2203), corresponding to a second exposed hydrophobic β turn containing M2199 and F2200, exhibits more extensive van der Waals contact with the Fab surface, and there are more polar interactions than in the first epitope (B). Two R residues from the C2 domain, both of which are proposed to interact with anionic lipid head groups when factor VIII binds to platelet membranes, form salt links with D residues on the Fab surface. R2220 lies within a cleft between the 2 hydrophobic hairpin turns (C) and interacts with D102 of the heavy chain. R2215 resides on the third loop at this end of the C2 molecule and interacts with D52 of the heavy chain (D). An adjacent loop contains residues Q2222 and V2223, which contact the Fab surface directly (E). V2223 is another hydrophobic residue that was solvent-exposed in the free C2 structure, and it was proposed that it may make additional contacts with membrane surfaces. Finally, residues H2315 and Q2316 participate in polar interactions with specific residues and buried water molecules at the C2-Fab interface (F). See Table 2 for a list of all contacts in this interface.
Atomic contacts between factor VIII C2 domain and BO2C11
| C2-domain residue . | BO2C11 residue . | Fab chain . | Distance (Å) . |
|---|---|---|---|
| Peptide 1 | |||
| S2250 OG | D100 OD2 | Heavy | 2.5 |
| P101 N | Heavy | 3.7 | |
| L2251 N | D100 OD2 | Heavy | 3.0 |
| L2251 CG | L32 CD1 | Heavy | 3.4 |
| L2251 CD2 | Y27 CD2 | Heavy | 3.8 |
| V2 CG2 | Heavy | 3.5 | |
| L2252 CD2 | V2 CG2 | Heavy | 3.7 |
| L2252 N | D100 OD2 | Heavy | 2.7 |
| T2253 N | D100 OD1 | Heavy | 3.4 |
| T2253 CG2 | P101 CD | Heavy | 3.4 |
| Peptide 2 | |||
| F2196 CE1 | S32 OG | Light | 3.6 |
| Y33 CE1 | Light | 3.5 | |
| F2196 CE2 | D102 OD1 | Heavy | 3.4 |
| T2197 OG1 | Y33 OH | Light | 3.4 |
| T2197 O | Y33 OH | Light | 2.7 |
| N2198 OD1 | D102 O | Heavy | 3.0 |
| N2198 ND2 | G93 O | Light | 3.4 |
| N2198 O | T94 OG1 | Light | 3.6 |
| M2199 N | G93 O | Light | 3.2 |
| M2199 CB | G93 O | Light | 3.1 |
| M2199 CG | T94 O | Light | 3.7 |
| M2199 SD | I59 CD1 | Heavy | 3.1 |
| S50 CB | Heavy | 3.4 | |
| H35 CE1 | Heavy | 3.8 | |
| F2200 CD1 | P33 CB | Heavy | 3.5 |
| F2200 CD1 | D52 CB | Heavy | 3.7 |
| F2200 CZ | E57 OE2 | Heavy | 3.4 |
| I59 CG2 | Heavy | 3.6 | |
| Peptide 3 | |||
| R2220 NH1 | D102 OD2 | Heavy | 2.9 |
| R2220 NH2 | D102 OD1 | Heavy | 3.0 |
| Peptide 4 | |||
| R2215 NE | E31 O | Heavy | 3.0 |
| R2215 NH1 | D52 OD2 | Heavy | 2.8 |
| R2215 NH2 | D52 OD1 | Heavy | 3.0 |
| T30 O | Heavy | 2.9 | |
| Peptide 5 | |||
| Q2222 OE1 | Y33 OH | Light | 3.2 |
| Q2222 NE1 | S30 OG | Light | 3.6 |
| S32 OG | Light | 3.2 | |
| Q2222 O | S32 OG | Light | 3.3 |
| V2223 CG1 | S30 OG | Light | 3.6 |
| Peptide 6 | |||
| H2315 CE1 | T57 OG1 | Light | 3.7 |
| H2315 O | T57 OG1 | Light | 3.5 |
| Q2316 NE2 | Y50 OH | Light | 3.5 |
| Y50 CE1 | Light | 3.3 |
| C2-domain residue . | BO2C11 residue . | Fab chain . | Distance (Å) . |
|---|---|---|---|
| Peptide 1 | |||
| S2250 OG | D100 OD2 | Heavy | 2.5 |
| P101 N | Heavy | 3.7 | |
| L2251 N | D100 OD2 | Heavy | 3.0 |
| L2251 CG | L32 CD1 | Heavy | 3.4 |
| L2251 CD2 | Y27 CD2 | Heavy | 3.8 |
| V2 CG2 | Heavy | 3.5 | |
| L2252 CD2 | V2 CG2 | Heavy | 3.7 |
| L2252 N | D100 OD2 | Heavy | 2.7 |
| T2253 N | D100 OD1 | Heavy | 3.4 |
| T2253 CG2 | P101 CD | Heavy | 3.4 |
| Peptide 2 | |||
| F2196 CE1 | S32 OG | Light | 3.6 |
| Y33 CE1 | Light | 3.5 | |
| F2196 CE2 | D102 OD1 | Heavy | 3.4 |
| T2197 OG1 | Y33 OH | Light | 3.4 |
| T2197 O | Y33 OH | Light | 2.7 |
| N2198 OD1 | D102 O | Heavy | 3.0 |
| N2198 ND2 | G93 O | Light | 3.4 |
| N2198 O | T94 OG1 | Light | 3.6 |
| M2199 N | G93 O | Light | 3.2 |
| M2199 CB | G93 O | Light | 3.1 |
| M2199 CG | T94 O | Light | 3.7 |
| M2199 SD | I59 CD1 | Heavy | 3.1 |
| S50 CB | Heavy | 3.4 | |
| H35 CE1 | Heavy | 3.8 | |
| F2200 CD1 | P33 CB | Heavy | 3.5 |
| F2200 CD1 | D52 CB | Heavy | 3.7 |
| F2200 CZ | E57 OE2 | Heavy | 3.4 |
| I59 CG2 | Heavy | 3.6 | |
| Peptide 3 | |||
| R2220 NH1 | D102 OD2 | Heavy | 2.9 |
| R2220 NH2 | D102 OD1 | Heavy | 3.0 |
| Peptide 4 | |||
| R2215 NE | E31 O | Heavy | 3.0 |
| R2215 NH1 | D52 OD2 | Heavy | 2.8 |
| R2215 NH2 | D52 OD1 | Heavy | 3.0 |
| T30 O | Heavy | 2.9 | |
| Peptide 5 | |||
| Q2222 OE1 | Y33 OH | Light | 3.2 |
| Q2222 NE1 | S30 OG | Light | 3.6 |
| S32 OG | Light | 3.2 | |
| Q2222 O | S32 OG | Light | 3.3 |
| V2223 CG1 | S30 OG | Light | 3.6 |
| Peptide 6 | |||
| H2315 CE1 | T57 OG1 | Light | 3.7 |
| H2315 O | T57 OG1 | Light | 3.5 |
| Q2316 NE2 | Y50 OH | Light | 3.5 |
| Y50 CE1 | Light | 3.3 |
All atoms in the interface from opposing protein molecules displaying contact distances of less than 3.8 Å (corresponding to a conservative estimate of the upper range of distance for van der Waals interactions) are included. The six separate peptides from the factor VIII C2 domain correspond to those forming the observed epitope for BO2C11 binding, as described in “Results” and “Discussion” and illustrated in Figure 2.
Molecular surface representation of the surfaces that interact in the BO2C11–C2-domain complex.
The individual proteins have been separated and rotated to allow the reader to view the complementary, buried protein interfaces. The predominantly negative charge (red) of the Fab fragment interacts favorably with the positively charged patches (blue) on the surface of the C2 domain.
Molecular surface representation of the surfaces that interact in the BO2C11–C2-domain complex.
The individual proteins have been separated and rotated to allow the reader to view the complementary, buried protein interfaces. The predominantly negative charge (red) of the Fab fragment interacts favorably with the positively charged patches (blue) on the surface of the C2 domain.
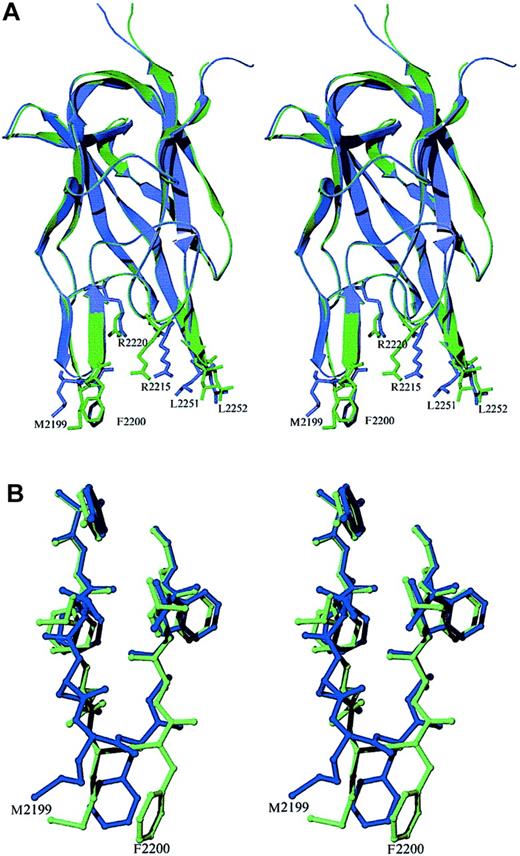
The fold of the C2 domain is similar in the free and Fab-bound forms, with an all-atom rms difference of 0.33 Å. However, there is a shift in the position of the hydrophobic β–hairpin loop containing M2199 and F2200. The backbone of this loop region twists significantly, allowing the side chains to form specific interactions at the binding interface (Figure 4). The analogous region of the factor V C2 domain has been visualized in 2 different conformations,36 indicating that the membrane-binding region has some inherent flexibility.
Stereo diagram of superimposed structures of the recombinant factor VIII C2 domain in the free and antibody-bound crystal forms.
(A) The free and Fab-complexed C2 proteins are shown in blue and green, respectively. The structure of the β core is virtually identical in the 2 structures. Most of the deviations between the structures occur at or near the 2 hydrophobic β-hairpin turns at the bottom of the figure. The plane formed by the 2 strands of the second β hairpin (residues 2197 to 2203) moves to a position perpendicular to that seen in the unbound C2-domain structure. The ψ angle of M2199 rotates from −11° to +122°, whereas in F2200, the φ and ψ angles rotate from −99° to +56° and from −23° to +26°, respectively. This region interacts with residues in CDR-H1, CDR-H2, and CDR-H3 of the heavy chain as well as with sites on CDR-L1 and CDR-L3 of the light chain. (B) Rotations about backbone dihedral angles in the β hairpin containing residues N2198 through A2201 shift the orientation of these side chains by up to 5 Å.
Stereo diagram of superimposed structures of the recombinant factor VIII C2 domain in the free and antibody-bound crystal forms.
(A) The free and Fab-complexed C2 proteins are shown in blue and green, respectively. The structure of the β core is virtually identical in the 2 structures. Most of the deviations between the structures occur at or near the 2 hydrophobic β-hairpin turns at the bottom of the figure. The plane formed by the 2 strands of the second β hairpin (residues 2197 to 2203) moves to a position perpendicular to that seen in the unbound C2-domain structure. The ψ angle of M2199 rotates from −11° to +122°, whereas in F2200, the φ and ψ angles rotate from −99° to +56° and from −23° to +26°, respectively. This region interacts with residues in CDR-H1, CDR-H2, and CDR-H3 of the heavy chain as well as with sites on CDR-L1 and CDR-L3 of the light chain. (B) Rotations about backbone dihedral angles in the β hairpin containing residues N2198 through A2201 shift the orientation of these side chains by up to 5 Å.
Epitope mapping using fragments of the factor VIII C2 domain indicated that BO2C11 does not recognize a linear epitope corresponding to a single stretch of amino acid residues.30 Rather, it interacts with a conformational epitope composed of side chains from various regions within the C2 sequence that are adjacent to each other in the folded protein. These results are in agreement with the interactions present in this crystal structure, as illustrated in Figure 2 and listed in Table 2. Two β-turn regions (residues 2250 to 2253 and 2197 to 2203) and an additional loop containing residues Q2222 and V2223 present hydrophobic residues at the protein surface. These residues interact with residues from multiple CDRs on the heavy and light chains of the Fab and the amino terminus of the heavy chain. These loop regions of factor VIII represent the largest amount of surface area buried within the antibody interface. Two basic residues from the C2 protein, R2215 and R2220, are postulated to interact with phospholipid head groups. These residues form salt links with D52 and D102 of the heavy chain, respectively. Finally, residues H2315 and Q2316 participate in polar interactions with specific residues and buried water molecules at the antibody interface.
Discussion
The BO2C11 IgG antibody binds to the C2 domain of factor VIII, inhibiting its ability to bind negatively charged phospholipids and vWF.30 The evidence strongly suggests that the BO2C11 antibody masks the membrane-binding surface of the C2 domain and thus prevents binding to negatively charged phospholipids. The membrane-binding surface of factor VIII has been proposed to consist of 2 β-hairpins turns and an adjacent loop. This region displays 5 exposed hydrophobic residues that are available to participate in membrane binding (L2251, L2252, M2199, F2200, and possibly V2223). Four of these hydrophobic residues are buried in the Fab complex, whereas V2223 is located immediately adjacent to the interface and is in contact with the Fab molecule. A ring of positively charged basic residues is positioned to interact with charged phospholipid head groups upon burial of the hydrophobic side chains in the lipid bilayer. Two of these residues, R2215 and R2220, are buried in the Fab interface, where they form salt bridges with D residues in BO2C11 (Figure 2).
The large contact surface between the C2 domain and BO2C11 is in agreement with the high affinity of factor VIII binding to BO2C11, as measured by surface plasmon resonance. The latter showed an antibody–factor VIII association rate constant of 7.4 × 105 · M−1 · s−1 and dissociation rate constant of less than or equal to 1 × 10−5 · s−1, with a calculated apparent dissociation constant of 1.4 × 10−11 · M−1.30
Anti-factor VIII antibodies were initially distinguished according to the kinetics of factor VIII inactivation.37,38 In 1982, Gawryl and Hoyer39 delineated 2 populations of such antibodies: type I antibodies inactivate factor VIII completely, following second-order kinetics; whereas type II inhibitors inactivate factor VIII only partially, even when added in large excess over factor VIII, and follow more complex kinetics. Later, Biggs40observed that even antibodies completely inactivating plasma factor VIII frequently follow complex kinetics of interaction with factor VIII, thereby identifying an intermediate category between type I and type II inhibitor antibodies. As a consequence of their particular kinetics of factor VIII inhibition, such antibodies can be difficult to quantify in plasma using the conventional Bethesda method.41
Competition with vWF for factor VIII binding was first described as the mechanism responsible for the type II kinetic pattern of many type II inhibitor antibodies, which in the absence of vWF completely inhibited factor VIII activity following second-order kinetics, much like type I inhibitors.39 However, some type II inhibitors inactivate factor VIII only when the latter is bound to vWF,21,42 and recent observations have indicated that some type II antibodies inhibit factor VIII partially even in the absence of vWF.27 The population of type II inhibitors therefore appears to be heterogeneous.
The data on the crystal structure of the C2 domain in combination with BO2C11 presented here shed light on the actual mechanisms by which the antibody inhibits factor VIII function. BO2C11 inactivates factor VIII following a kinetic pattern intermediate between that of type I and type II inhibitors. With excess of antibody, factor VIII inactivation by BO2C11 is complete.30 However, when analyzed as a function of time, the kinetics of the reaction is different from that of type I inhibitor antibodies; that is, the relation between the logarithm of residual factor VIII activity and time is not linear (M.J., unpublished results, September 1997). The likely explanation for this observation is that BO2C11 is in competition with vWF for factor VIII binding.30 In plasma, factor VIII is complexed to vWF, which prevents BO2C11 binding. Over time, however, the factor VIII/vWF complex dissociates, and BO2C11 binds to factor VIII in a nearly irreversible manner.30 Indeed, although the association rate constant of BO2C11 is only 8-fold lower than that of vWF for factor VIII, the dissociation rate constant of BO2C11 from factor VIII is 100-fold lower than that of the vWF–factor VIII complex as estimated by Vlot et al.43 Therefore, given the rapid spontaneous dissociation of the factor VIII/vWF complex, the protection toward BO2C11-mediated inactivation provided by vWF is significant only for a short time.44 Moreover, BO2C11 can bind and inactivate factor VIIIa upon its dissociation from vWF, thereby preventing the binding of factor VIIIa to phospholipids. This phenomenon was previously shown to allow rapid inactivation of factor VIII when BO2C11 was present at concentrations of several hundred Bethesda units.30,44 In addition, given the importance of vWF for factor VIII stability in plasma, it is possible that factor VIII bound to BO2C11 is cleared more rapidly from the circulation.45-47 It is noteworthy that polyclonal anti-factor VIII antibodies of the patient from whom the cell line producing BO2C11 was derived inactivate factor VIII following a type I pattern (M.J., unpublished data, September 1997). This probably occurs because in addition to antibodies toward the C2 domain, this patient's plasma contains antibodies recognizing antigenic determinants in other regions of the factor VIII molecule, including the heavy chain.48
The observed competition between the BO2C11 antibody and vWF for binding to factor VIII suggests the straightforward explanation that the C2 domain visualized here either includes or overlaps with the factor VIII binding site for vWF. However, caution must be exercised in proposing structural explanations for the various binding experiments because there are as yet no direct data on the structure of the vWF–factor VIII complex. Two additional hypotheses, which do not exclude the possibility that the C2-domain epitope identified here may overlap the binding site for vWF, can be presented to account for the competition between BO2C11 and vWF for binding to factor VIII. The bound BO2C11 antibody molecule may cause steric interference with the binding of vWF, which is itself an extremely large, multimeric protein. In other words, occlusion of the binding of 2 large proteins may not involve a direct competition for the same surface on the C2 domain, but could be due to interference elsewhere between the factor VIII or vWF proteins. Alternatively, the binding of BO2C11 to the C2 domain may prevent a conformational change within factor VIII that is necessary for vWF binding. In support of the first possibility, indirect steric interference between bound vWF and bound BO2C11, we note that the binding of vWF involves other regions of factor VIII in addition to the C2 domain. An acidic stretch of 41 amino acid residues at the amino terminal region of the light chain (A3-C1-C2) increases the affinity of factor VIII binding to vWF. This peptide is removed during the proteolytic activation of factor VIII. The proximity of this acidic stretch to regions on the C2-domain surface is not yet known.
Several C2-domain missense mutations associated with hemophilia A appear to affect vWF binding, indicating that these residues may represent part of the vWF-binding surface. However, the effects of these mutations, some of which affect bulky side chains (W2229, R2307, and R2304), on the structure of factor VIII have not yet been characterized. Therefore, it is not yet known whether the substitutions alter merely the surface characteristics of the C2 domain or whether they result in more profound effects on folding or secretion of the factor VIII protein. The value of these mutations as a tool for mapping the association surface of C2 with vWF awaits a more complete characterization of the mutations themselves, and it may be possible to investigate this further using recombinant factor VIII constructs. The picture of the C2-domain epitope presented here can now be used to inform additional, detailed mutational and biochemical studies that will test the possible involvement of the epitope region in the binding of vWF to factor VIII.
It is likely that the complex described here is similar to those formed by the C2 domain with other anti-C2 inhibitor antibodies. The equivalence of the epitope with the proposed membrane-binding region is completely consistent with the observed blockage of membrane binding by other anti-C2 inhibitor antibodies. Studies of unrelated antibody complexes have shown that antigenic determinants on protein surfaces may correspond to peptide regions having dynamic flexibility, hydrophobicity, and/or significant solvent accessibility of side chains.49-51 The C2 residues in contact with BO2C11 meet all of these criteria. Crystal structures of the C2 domains of factors V and VIII have shown unequivocally that the β-hairpin turns can assume various orientations under different conditions,11,36 and the crystal structures set only a lower limit on the accessible range of motion available to these loops as they interact with various surfaces. The epitope region is also highly hydrophobic and presents a side-chain surface area that is larger than that of other surface loops of similar length on the C2 domain. The membrane-binding surface of the C2 domain thus appears to represent an antigenic “hotspot” on the factor VIII surface. A sequence alignment of human and porcine factor VIII supports this conclusion (Figure 5). Chimeric human/porcine factor VIII molecules have been constructed that exhibit less antigenicity against a variety of patient-derived antibody inhibitors. Such chimeric proteins represent a promising approach to improve treatment options for patients who develop inhibitory antibodies.52 Inspection of the sequence alignment reveals that there are significant differences between the human and porcine sequences at and around the β-hairpin turn containing M2199 and F2200. Indeed, recent experiments using recombinant factor VIII proteins incorporating point mutations in this region have shown that the substitutions affected the antigenicity of factor VIII against both polyclonal and monoclonal antibodies. These results are consistent with the BO2C11 epitope identified here.53
Sequence alignment of the C2 domains of human and porcine factor VIII.
The regions corresponding to those in the C2-domain epitope in contact with the BO2C11 Fab are underlined.
Sequence alignment of the C2 domains of human and porcine factor VIII.
The regions corresponding to those in the C2-domain epitope in contact with the BO2C11 Fab are underlined.
Only 2 point mutations associated with hemophilia A, A2201P and V2223M, have been identified within the antibody interface identified here. The patients carrying these mutations suffered from relatively mild bleeding disorders.10 It was proposed previously that the relative dearth of deleterious point mutations near the exposed hydrophobic surface of the C2 domain reflected a redundant, protective evolution of the membrane-binding region.10 11 The binding energy to membrane surfaces was proposed to derive from a combination of favorable solvation changes upon insertion of hydrophobic residues into a nonpolar lipid environment. Favorable electrostatic interactions were also proposed between several basic residues and polar phospholipid head groups. At least 10 to 12 amino acid side chains were deemed likely to contribute to the interface with the membrane. Because the free energy of solvation changes involving hydrophobic moieties is relatively insensitive to precise side-chain orientation, the attachment of the C2 domain to membranes should readily accommodate minor structural changes due to movement of the loops or substitutions of individual side chains. Similarly, the large number of residues involved in the membrane–protein interface would tend to minimize the energetic penalty of mutating any individual side chain in this region. In contrast, the binding of this C2 surface to protein ligands, including vWF or inhibitor antibodies, may be more vulnerable to conformational changes and to disruptions caused by point mutations. This may have positive implications for patients with hemophilia A with an antibody inhibitor response. It should be possible to introduce minor modifications to these loops that preclude an effective antibody/antigen interaction but that result in a molecule that is still competent to bind membranes and to carry out the critical cofactor function of factor VIII in coagulation. It is well established that effective hemostasis is possible even with a plasma concentration of factor VIII as low as 10% of average levels. Below this level, an increase in factor VIII concentration of even a few percentage points can have a profound impact on the quality of life for the patient. Thus, the design of a “hobbled” factor VIII, which may sacrifice some membrane-binding capacity to decrease the antigenicity of the infusion, may be a desirable and achievable goal. The identification here of specific amino acid residues mediating an inhibitor antibody response presents a compelling opportunity to further fine tune the production of improved, recombinant “designer” factor VIII proteins that are tolerated by a larger fraction of the patient population.
We thank Betty W. Shen for assistance at all stages of structure determination; Eric Galburt, Roland Strong, Kam Zhang, and Adrian Ferre-D'Amare for advice and discussion; Earl Davie and Kazuo Fujikawa for assistance with the C2-domain purification and analysis; Benoı̂t Desqueper for production of BO2C11 Fab fragment; and Thomas Earnest and staff at ALS beamline 5.0.2 for assistance with data collection.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Barry L. Stoddard, Fred Hutchinson Cancer Research Center, Division of Basic Sciences, 1100 Fairview Ave North A3-023, Seattle, WA 98109; e-mail: bstoddar@fhcrc.org.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal