Abstract
The aim of this study was to characterize individual-segment and overall patterns of VH gene usage in adult B-lineage acute lymphoblastic leukemia (ALL). Theoretical values of VH segment usage were calculated with the assumption that all VH segments capable of undergoing rearrangement have an equal probability of selection for recombination. Leukemic clones from 127 patients with adult B-lineage acute leukemias were studied by fingerprinting by means of primers for the framework 1 and joining segments. Clones from early preimmune B cells (245 alleles identified) show a predominance of VH6 family rearrangements and, consequently, do not conform to this hypothesis. However, profiles of VH gene family usage in mature B cells, as investigated in peripheral blood (6 samples), B-cell lymphomas (36 clones) and chronic lymphocytic leukemia (56 clones), are in agreement with this theoretical profile. Sequence analyses of 64 VH clones in adult ALL revealed that the rate of VH usage is proportional to the proximity of the VH gene to the JH locus and that the relationship can be mathematically defined. Except for VH6, no other VH gene is excessively used in adult ALL. VH pseudogenes are rarely used (n = 2), which implies the existence of early mechanisms in the pathway to B-cell maturation to reduce wasteful VH-(DH)-JHrecombination. Finally, similar to early immunoglobulin-H rearrangement patterns in the mouse, B cells of ALL derive from a pool of cells more immature than the cells in chronic lymphoid B-cell malignancies.
Introduction
Immunoglobulin heavy chain gene rearrangement involves the joining of 1 variable (VH), 1 diversity (D), and 1 joining segment (JH) and precedes light chain gene rearrangement in a hierarchical pattern. This process is essential for the progression of early B cells to maturity and functional antibody assembly.1,2 A total of 123 VHsegments3,4 (depending on haplotype), 26 D segments,4,5 and 9 JH segments5are organized in a telomeric-to-centromeric orientation on chromosome 14 band q32.2 VH segments are classified into 7 families (VH1 through VH7) on the basis of amino acid sequence homology6 (Table1). VH3 is the largest family, followed by VH4 and VH1 (64, 32, and 19 members, respectively), whereas VH2, VH5, and VH6 contain only 4, 2, and 1 member, respectively. The unique VH6 is the most 3′ element and is closest to the D and JH loci.4
VH gene segments on chromosome 14q32.2
| Type of VH gene segment . | No. segments in VH families . | Total . | |||||
|---|---|---|---|---|---|---|---|
| VH1/VH7 . | VH2 . | VH3 . | VH4 . | VH5 . | VH6 . | ||
| Total germline segments (rows A, B, C) | 19 | 4 | 65 | 32 | 2 | 1 | 123 |
| (%) | (15.4) | (3.3) | (52.8) | (26.0) | (1.6) | (0.8) | |
| Rearrangable (rows A + B) | 14 | 4 | 27 | 8 | 1 | 1 | 55 |
| (%) | (25.5) | (7.3) | (49.1) | (14.5) | (1.8) | (1.8) | |
| A) Functional | 10 | 3 | 20 | 7 | 1 | 1 | 42 |
| (%) | (23.8) | (7.1) | (47.6) | (16.7) | (2.4) | (2.4) | |
| Functional protein equivalent found | 9 | 3 | 19 | 6 | 1 | 1 | |
| Transcribed mRNA equivalent found | — | — | — | 1 | — | — | |
| Other VH with ORF, intact promoter, and RSS | 1 | — | 1 | — | — | — | |
| B) Pseudogenes with intact promoter and RSS | 4 | 1 | 7 | 1 | — | — | 13 |
| C) Nonrearrangeable pseudogenes | 5 | — | 38 | 24 | 1 | — | 68 |
| Type of VH gene segment . | No. segments in VH families . | Total . | |||||
|---|---|---|---|---|---|---|---|
| VH1/VH7 . | VH2 . | VH3 . | VH4 . | VH5 . | VH6 . | ||
| Total germline segments (rows A, B, C) | 19 | 4 | 65 | 32 | 2 | 1 | 123 |
| (%) | (15.4) | (3.3) | (52.8) | (26.0) | (1.6) | (0.8) | |
| Rearrangable (rows A + B) | 14 | 4 | 27 | 8 | 1 | 1 | 55 |
| (%) | (25.5) | (7.3) | (49.1) | (14.5) | (1.8) | (1.8) | |
| A) Functional | 10 | 3 | 20 | 7 | 1 | 1 | 42 |
| (%) | (23.8) | (7.1) | (47.6) | (16.7) | (2.4) | (2.4) | |
| Functional protein equivalent found | 9 | 3 | 19 | 6 | 1 | 1 | |
| Transcribed mRNA equivalent found | — | — | — | 1 | — | — | |
| Other VH with ORF, intact promoter, and RSS | 1 | — | 1 | — | — | — | |
| B) Pseudogenes with intact promoter and RSS | 4 | 1 | 7 | 1 | — | — | 13 |
| C) Nonrearrangeable pseudogenes | 5 | — | 38 | 24 | 1 | — | 68 |
Based on Matsuda et al.4
mRNA indicates messenger RNA; ORF, open-reading frames; RSS, recombination signal sequence.
Only half of all VH genes carry intact recombination signal sequences (RSS), which are important cis-acting elements required for VH-(D)-JHrecombination.7 The remaining are consequently “nonfunctional,” as are VH genes with point mutations resulting in frame shifts and stop codons (28 VH genes). Forty-two VH segments either are known to be functional (39 segments) or have been found transcribed (1 segment) or with open-reading frames (2 genes) but not yet in a rearranged form. A putative requirement for recombination is low-level tissue-specific transcription8-10 from VH promoters (one per segment), coordinated with chromatin accessibility to recombination enzymes,11 in both human and murine B cells.12-15 When these factors are taken into account, 55 out of 123 (44.7%) VH segments are capable of undergoing rearrangement, but only 42 of 55 (76%) are potentially functional (Table 1). Under the assumption that 2 of 3 recombinations will be out of frame, the probability that any rearrangement will be functional is 1/3 × (42/55) = 0.25 or 25%.
The final immunoglobulin repertoire seen in B cells will reflect diversity generated through VH-(D)-JHrecombinations, and any positive or negative selections throughout development and maturation, prior to contact with antigens.5 Although it is clear that the repertoire shifts throughout ontogeny and maturation, the early controlling events are not clear. The final repertoire in mature adult B cells are unrestricted and unbiased, as shown by studies in peripheral blood, spleen B cells, or tonsil B cells.16-21 Many groups have reported that mammalian fetal VH-(D)-JHrepertoires (in liver, spleen, bone marrow, or peripheral blood) are highly restricted, and only a few rearranged VH genes (VH3 or VH5 or VH6) dominate.16,17,22-28 Other researchers disagree.29-31 Murine studies show a consistent overuse of JH-proximal VH segments in fetal22,23,25-27 and adult23,26,27 32 B-cell progenitor cells.
Few studies have been done in adult human preimmune B cells. Immature splenic B cells, but not mature peripheral B cells, have been shown to exhibit fetal-like VH6 overusage.17In post– allogeneic marrow transplantation patients, it has been shown that the B cells repopulating the periphery early (at 2 to 5 months) have overexpression of VH6 and a pattern of VH usage similar to those of neonatal and infant blood cells. By 6 to 12 months, the pattern becomes less marked.33 Others studies disagree34 and suggest a limited clonal diversity at 6 to 10 weeks and a normal repertoire by 6 to 9 months. This was supported by a group who showed that VH3 is the predominant family in the periphery at 6 months.35 It has been suggested that B-cell development during reconstitution of the periphery might follow a wavelike shifting pattern.36
Acute lymphoblastic leukemia (ALL) is a good model for the study of VH-(D)-JH rearrangements, as it allows examination of individual recombination events from clonally expanded pre-immune B-cell populations. Previous work37,38including our own,39 demonstrated essentially normal VH usage profiles in B-lineage ALL in both adults and children, but with a noticeable increase in VH6 usage (between 8.5% and 12%). These studies were carried out on mixed age groups with limited numbers, a situation that restricts firm statistical comparisons.
Here we expand our preliminary study39 and report on the immunoglobulin IGH gene analysis carried out on 127 adult ALL patients (245 alleles) with the use of IGH fingerprinting to define the VH gene pattern of rearrangement. Our aim was to investigate the pattern of VH usage in ALL patients, free of antigen-driven selection pressure, and test the hypothesis that preimmune B cells (as in adult B-lineage ALL clones) undergo random, nonbiased, nonrestricted VH segment usage.
Patients, materials, and methods
Patient and control samples
Bone marrow (BM) specimens from 159 adults (aged 15 to 55) with B-lineage ALL were investigated as part of the UKALL XII adult leukemia trial. Between 1992 and 1999, samples were collected from patients at presentation (147 cases) or first relapse (12 cases). The majority of cases were common ALL (cALL) (93 of 159; 58%) or pre-B ALL (48 of 159; 30%); fewer had null ALL (14 of 159; 9%) or L3 B ALL (4 of 159; 2.5%).40
Control samples used for comparison consisted of 6 BM (BM1 through BM6) and 6 peripheral blood (PB) (PB1 through PB6) samples from healthy donors, PB from 56 patients with chronic lymphocytic leukemia (CLL), diagnostic biopsies from 36 B-cell lymphoma patients, and BM from 63 childhood ALL patients.
DNA extraction procedures
Where fresh material was available (including all normal BM and PB, approximately 60% of adult B-lineage ALL, and all CLL and B-cell lymphoma samples), mononuclear cell separation, DNA extraction, and qualitative assessment of DNA prior to polymerase chain reaction (PCR) amplification were carried out as previously described.41Where fresh material was not available (about 40% of adult B-lineage ALL), DNA was extracted from archival material (stained or unstained slides) as follows. Slides were washed twice, briefly, with sterile water. Cells were scraped off damp slides into 0.5 mL Dexpat suspension (Biowhittaker, Workingham, Berkshire, United Kingdom) and boiled for 10 minutes in a dry heating block. After this was allowed to cool to room temperature, a suspension was spun in a microfuge for 10 minutes at 4°C. Supernatant was then transferred to a new tube and extracted twice with an equal volume of phenol/chloroform/isopropanol (25:24:1 vol/vol), followed by one chloroform/isopropanol (24:1 vol/vol) extraction. DNA was then recovered by ethanol precipitation. Qualitative assessment of DNA prior to PCR amplification was carried out as before.41
PCR assay of VH-(D)-JH rearrangement
For material from B-cell lymphoma, CLL, and childhood ALL, assessment of IgH rearrangement (fingerprint analysis) was carried out by PCR as previously described.39 In patients with less than 5 μg DNA total (fewer than 30% of adult ALL patients) the IgH pattern was analyzed by radionucleotide-incorporated PCR.41 In brief, 6 PCR reactions, spiked with the radionucleotide α32P–deoxycytidine triphosphate, were set up per sample, by means of 1 each of 6 sense family-specific (VH1 through VH6) framework 1 primers in combination with an antisense JH primer, as previously described.41 The VH1 forward primer was designed to also amplify VH7 sequences. The B-cell lymphoma samples were assayed with sense family-specific (VH1 through VH6) primers for the leader sequences42 and the JH primer described above. CLL samples were amplified as described for the adult ALL, as were samples from childhood ALL.
Cloning and DNA sequencing
After agarose or polyacrylamide gel separation, the DNA bands were excised and purified through Sephadex G50-medium columns or Jetsorb gel extraction kit (Genomed, Bad Oeynhausen, Germany), respectively. DNA was digested with the restriction enzymes included in the designed primers (HindIII and EcoRI) (Biolabs, Hitchin, Herts, United Kingdom) and then cloned into Bluescript plasmid KS+ (Stratagene, Amsterdam, The Netherlands), and DNA from colonies was prepared by means of the QIAprep Spin Plasmid Kit (Qiagen, Crawley, West Sussex, United Kingdom), following the manufacturer's recommendations. The relevant DNA fragments were sequenced by means of an automated sequencer (ABI PRISM 377, Warrington, Cheshire, United Kingdom) according to the manufacturer's specifications.
Densitometry
Normal BM, normal PB, and a small proportion of adult B-lineage ALL PCR products were quantitated by densitometry. Visible PCR bands on photosensitive films were scanned on an imaging densitometer (Biorad GS-700, Hemel Hempstead, Herts, United Kingdom) and analyzed for optical density (Quantity One version 4.0).
Sequence analysis
The VH-(D)-JH sequences were analyzed on the Internet by the Immunogenetics database (http://imgt.cines.fr: 8104),43 which identified the closest matching functional VH and JH segment. Those sequences that were not successfully matched were sent to BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) on the Internet for possible matches to pseudogenes. The human VH locus as published by Matsuda et al4 was taken to be the main reference for comparison of sequences.
Statistical analysis
Standard statistical tests were carried out (χ2contingency tests, parametric and nonparametric t tests, and parametric and nonparametric correlation) by means of the statistical programs Graphpad Prism (Software Inc, San Diego, CA) and Statistical Product and Service Solutions (SPSS) 10.0 for Windows. Poisson regression statistics for analyzing rare frequencies were calculated on SAS, and nonlinear regression curve fitting was carried out on GraphPad Prism.
Results
VH-(D)-JH rearrangement pattern in normal BM and normal PB
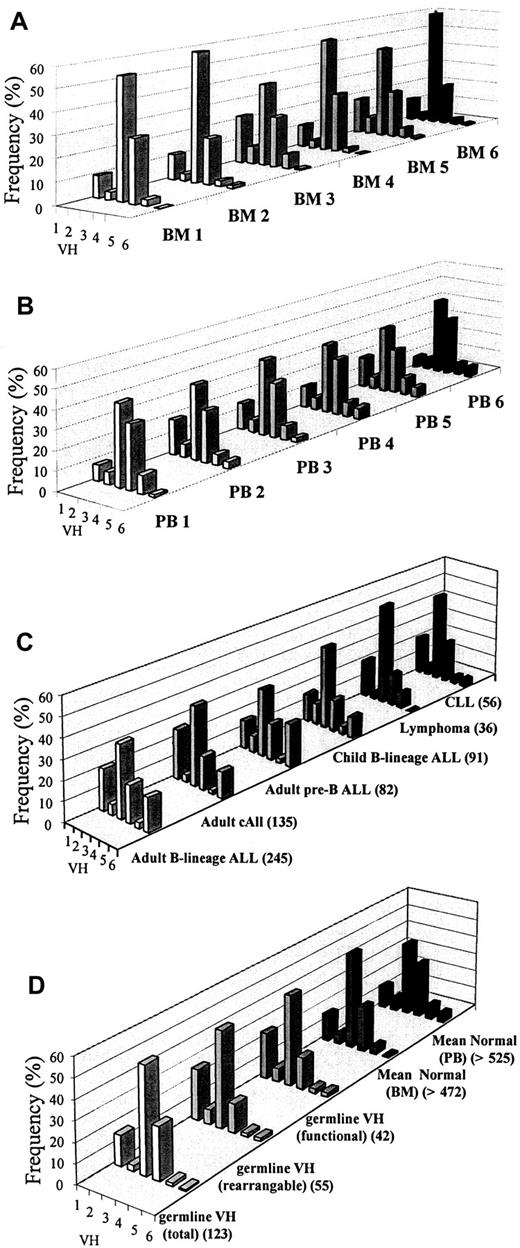
We used 6 normal adult BM samples and 6 normal adult PB samples for the amplification of VH-(D)-JHrearrangements by radiolabeled PCR followed by high-resolution polyacrylamide gel electrophoresis (PAGE). Amplified DNA products were found to be highly polyclonal, and evenly distributed DNA bands or ladders (average of 14 visible bands per ladder) were observed in both normal BM and PB (Figure 1), with each band within a ladder differing from the next by an estimated 1 base pair (bp). The total number of VH-(DH)-JH rearrangements were estimated to be equal to or greater than the total number of PCR bands observed, since each band can be expected to carry more than one clone (total estimate: greater than 472 for normal BM and greater than 525 for normal PB specimens). Bands were quantitated by densitometry, and data were pooled for every ladder to obtain the relative frequency of usage for each VH family (Table2; Figure2A-B). The intensity of bands for VH3 and VH4 families was always higher than those of other VH amplifications (particularly in the BM), accounting for 50% to 70% of all VH usage. VH1 followed with bands of intermediate to strong intensity (12% of all products), except for PB6 (less than 4%). In contrast, VH2, VH5, and VH6 bands were consistently weak (6%, 6%, and 2.6% of all products, respectively) and were often visualized only after prolonged exposure. In normal BM, the latter VH families always ranked in the following order: VH2 was greater than VH5, which was greater than VH6, and VH5 was sometimes slightly stronger in PB (PB1 and PB5), compared with VH2 and VH6. VH6 usage showed the faintest signals in all controls. The mean profiles are shown in Figure 2D. The overall ranking order of VH family usage for normal BM and PB was in broad agreement (VH3 was greater than VH4, which was greater than VH1, which was greater than VH2, which was greater than VH5, which was greater than VH6), with expected theoretical profiles calculated from germline segments (rearrangeable or functional) (Table1; Figure 2D) that have the same ranking order, except for VH1, which is expected to be stronger than VH4.
Typical PAGE fingerprints showing usage of VH families in VH-(D)-JHrearrangements.
(A) Full fingerprint of normal BM; film exposed for 24 hours. (B) Darkened sections from panel A for visualization of faint bands. (C) Film exposed for 10 days and showing blank control. (D) Adult ALL consisting of a major VH3-positive clonal population; 24 hours' exposure.
Typical PAGE fingerprints showing usage of VH families in VH-(D)-JHrearrangements.
(A) Full fingerprint of normal BM; film exposed for 24 hours. (B) Darkened sections from panel A for visualization of faint bands. (C) Film exposed for 10 days and showing blank control. (D) Adult ALL consisting of a major VH3-positive clonal population; 24 hours' exposure.
VH segment usage in normal bone marrow and normal peripheral blood
| . | VH families . | Total VH . | |||||
|---|---|---|---|---|---|---|---|
| VH1/VH7 . | VH2 . | VH3 . | VH4 . | VH5 . | VH6 . | ||
| Normal BM (6 donors) | |||||||
| Total reflected density of PCR ladders | 575.3 | 210.2 | 2072.6 | 962.3 | 162.6 | 23.0 | 4005.9 |
| Mean reflected density per donor | 95.9 | 35.0 | 345.4 | 160.4 | 27.1 | 3.8 | 667.6 |
| Mean relative frequencies (%) | (14.4) | (5.2) | (51.7) | (24) | (4.1) | (0.6) | |
| SD (CV %) | 51 (53) | 24 (68) | 22 (6) | 29 (18) | 17 (61) | 3 (78) | |
| Total no. PCR-amplified bands | 90 | 60 | 126 | 116 | 60 | 20 | 472 |
| Normal PB (6 donors) | |||||||
| Total reflected density of PCR ladders | 980.8 | 550.7 | 3336.1 | 2501.0 | 644.0 | 390.8 | 8403.4 |
| Mean reflected density per donor | 163.5 | 91.8 | 556.0 | 416.8 | 107.3 | 65.1 | 1400.6 |
| Mean relative frequencies (%) | (11.7) | (6.6) | (39.7) | (29.8) | (7.7) | (4.7) | |
| SD (CV %) | 59 (36) | 38 (41) | 247 (44) | 197 (47) | 41 (38) | 48 (74) | |
| Total no. PCR-amplified bands | 85 | 60 | 137 | 129 | 69 | 45 | 525 |
| . | VH families . | Total VH . | |||||
|---|---|---|---|---|---|---|---|
| VH1/VH7 . | VH2 . | VH3 . | VH4 . | VH5 . | VH6 . | ||
| Normal BM (6 donors) | |||||||
| Total reflected density of PCR ladders | 575.3 | 210.2 | 2072.6 | 962.3 | 162.6 | 23.0 | 4005.9 |
| Mean reflected density per donor | 95.9 | 35.0 | 345.4 | 160.4 | 27.1 | 3.8 | 667.6 |
| Mean relative frequencies (%) | (14.4) | (5.2) | (51.7) | (24) | (4.1) | (0.6) | |
| SD (CV %) | 51 (53) | 24 (68) | 22 (6) | 29 (18) | 17 (61) | 3 (78) | |
| Total no. PCR-amplified bands | 90 | 60 | 126 | 116 | 60 | 20 | 472 |
| Normal PB (6 donors) | |||||||
| Total reflected density of PCR ladders | 980.8 | 550.7 | 3336.1 | 2501.0 | 644.0 | 390.8 | 8403.4 |
| Mean reflected density per donor | 163.5 | 91.8 | 556.0 | 416.8 | 107.3 | 65.1 | 1400.6 |
| Mean relative frequencies (%) | (11.7) | (6.6) | (39.7) | (29.8) | (7.7) | (4.7) | |
| SD (CV %) | 59 (36) | 38 (41) | 247 (44) | 197 (47) | 41 (38) | 48 (74) | |
| Total no. PCR-amplified bands | 85 | 60 | 137 | 129 | 69 | 45 | 525 |
BM indicates bone marrow; PCR, polymerase chain reaction; CV, coefficient of variation; PB, peripheral blood.
VH family usage profiles.
(A) Six normal adult BM samples. (B) Six normal adult PB samples. (C) Adult B-lineage ALL and other lymphoid malignancies (total alleles analyzed in brackets). (D) Expected usage (germline segments in brackets) and mean usage in normal BM and PB (estimated minimum number of clones in brackets).
VH family usage profiles.
(A) Six normal adult BM samples. (B) Six normal adult PB samples. (C) Adult B-lineage ALL and other lymphoid malignancies (total alleles analyzed in brackets). (D) Expected usage (germline segments in brackets) and mean usage in normal BM and PB (estimated minimum number of clones in brackets).
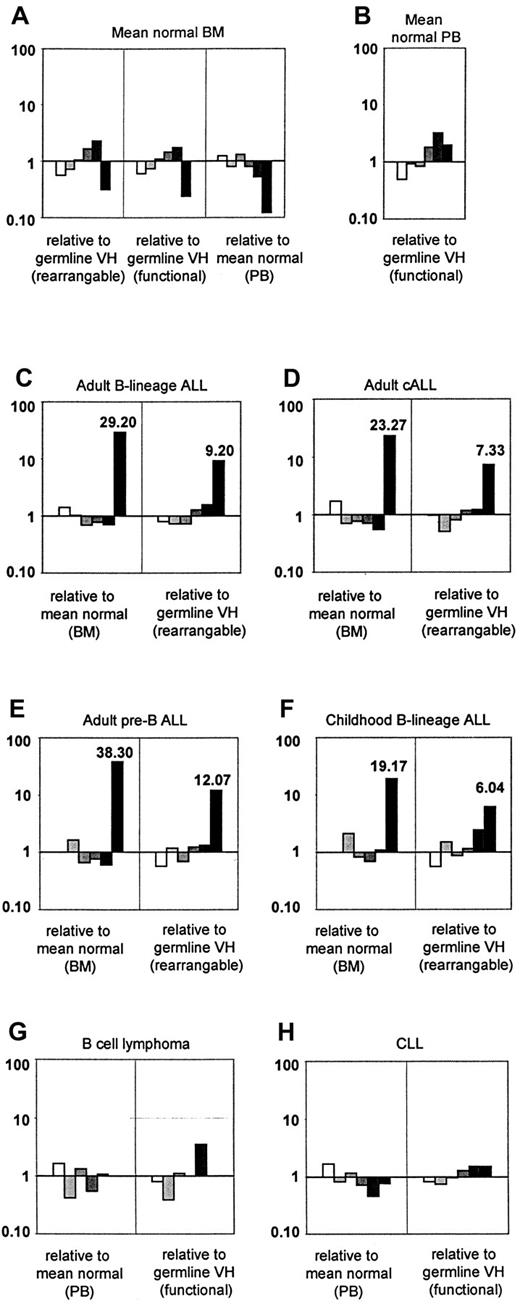
To compare VH family usage profiles with expected theoretical profiles, the former were expressed as a ratio relative to the latter, with ratios near to 1 indicating similarity (Figure3A-B). Normal BM had smaller differences from germline VH (rearrangeable or functional) for VH families VH1 through VH5, with differences not exceeding 2.2-fold in either direction.VH6 usage was 3- to 4-fold lower than expected although original values for actual and expected usage were low (less than 2.5%). Normal PB matched germline VH(functional) well, with the largest deviation from the expected occurring at VH5 (3.2-fold) although all original values were small (less than 5%). Normal BM and normal PB were also compared with each other, and profiles were found to be similar, with the largest deviations occurring at VH6 (BM samples were 8.3-fold lower) although again all original values were low at less than 5%.
Relative VH family usage profiles (ratios).
Log-scale used throughout. (A) Mean normal BM relative to germline VH (rearrangeable), germline VH (functional), or mean normal PB. (B) Mean normal PB relative to germline VH (functional). (C) Adult B-lineage ALL relative to mean normal BM or germline VH (rearrangeable). (D) Adult cALL relative to mean normal BM or germline VH (rearrangeable). (E) Adult pre-B ALL relative to mean normal BM or germline VH (rearrangeable). (F) Childhood B-lineage ALL relative to mean normal BM or germline VH (rearrangeable). (G) B-cell lymphoma relative to mean normal PB and germline VH(functional). (H) CLL relative to mean normal PB and germline VH (functional). ▭, VH1; , VH2;
, VH2; , VH3;
, VH3; , VH4;
, VH4; , VH5;
, VH5; , VH6.
, VH6.
Relative VH family usage profiles (ratios).
Log-scale used throughout. (A) Mean normal BM relative to germline VH (rearrangeable), germline VH (functional), or mean normal PB. (B) Mean normal PB relative to germline VH (functional). (C) Adult B-lineage ALL relative to mean normal BM or germline VH (rearrangeable). (D) Adult cALL relative to mean normal BM or germline VH (rearrangeable). (E) Adult pre-B ALL relative to mean normal BM or germline VH (rearrangeable). (F) Childhood B-lineage ALL relative to mean normal BM or germline VH (rearrangeable). (G) B-cell lymphoma relative to mean normal PB and germline VH(functional). (H) CLL relative to mean normal PB and germline VH (functional). ▭, VH1; , VH2;
, VH2; , VH3;
, VH3; , VH4;
, VH4; , VH5;
, VH5; , VH6.
, VH6.
Clonality of VH-(D)-JH rearrangements in adult B-lineage ALL
We assayed 159 adult patients with B-lineage ALL for the presence of VH-(D)-JH rearrangements as described above. We found 127 patients to have one or more discrete clonal VH-(D)-JH rearrangements (Figure 1D), unlike the normal BM ladders previously described. In 32 ALL patients (20%), no discrete bands were detected. A clonal pattern was highest in the subtypes L3 B ALL (4 of 4; 100%), followed by pre-B ALL (43 of 48; 91%), null ALL (12 of 14; 86%), and finally cALL (68 of 93; 74%).
Of the 127 clonal patients, 60 had a single clone (47.5%), 37 had 2 clones (29%), and 30 had 3 to 6 clones (23.5%), with an average of 1.9 clones per patient (excluding those with no clones). Among patients with 2 or more discrete bands, 58 out of 67 (86.5%) had signals of different intensities (greater than 4-fold differences by densitometry), suggestive of oligoclonality, as recently described.44 In the remaining 9 patients, 2 or more bands with similar densities were observed, and bi-allelic rearrangement could not be ruled out. Of the 245 total clones, 60 (24%) were monoclonal, 74 (30%) were biclonal, and 111 (45%) were from multiclonal cell populations. Each discrete band was taken to represent an independent clone of leukemia and was allocated to a VHfamily and scored as 1.
VH family usage in adult B-lineage ALL
As expected from the germline configuration and patterns seen in normal BM and PB, the largest number of clones identified were VH3 clones (88 of 245; 36%), followed by VH1 (50 of 245; 20.4%) and VH4 (46 of 245; 18.8%) (Table3). However, we observed that there were considerably more VH6 clones (41 of 245; 16.7%) than VH2 (13 of 245; 5.3%) and VH5 (7 of 245; 2.9%), contrary to a theoretical expectation of only 0.6% to 1.6%. Information on 102 alleles (in adults) and 91 alleles in children had been previously reported in a preliminary study.39 Except for VH6, the profile appears to be similar to the germline VH (rearrangable) profile.
VH segment usage in adult B-lineage acute lymphoblastic leukemia
| . | No. patients with clones . | No. clones in VH families . | Total no. clones . | |||||
|---|---|---|---|---|---|---|---|---|
| VH1/VH7 . | VH2 . | VH3 . | VH4 . | VH5 . | VH6 . | |||
| Adult B-lineage ALL | 127 | 50 | 13 | 88 | 46 | 7 | 41 | 245 |
| (%) | (20.4) | (5.3) | (36.0) | (18.8) | (2.9) | (16.7) | ||
| Subtypes | ||||||||
| Null ALL | 12 | 4 | 0 | 4 | 6 | 2 | 4 | 20 |
| (%) | (20) | (0) | (20) | (30) | (10) | (20) | ||
| Common ALL | 68 | 33 | 5 | 53 | 23 | 3 | 18 | 135 |
| (%) | (24.4) | (3.7) | (39.3) | (17) | (2.2) | (13.3) | ||
| Pre-B ALL | 43 | 12 | 7 | 28 | 15 | 2 | 18 | 82 |
| (%) | (14.6) | (8.5) | (34.1) | (18.3) | (2.4) | (22) | ||
| B ALL | 4 | 1 | 1 | 3 | 2 | 0 | 1 | 8 |
| (%) | (12.5) | (12.5) | (37.5) | (25) | (0) | (12.5) | ||
| . | No. patients with clones . | No. clones in VH families . | Total no. clones . | |||||
|---|---|---|---|---|---|---|---|---|
| VH1/VH7 . | VH2 . | VH3 . | VH4 . | VH5 . | VH6 . | |||
| Adult B-lineage ALL | 127 | 50 | 13 | 88 | 46 | 7 | 41 | 245 |
| (%) | (20.4) | (5.3) | (36.0) | (18.8) | (2.9) | (16.7) | ||
| Subtypes | ||||||||
| Null ALL | 12 | 4 | 0 | 4 | 6 | 2 | 4 | 20 |
| (%) | (20) | (0) | (20) | (30) | (10) | (20) | ||
| Common ALL | 68 | 33 | 5 | 53 | 23 | 3 | 18 | 135 |
| (%) | (24.4) | (3.7) | (39.3) | (17) | (2.2) | (13.3) | ||
| Pre-B ALL | 43 | 12 | 7 | 28 | 15 | 2 | 18 | 82 |
| (%) | (14.6) | (8.5) | (34.1) | (18.3) | (2.4) | (22) | ||
| B ALL | 4 | 1 | 1 | 3 | 2 | 0 | 1 | 8 |
| (%) | (12.5) | (12.5) | (37.5) | (25) | (0) | (12.5) | ||
ALL indicates acute lymphoblastic leukemia.
The largest immunophenotypic sub-group of adult B-lineage ALL in this study was represented by cALL (135 of 245; 55%). VH family usage for this group almost exactly mirrored the profile observed in ALL patients as a whole. VH family usage in pre-B ALL (82 of 245; 33%) showed a smaller proportional representation of VH1 (12 of 82; 14.6%) and an even greater bias toward VH6 usage (18 of 82; 22%). Comparable patterns were observed in null ALL (20 of 245; 8%) and L3 B ALL (8 of 245; 3%).
Statistical analyses were carried out (χ2 contingency test) comparing the actual frequencies of VH family usage in adult B-lineage ALL with expected frequencies. The expected frequencies were calculated by redistributing the total number of ALL clones in the same proportions as either (1) mean normal BM or (2) germline VH (rearrangeable). In both comparisons, adult B-lineage ALL was found to have a significantly different overall profile from what was expected (χ2 = 50,P < .0001; χ2 = 39,P < .0001, respectively), with deviation from the expected VH6 usage being the greatest contributory factor for both. The profile for adult B-lineage ALL was plotted as a ratio relative to mean normal BM or germline VH (rearrangeable) (Figure 3C). VH6 usage was higher than expected in both comparisons (29.2-fold and 9.2-fold, respectively). This observation remained true in the cALL and pre-B ALL sub-groups (Figure 3D-E), in which the latter showed greater deviation in VH6 usage from the expected (38.3-fold and 12.07-fold higher, respectively) than did the former (23.27-fold and 7.33-fold higher, respectively).
VH family usage in other lymphoid malignancies
For comparison, previous data on VH family usage in childhood B-lineage ALL (91 clones),39 B-cell lymphoma (36 clones), and CLL (56 clones) have been displayed in Table4 and Figure 2C. The childhood B-lineage ALL profile appears to be most similar to the adult pre-B ALL profile, with the exception that VH6, though overrepresented (10 of 91; 10%), is not as greatly elevated as in the adult ALL group. Both the lymphoma and CLL profiles closely resembled the germline VH functional profile (Figure 2D), with very low usage of VH6, in B-cell lymphoma (0 of 36; 0%), compared with an expected frequency of 2.4%, whereas VH6 usage in CLL was 3.6% (2 of 56). As with adult B-lineage ALL, all 3 profiles were plotted as ratios relative to the most appropriate expected profiles (Figure 3F-H). The childhood B-lineage ALL profile had higher-than-expected VH6 (19.17 relative to mean normal BM; 6.04 relative to germline rearrangeable VH) although, owing to lack of a large enough data set, statistical analysis could not be carried out. Both B-cell lymphoma and CLL had ratios close to 1 for all VH families.
VH gene segment usage in other lymphoid malignancies
| . | No. patients with clones . | No. clones in VH families . | Total no. clones . | |||||
|---|---|---|---|---|---|---|---|---|
| VH1/VH7 . | VH2 . | VH3 . | VH4 . | VH5 . | VH6 . | |||
| Childhood B-lineage ALL | 63 | 13 | 10 | 39 | 15 | 4 | 10 | 91 |
| (%) | (14.3) | (11) | (42.9) | (16.5) | (4.4) | (11) | ||
| B-cell lymphoma | 36 | 7 | 1 | 19 | 6 | 3 | 0 | 36 |
| (%) | (19.4) | (2.8) | (52.8) | (16.7) | (8.3) | (0) | ||
| CLL | 56 | 11 | 3 | 26 | 12 | 2 | 2 | 56 |
| (%) | (19.6) | (5.4) | (46.4) | (21.4) | (3.6) | (3.6) | ||
| . | No. patients with clones . | No. clones in VH families . | Total no. clones . | |||||
|---|---|---|---|---|---|---|---|---|
| VH1/VH7 . | VH2 . | VH3 . | VH4 . | VH5 . | VH6 . | |||
| Childhood B-lineage ALL | 63 | 13 | 10 | 39 | 15 | 4 | 10 | 91 |
| (%) | (14.3) | (11) | (42.9) | (16.5) | (4.4) | (11) | ||
| B-cell lymphoma | 36 | 7 | 1 | 19 | 6 | 3 | 0 | 36 |
| (%) | (19.4) | (2.8) | (52.8) | (16.7) | (8.3) | (0) | ||
| CLL | 56 | 11 | 3 | 26 | 12 | 2 | 2 | 56 |
| (%) | (19.6) | (5.4) | (46.4) | (21.4) | (3.6) | (3.6) | ||
ALL indicates acute lymphoblastic leukemia; CLL, chronic lymphocytic leukemia.
Sequence analysis of VH-(D)-JHrearrangements in adult B-lineage ALL
We further analyzed 77 of 254 adult B-lineage ALL clones by DNA sequencing (Table 5). The germline VH segment counterpart of the VHrearranged genes were identified in 65 clones. The remaining 12 could not be identified precisely, though they were clearly true VH-(D)-JH rearrangements (listed at bottom of Table 5). This was because the clone either (1) matched more than 3 known VH segments equally well, so no single segment could be assigned, or (2) did not match any of the published VHsegments well (less than 90% homology) and thus could have been a derivative of an unpublished polymorphic variable. Out of the 65 identified clones, 1 (clone 17) was allocated to the polymorphic variant VH (5-a), which has yet to be mapped, leaving 64 clones whose matching VH segments have been precisely mapped on the human VH locus. Five of these matched 2 to 3 different segments equally, and all possible matches have been listed. Three clones (323, 243c, 362b) matched polymorphic segments that occur in an as-yet-unsized proportion of the human population. Where this polymorphism occurs, the locus carries 5 extra VH segments between segments 3-30 and 4-31, widening the gap between the 2 by approximately 250 kilobases (kb). We found 2 clones that matched 2 rearrangeable pseudogenes (4-55 and 3-65).
Properties of sequenced VDJ rearrangements from 78 adult B-lineage acute lymphoblastic leukemia clones
| Patient (clone) . | VH family . | Closest matching VHsegment5-150 . | Distance of VH from JH1 (kb) . | Closest matching JHsegment . | Age . | Sex . | Immunophenotype . | Karyotype . | WBC (log) . | Time to first CR5-151 . | Clinical outcome5-152 . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 57 | VH6 | 6 -1 | 74.31 | JH1 | 24 | M | null ALL | t(4;11) | 2.75 | standard | CCR |
| 355 | VH6 | 6 -1 | 74.31 | JH2 | 40 | F | null ALL | t(4;11) | 2.38 | standard | relapse |
| 406 | VH6 | 6 -1 | 74.31 | JH4 | 20 | F | null ALL | t(4;11) | 2.58 | standard | CCR |
| 5 | VH6 | 6 -1 | 74.31 | JH5 | 17 | M | CALL | other | standard | relapse | |
| 110 | VH6 | 6 -1 | 74.31 | JH4 | 25 | M | CALL | normal | 0.23 | standard | CCR |
| 181 | VH6 | 6 -1 | 74.31 | Unclear5-153 | 29 | M | CALL | normal | 0.77 | standard | CCR |
| 243(a) | VH6 | 6 -1 | 74.31 | JH4 | 27 | M | CALL | other | 0.32 | standard | relapse |
| 373 | VH6 | 6 -1 | 74.31 | JH4 | 48 | F | CALL | t(1;19) | |||
| 378(a) | VH6 | 6 -1 | 74.31 | JH5 | 20 | M | CALL | 9p abn | relapse | ||
| 80(a) | VH6 | 6 -1 | 74.31 | Unclear5-153 | 31 | F | pre-B ALL | 1.03 | standard | CCR | |
| 192(a) | VH6 | 6 -1 | 74.31 | JH4 | 20 | F | pre-B ALL | t(4;11) | 2.53 | standard | CCR |
| 192(b) | VH6 | 6 -1 | 74.31 | JH5 | 20 | F | pre-B ALL | t(4;11) | 2.53 | standard | CCR |
| 223 | VH6 | 6 -1 | 74.31 | JH4 | 23 | F | pre-B ALL | 11q23 [non t(4;11)] | 1.23 | late | relapse |
| 392 | VH6 | 6 -1 | 74.31 | JH4 | 39 | F | pre-B ALL | other | standard | CCR | |
| 263(a) | VH1 | 1 -2 | 121.36 | JH4 | 24 | F | cALL | 1.03 | standard | CCR | |
| 321 | VH1 | 1 -2 | 121.36 | JH5 | 18 | M | cALL | HeH | 1.34 | standard | relapse |
| 338 | VH1 | 1 -2 | 121.36 | JH4 | 27 | F | cALL | 9p abn | 0.93 | standard | CCR |
| 378(b) | VH1 | 1 -2 | 121.36 | JH5 | 20 | M | cALL | 9p abn | relapse | ||
| 121 | VH1 | 1 -3 | 139.94 | JH4 | 19 | M | cALL | other | 1.40 | standard | relapse |
| 1235-155 | VH1 | 1 -35-155 | 139.94 | JH6 | 16 | F | pre-B ALL | 0.36 | relapse | ||
| 1265-155 | VH1 | 1 -35-155 | 139.94 | JH6 | 17 | F | pre-B ALL | 9p abn | 1.42 | standard | relapse |
| 362(a) | VH4 | 4 -4 | 146.80 | JH5 | 29 | F | cALL | other | 0.70 | standard | relapse |
| 26 | VH4 | 4 -4 | 146.80 | JH4 | 25 | M | pre-B ALL | t(12;21) | 0.95 | standard | relapse |
| 410 | VH4 | 4 -4 | 146.80 | JH4 | 35 | F | B ALL | 2.11 | CCR | ||
| 7(a) | VH2 | 2 -5 | 162.83 | JH6 | 31 | M | pre-B ALL | t(9;22) | 1.96 | late | |
| 222(a) | VH2 | 2 -5 | 162.83 | JH4 | 40 | F | pre-B ALL | t(9;22) | 2.05 | standard | CCR |
| 243(b) | VH1 | 1 -8 | 207.77 | JH5 | 27 | M | cALL | other | 0.32 | standard | relapse |
| 263(b) | VH1 | 1 -8 | 207.77 | JH6 | 24 | F | cALL | 1.03 | standard | CCR | |
| 374 | VH3 | 3 -9 | 220.00 | JH4 | 19 | M | cALL | relapse | |||
| 72 | VH3 | 3 -11 | 221.00 | JH4 | 20 | M | cALL | relapse | |||
| 204 | VH3 | 3 -11 | 221.00 | JH6 | 19 | M | cALL | HeH | 0.52 | standard | CCR |
| 2835-155 | VH3 | 3 -135-155 | 254.84 | Unclear5-153 | 55 | M | null ALL | normal | 0.70 | standard | CCR |
| 76 | VH3 | 3 -13 | 254.84 | JH6 | 21 | M | cALL | normal | 1.58 | standard | CCR |
| 255(a) | VH3 | 3 -13 | 254.84 | JH4 | 16 | M | cALL | relapse | |||
| 7(b) | VH3 | 3 -13 | 254.84 | JH6 | 31 | M | pre-B ALL | t(9;22) | 1.96 | late | |
| 401 | VH3 | 3 -13 | 254.84 | JH5 | 49 | F | pre-B ALL | t(9;22) | standard | relapse | |
| 112 | VH3 | 3 -23 | 393.91 | JH5 | 22 | M | pre-B ALL | t(1;19) | 1.59 | late | relapse |
| 177 | VH3 | 3 -23 | 393.91 | JH5 | 44 | F | pre-B ALL | 14q32 abn | 0.85 | ||
| 40 | VH3 | 3 -30 | 459.71 | JH6 | 19 | M | cALL | 1.59 | standard | ||
| 198 | VH3 | 3 -30 | 459.71 | JH3 | 43 | M | pre-B ALL | t(9;22) | 0.61 | standard | relapse |
| 323 | VH4 | 4-30-4(poly) | See text | JH4 | 20 | M | cALL | other | 1.74 | standard | CCR |
| 243(c) | VH3 | 3-30/3-30-5(poly) | See text | JH1 | 27 | M | cALL | other | 0.32 | standard | relapse |
| 389 | VH4 | 4 -31 | 473.90 | JH5 | 22 | F | null ALL | t(4;11) | 2.30 | standard | relapse |
| 77(a) | VH4 | 4 -31 | 473.90 | JH6 | 24 | F | pre-B ALL | 6q del | 1.19 | standard | CCR |
| 255(b) | VH4 | 4 -39 | 546.31 | JH5 | 16 | M | cALL | relapse | |||
| 272 | VH3 | 3 -43 | 594.90 | JH4 | 28 | F | cALL | 6q del | 1.65 | standard | CCR |
| 255(c) | VH3 | 3 -48 | 662.52 | JH5 | 16 | M | cALL | relapse | |||
| 408 | VH3 | 3 -53 | 717.67 | JH6 | 18 | M | cALL | other | 1.99 | late | relapse |
| 299(a) | VH3 | 3 -53 | 717.67 | Unclear5-153 | 18 | F | pre-B ALL | standard | CCR | ||
| 18 | VH4 | 4-55(Ps) | 730.82 | JH4 | 16 | F | pre-B ALL | 1.36 | standard | CCR | |
| 56 | VH4 | 4 -61 | 763.82 | JH5 | 16 | M | cALL | 14q32 abn | 0.79 | standard | CCR |
| 376(a) | VH3 | 3 -64 | 782.45 | JH4 | 50 | F | cALL | t(9;22) | 1.39 | standard | relapse |
| 327 | VH3 | 3-65(Ps) | 790.80 | JH4 | 36 | M | cALL | 0.36 | standard | CCR | |
| 289 | VH1 | 1 -69 | 838.62 | JH5 | 20 | M | cALL | 1.08 | late | CCR | |
| 376(b) | VH1 | 1 -69 | 838.62 | JH4 | 50 | F | cALL | t(9;22) | 1.39 | standard | relapse |
| 7(c) | VH1 | 1 -69 | 838.62 | JH6 | 31 | M | pre-B ALL | t(9;22) | 1.96 | late | |
| 296 | VH2 | 2 -70 | 847.52 | Unclear5-153 | 40 | F | pre-B ALL | 12p abn | 1.05 | late | CCR |
| 11 | VH2 | 2 -70 | 847.52 | JH4 | 19 | M | B ALL | 2.19 | standard | relapse | |
| 134 | VH3 | 3 -73 | 879.65 | JH3 | 48 | M | B ALL | 14q32 abn | 2.74 | late | relapse |
| 475-154 | VH4 | 4 -45-154 | 146.80 | JH5 | 26 | M | cALL | other | 1.01 | late | CCR |
| 4 -615-154 | 763.82 | ||||||||||
| 1875-154 | VH2 | 2 -55-154 | 162.83 | JH4 | 24 | F | cALL | relapse | |||
| 2 -265-154 | 426.35 | ||||||||||
| 2 -705-154 | 847.52 | ||||||||||
| 1955-154 | VH4 | 4 -345-154 | 498.28 | JH6 | 35 | M | cALL | t(9;22) | 1.60 | never | |
| 4 -395-154 | 546.31 | ||||||||||
| 2625-154 | VH4 | 4 -395-154 | 546.31 | JH5 | 54 | M | null ALL | t(4;11) | 2.34 | late | CCR |
| 4 -595-154 | 751.94 | ||||||||||
| 362(b)5-154 | VH3 | 3 -305-154 | 459.71 | JH4 | 29 | F | cALL | other | 0.70 | standard | relapse |
| 3-30-3 (poly)5-154 | See text | ||||||||||
| 3-30/3-30-5(poly)5-154 | See text | ||||||||||
| 17 | VH5 | 5-a(poly)# | — | JH6 | 16 | F | pre-B ALL | relapse | |||
| 71 | VH | Unclear5-160 | — | JH4 | 16 | M | cALL | 0.63 | standard | CCR | |
| 241 | VH | Unclear5-160 | — | Unclear5-154 | 16 | F | cALL | 12p abn | 1.91 | standard | relapse |
| 248 | VH4 | Unclear5-160 | — | JH4 | 21 | F | cALL | HeH | 0.40 | standard | CCR |
| 347(a) | VH1 | Unclear5-160 | — | JH5 | 27 | M | cALL | t(12;21) | 1.60 | standard | relapse |
| 347(b) | VH3 | Unclear5-160 | — | JH4 | 27 | M | cALL | t(12;21) | 1.60 | standard | relapse |
| 77(b) | VH3 | Unclear5-160 | — | JH6 | 24 | F | pre-B ALL | 6q del | 1.19 | standard | CCR |
| 80(b) | VH1 | Unclear5-160 | — | JH5 | 31 | F | pre-B ALL | 1.03 | standard | CCR | |
| 130 | VH | Unclear5-160 | — | JH5 | 30 | F | pre-B ALL | t(9;22) | 1.89 | late | CCR |
| 149 | VH | Unclear5-160 | — | Unclear5-154 | 19 | M | pre-B ALL | 11q23 [non t(4;11)] | 2.07 | standard | relapse |
| 222(b) | VH3 | Unclear5-160 | — | JH4 | 40 | F | pre-B ALL | t(9;22) | 2.05 | standard | CCR |
| 299(b) | VH4 | Unclear5-160 | — | JH6 | 18 | F | pre-B ALL | standard | CCR | ||
| 375 | VH4 | Unclear5-160 | — | JH6 | 19 | M | pre-B ALL | HeH | 0.48 | standard | relapse |
| 244 | False†† | False†† | — | False†† | 50 | F | null ALL | t(4;11) | 1.99 | standard | CCR |
| Patient (clone) . | VH family . | Closest matching VHsegment5-150 . | Distance of VH from JH1 (kb) . | Closest matching JHsegment . | Age . | Sex . | Immunophenotype . | Karyotype . | WBC (log) . | Time to first CR5-151 . | Clinical outcome5-152 . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 57 | VH6 | 6 -1 | 74.31 | JH1 | 24 | M | null ALL | t(4;11) | 2.75 | standard | CCR |
| 355 | VH6 | 6 -1 | 74.31 | JH2 | 40 | F | null ALL | t(4;11) | 2.38 | standard | relapse |
| 406 | VH6 | 6 -1 | 74.31 | JH4 | 20 | F | null ALL | t(4;11) | 2.58 | standard | CCR |
| 5 | VH6 | 6 -1 | 74.31 | JH5 | 17 | M | CALL | other | standard | relapse | |
| 110 | VH6 | 6 -1 | 74.31 | JH4 | 25 | M | CALL | normal | 0.23 | standard | CCR |
| 181 | VH6 | 6 -1 | 74.31 | Unclear5-153 | 29 | M | CALL | normal | 0.77 | standard | CCR |
| 243(a) | VH6 | 6 -1 | 74.31 | JH4 | 27 | M | CALL | other | 0.32 | standard | relapse |
| 373 | VH6 | 6 -1 | 74.31 | JH4 | 48 | F | CALL | t(1;19) | |||
| 378(a) | VH6 | 6 -1 | 74.31 | JH5 | 20 | M | CALL | 9p abn | relapse | ||
| 80(a) | VH6 | 6 -1 | 74.31 | Unclear5-153 | 31 | F | pre-B ALL | 1.03 | standard | CCR | |
| 192(a) | VH6 | 6 -1 | 74.31 | JH4 | 20 | F | pre-B ALL | t(4;11) | 2.53 | standard | CCR |
| 192(b) | VH6 | 6 -1 | 74.31 | JH5 | 20 | F | pre-B ALL | t(4;11) | 2.53 | standard | CCR |
| 223 | VH6 | 6 -1 | 74.31 | JH4 | 23 | F | pre-B ALL | 11q23 [non t(4;11)] | 1.23 | late | relapse |
| 392 | VH6 | 6 -1 | 74.31 | JH4 | 39 | F | pre-B ALL | other | standard | CCR | |
| 263(a) | VH1 | 1 -2 | 121.36 | JH4 | 24 | F | cALL | 1.03 | standard | CCR | |
| 321 | VH1 | 1 -2 | 121.36 | JH5 | 18 | M | cALL | HeH | 1.34 | standard | relapse |
| 338 | VH1 | 1 -2 | 121.36 | JH4 | 27 | F | cALL | 9p abn | 0.93 | standard | CCR |
| 378(b) | VH1 | 1 -2 | 121.36 | JH5 | 20 | M | cALL | 9p abn | relapse | ||
| 121 | VH1 | 1 -3 | 139.94 | JH4 | 19 | M | cALL | other | 1.40 | standard | relapse |
| 1235-155 | VH1 | 1 -35-155 | 139.94 | JH6 | 16 | F | pre-B ALL | 0.36 | relapse | ||
| 1265-155 | VH1 | 1 -35-155 | 139.94 | JH6 | 17 | F | pre-B ALL | 9p abn | 1.42 | standard | relapse |
| 362(a) | VH4 | 4 -4 | 146.80 | JH5 | 29 | F | cALL | other | 0.70 | standard | relapse |
| 26 | VH4 | 4 -4 | 146.80 | JH4 | 25 | M | pre-B ALL | t(12;21) | 0.95 | standard | relapse |
| 410 | VH4 | 4 -4 | 146.80 | JH4 | 35 | F | B ALL | 2.11 | CCR | ||
| 7(a) | VH2 | 2 -5 | 162.83 | JH6 | 31 | M | pre-B ALL | t(9;22) | 1.96 | late | |
| 222(a) | VH2 | 2 -5 | 162.83 | JH4 | 40 | F | pre-B ALL | t(9;22) | 2.05 | standard | CCR |
| 243(b) | VH1 | 1 -8 | 207.77 | JH5 | 27 | M | cALL | other | 0.32 | standard | relapse |
| 263(b) | VH1 | 1 -8 | 207.77 | JH6 | 24 | F | cALL | 1.03 | standard | CCR | |
| 374 | VH3 | 3 -9 | 220.00 | JH4 | 19 | M | cALL | relapse | |||
| 72 | VH3 | 3 -11 | 221.00 | JH4 | 20 | M | cALL | relapse | |||
| 204 | VH3 | 3 -11 | 221.00 | JH6 | 19 | M | cALL | HeH | 0.52 | standard | CCR |
| 2835-155 | VH3 | 3 -135-155 | 254.84 | Unclear5-153 | 55 | M | null ALL | normal | 0.70 | standard | CCR |
| 76 | VH3 | 3 -13 | 254.84 | JH6 | 21 | M | cALL | normal | 1.58 | standard | CCR |
| 255(a) | VH3 | 3 -13 | 254.84 | JH4 | 16 | M | cALL | relapse | |||
| 7(b) | VH3 | 3 -13 | 254.84 | JH6 | 31 | M | pre-B ALL | t(9;22) | 1.96 | late | |
| 401 | VH3 | 3 -13 | 254.84 | JH5 | 49 | F | pre-B ALL | t(9;22) | standard | relapse | |
| 112 | VH3 | 3 -23 | 393.91 | JH5 | 22 | M | pre-B ALL | t(1;19) | 1.59 | late | relapse |
| 177 | VH3 | 3 -23 | 393.91 | JH5 | 44 | F | pre-B ALL | 14q32 abn | 0.85 | ||
| 40 | VH3 | 3 -30 | 459.71 | JH6 | 19 | M | cALL | 1.59 | standard | ||
| 198 | VH3 | 3 -30 | 459.71 | JH3 | 43 | M | pre-B ALL | t(9;22) | 0.61 | standard | relapse |
| 323 | VH4 | 4-30-4(poly) | See text | JH4 | 20 | M | cALL | other | 1.74 | standard | CCR |
| 243(c) | VH3 | 3-30/3-30-5(poly) | See text | JH1 | 27 | M | cALL | other | 0.32 | standard | relapse |
| 389 | VH4 | 4 -31 | 473.90 | JH5 | 22 | F | null ALL | t(4;11) | 2.30 | standard | relapse |
| 77(a) | VH4 | 4 -31 | 473.90 | JH6 | 24 | F | pre-B ALL | 6q del | 1.19 | standard | CCR |
| 255(b) | VH4 | 4 -39 | 546.31 | JH5 | 16 | M | cALL | relapse | |||
| 272 | VH3 | 3 -43 | 594.90 | JH4 | 28 | F | cALL | 6q del | 1.65 | standard | CCR |
| 255(c) | VH3 | 3 -48 | 662.52 | JH5 | 16 | M | cALL | relapse | |||
| 408 | VH3 | 3 -53 | 717.67 | JH6 | 18 | M | cALL | other | 1.99 | late | relapse |
| 299(a) | VH3 | 3 -53 | 717.67 | Unclear5-153 | 18 | F | pre-B ALL | standard | CCR | ||
| 18 | VH4 | 4-55(Ps) | 730.82 | JH4 | 16 | F | pre-B ALL | 1.36 | standard | CCR | |
| 56 | VH4 | 4 -61 | 763.82 | JH5 | 16 | M | cALL | 14q32 abn | 0.79 | standard | CCR |
| 376(a) | VH3 | 3 -64 | 782.45 | JH4 | 50 | F | cALL | t(9;22) | 1.39 | standard | relapse |
| 327 | VH3 | 3-65(Ps) | 790.80 | JH4 | 36 | M | cALL | 0.36 | standard | CCR | |
| 289 | VH1 | 1 -69 | 838.62 | JH5 | 20 | M | cALL | 1.08 | late | CCR | |
| 376(b) | VH1 | 1 -69 | 838.62 | JH4 | 50 | F | cALL | t(9;22) | 1.39 | standard | relapse |
| 7(c) | VH1 | 1 -69 | 838.62 | JH6 | 31 | M | pre-B ALL | t(9;22) | 1.96 | late | |
| 296 | VH2 | 2 -70 | 847.52 | Unclear5-153 | 40 | F | pre-B ALL | 12p abn | 1.05 | late | CCR |
| 11 | VH2 | 2 -70 | 847.52 | JH4 | 19 | M | B ALL | 2.19 | standard | relapse | |
| 134 | VH3 | 3 -73 | 879.65 | JH3 | 48 | M | B ALL | 14q32 abn | 2.74 | late | relapse |
| 475-154 | VH4 | 4 -45-154 | 146.80 | JH5 | 26 | M | cALL | other | 1.01 | late | CCR |
| 4 -615-154 | 763.82 | ||||||||||
| 1875-154 | VH2 | 2 -55-154 | 162.83 | JH4 | 24 | F | cALL | relapse | |||
| 2 -265-154 | 426.35 | ||||||||||
| 2 -705-154 | 847.52 | ||||||||||
| 1955-154 | VH4 | 4 -345-154 | 498.28 | JH6 | 35 | M | cALL | t(9;22) | 1.60 | never | |
| 4 -395-154 | 546.31 | ||||||||||
| 2625-154 | VH4 | 4 -395-154 | 546.31 | JH5 | 54 | M | null ALL | t(4;11) | 2.34 | late | CCR |
| 4 -595-154 | 751.94 | ||||||||||
| 362(b)5-154 | VH3 | 3 -305-154 | 459.71 | JH4 | 29 | F | cALL | other | 0.70 | standard | relapse |
| 3-30-3 (poly)5-154 | See text | ||||||||||
| 3-30/3-30-5(poly)5-154 | See text | ||||||||||
| 17 | VH5 | 5-a(poly)# | — | JH6 | 16 | F | pre-B ALL | relapse | |||
| 71 | VH | Unclear5-160 | — | JH4 | 16 | M | cALL | 0.63 | standard | CCR | |
| 241 | VH | Unclear5-160 | — | Unclear5-154 | 16 | F | cALL | 12p abn | 1.91 | standard | relapse |
| 248 | VH4 | Unclear5-160 | — | JH4 | 21 | F | cALL | HeH | 0.40 | standard | CCR |
| 347(a) | VH1 | Unclear5-160 | — | JH5 | 27 | M | cALL | t(12;21) | 1.60 | standard | relapse |
| 347(b) | VH3 | Unclear5-160 | — | JH4 | 27 | M | cALL | t(12;21) | 1.60 | standard | relapse |
| 77(b) | VH3 | Unclear5-160 | — | JH6 | 24 | F | pre-B ALL | 6q del | 1.19 | standard | CCR |
| 80(b) | VH1 | Unclear5-160 | — | JH5 | 31 | F | pre-B ALL | 1.03 | standard | CCR | |
| 130 | VH | Unclear5-160 | — | JH5 | 30 | F | pre-B ALL | t(9;22) | 1.89 | late | CCR |
| 149 | VH | Unclear5-160 | — | Unclear5-154 | 19 | M | pre-B ALL | 11q23 [non t(4;11)] | 2.07 | standard | relapse |
| 222(b) | VH3 | Unclear5-160 | — | JH4 | 40 | F | pre-B ALL | t(9;22) | 2.05 | standard | CCR |
| 299(b) | VH4 | Unclear5-160 | — | JH6 | 18 | F | pre-B ALL | standard | CCR | ||
| 375 | VH4 | Unclear5-160 | — | JH6 | 19 | M | pre-B ALL | HeH | 0.48 | standard | relapse |
| 244 | False†† | False†† | — | False†† | 50 | F | null ALL | t(4;11) | 1.99 | standard | CCR |
ALL indicates acute lymphoblastic leukemia; cALL, common ALL.
All named matches have less than 97% homology with no more than 3 mismatching base pairs, except those marked with ∥.
Standard time to CR is up to 42 days.
Patients followed for at least 24 months or up to first relapse.
Clones were found to match more than 1 JH segment equally well; therefore, a JH segment was not assigned.
These matches were relatively poor (90% to 95% homology) with between 8 and 10 mismatching base pairs.
Clones were found to match 2 to 3 VH segments equally well; all possibilities are listed.
#This polymorphic VH segment has not been accurately mapped at present.
Clones were found to match more than 3 VH segments equally well; therefore, a VH segment was not assigned.
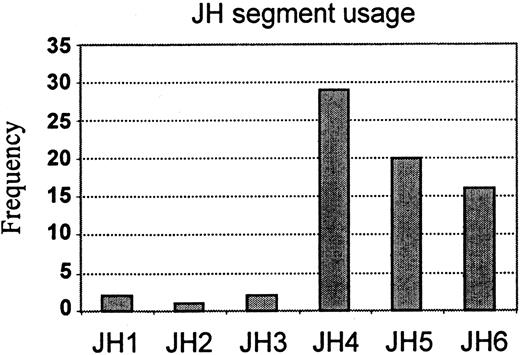
JH segment usage
We successfully assigned known JH segments to 70 clones (Table 5; Figure 4). In 7 other clones, the rearrangement process resulted in the deletion of a sizable section of the JH, making it difficult to precisely identify the germline sequence. JH4 occurred most commonly (29 of 70; 41.4%), followed by JH5 (20 of 70; 28.6%) and then JH6 (16 of 70; 23.9%). JH1, JH2, and JH3 rearrangements were rare at between 1.4% and 2.9% (2 of 70; 1 of 70; 2 of 70, respectively).
JH gene segment usage in 70 sequenced adult B-lineage ALL clones.
The JH usage is expressed as the percentage of all alleles studied and is illustrated by vertical bars.
JH gene segment usage in 70 sequenced adult B-lineage ALL clones.
The JH usage is expressed as the percentage of all alleles studied and is illustrated by vertical bars.
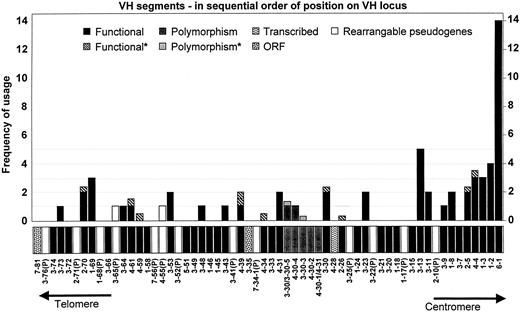
Positional analysis of VH gene usage in adult B-lineage ALL
Frequency of usage of specific germline VHsegments by adult B-lineage ALL (total, 64 clones) was scored (Figure5). Only segments capable of rearranging (including pseudogenes) were considered. These included VHgenes with intact RS and promoter sequences. Segments without these elements were never found rearranged and were therefore excluded from further analysis. Note that in addition to the 55 rearrangeable segments taken from Table 1, five polymorphic segments (not reported in Matsuda et al4) have also been included in Figure 5. Where a clone was assigned to 2 or 3 possible segments, each possibility was given a score of 0.5 or 0.33, accordingly. We observed that 29 out of 60 (48%) rearrangeable VH segments were used at least 0.33 times. Nonused VH segments were scattered across the locus, including a clump of 6 adjacent segments near the JH locus. The most JH-proximal segment(VH6) was overused (14 of 64; 22%) with a considerably higher frequency than the second most common segment (3-13; 5 of 64; 7.8%). Segments with a frequency of more than 2 tended to occur near to the JH locus, whereas those with frequencies of 2 or lower were more often found in the midsection and upstream.
VH gene segment usage in 63 sequenced adult B-lineage ALL clones.
Only rearrangeable segments have been shown. Asterisk indicates that clone matched 2 to 3 segments in total, and each segment has been given a score of 0.33 or 0.5, accordingly.
VH gene segment usage in 63 sequenced adult B-lineage ALL clones.
Only rearrangeable segments have been shown. Asterisk indicates that clone matched 2 to 3 segments in total, and each segment has been given a score of 0.33 or 0.5, accordingly.
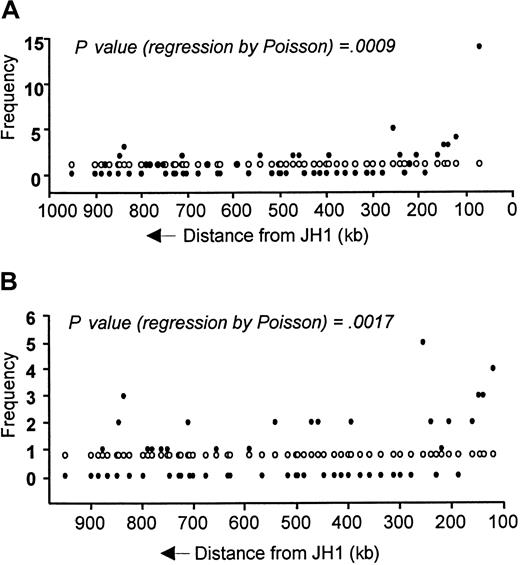
We estimated that if each segment had an equal chance of being selected for rearrangement, the expected frequency would be around 1 per segment, which differs from our observation. The hypothesis that the frequency of usage of any VH segment is a function of its distance from the JH locus was tested and confirmed by Poisson regression analysis and found to be correct (P = .0009; Figure 6A). This finding remains true, even when the extremeVH6 data were excluded from the calculation (P = .0017; Figure 6B). For simplicity, polymorphic segments were excluded from these calculations, and noninteger frequencies were truncated to the nearest integer (total frequencies equal 58 or 44 without VH6).
Comparison of expected and actual VH gene segment usage in sequenced adult B-lineage ALL clones in relation to distance upstream from JH locus.
(A) All clones. (B) Excluding VH6 clones. ●, actual frequency; ○, expected frequency.
Comparison of expected and actual VH gene segment usage in sequenced adult B-lineage ALL clones in relation to distance upstream from JH locus.
(A) All clones. (B) Excluding VH6 clones. ●, actual frequency; ○, expected frequency.
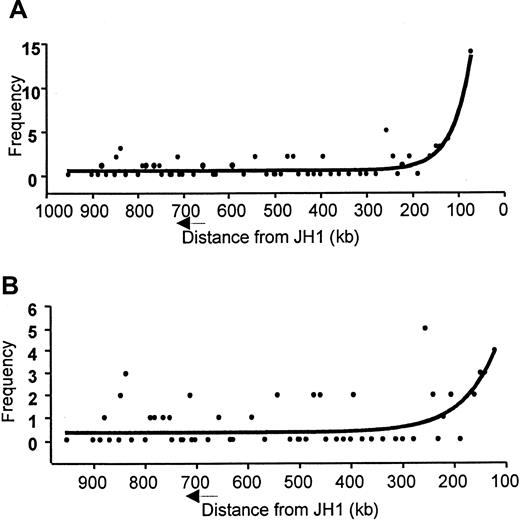
We attempted to fit a simple model of nonlinear regression to our data, using the principles of one-phase exponential decay (used for defining radioactive decay over time). This model is defined by the equation below and was found to represent our data reasonably well, with and without VH6 data (Figure7A-B). The equation for the curve is Y = span · e−K·X + plateau, where span is the highest possible frequency, plateau is the lowest frequency,K is a unique constant, and the half-life equals 0.69/K. For the curve shown in Figure 7B, the span equals 21, the plateau equals 0.5, K equals 0.0154, and the frequency decreased by half (half-life) every 45 kb upstream from the first segment. Thus, it has been shown that the relationship between frequency of VH gene segment usage and distance from JH locus can be mathematically defined.
Nonlinear regression model of VH usage across distance upstream of JH1, using the principles of one-phase exponential decay.
(A) Frequency of VH gene usage including the VH6. (B) Frequency of VH gene usage excluding the VH6 gene.
Nonlinear regression model of VH usage across distance upstream of JH1, using the principles of one-phase exponential decay.
(A) Frequency of VH gene usage including the VH6. (B) Frequency of VH gene usage excluding the VH6 gene.
Discussion
Mature B-cell populations
Our study analyzed VH gene segment usage during VH-(D)-JH rearrangement in B cells from adult and childhood B-lineage ALL populations. We also assessed the baseline patterns in normal BM and normal PB B-cell populations, which showed no difference from the predicted profiles. The patterns in chronic lymphoid malignancies (in CLL) and lymphomas were also assessed and found to conform to profiles derived from mature post–antigen-stimulation B-cell profiles.16-21,37 45-48
By contrast, adult B-lineage ALL clones, taken as models of progenitors and precursors of B cells, showed a statistically significant deviation from the expected pattern, with overrepresentation ofVH6 rearrangements and use of JH-proximal segments. We showed that the relationship between frequency of usage and location on VH locus can be mathematically defined by one-phase exponential decay whereby the frequency of usage halves with every 45-kb distance away from JH proximity. It could be argued that the unexpected usage of individual VH genes are a result of the leukemic process' driving an unnatural pattern of VH rearrangement, rather than a reflection of truly normal preimmune B-cell events. We believe that our data reflect what could be a normal pattern of IGH rearrangements in pre–antigen-stimulation B cells, as the data do not differ from those in studies of fetal human and mouse B cells. The difference observed between BM or PB and the ALL group reflects a biological rather than a disease-related difference, as we first suggested in a preliminary study.49 Comparing our results with previous reports, we did not find overuse of VH5 (previously described as VH251) in ALL50 or VH (4-34), VH(4-39), VH (4-59), VH (3-23), and VH (3-30) in fetal bone marrow.28
The VH6 gene
The highly polymorphic human VH region is thought to have arisen prior to racial divergence and has been stable for at least 30 000 years.51 However, VH6 is unusual in being the sole member of its family with no polymorphic variables in different individuals52 and racial groups.51 Lack of genetic polymorphism in the 30-kb area around the VH6 segment is in sharp contrast to the expected occurrence of a restriction fragment length polymorphism every 100 to 300 bp.53 A relative lack of repetitive elements in the same area is also unusual.4,54 It has been suggested that VH6 is protected from mutations by strong selection pressure to maintain this area intact.51 Compelling studies of the nature of theVH6 promoter have revealed that it has many unique features not shared by the other VH segment promoters. Promoters of VH genes contain one consistent transcription-factor–binding site, in addition to the TATA box. This octamer motif, with a consensus sequence ATGCAAAT, is found approximately 70 bases upstream of the transcription initiation site. The motif is perfectly conserved in all but 3 of the 42 functional VH segments (octamers for the segments 3-20, 3-53, andVH6 have 1- to 2-bp variations on the consensus). The presence of an octamer is an absolute requirement for tissue-specific transcription of VH segments in vivo and in vitro,55-57 and a single base change prevents the binding of the Octamer-2 (a B-lineage–specific transcription factor).58 Sun and Kitchingman59,60 have shown that the imperfect VH6 octamer, with a single base-pair change (AgGCAAAT), lacks B-cell specificity and transcribes in an enhancer-independent fashion, has bidirectional transcriptional capabilities, and has greater transcriptional strength. Furthermore, non–B-cell silencer activity, as found in one of the VH2 promoters, was not found in VH6. These data suggest that the VH6 promoter is under a different transcriptional control compared with other VH promoters. VH6 promoter is often the first VH promoter activated during ontogeny and is also overrepresented in artificially activated resting small B cells.19 61
Our study describes, with statistical relevance, the overusage of theVH6 segment during VH-(D)-JH rearrangement, which we had previously noted in ALL.39 Our work supports the idea thatVH6 has special properties, such as marking the cells ready to undergo true semi-random rearrangement. Of note, there is a non-VH gene (KIAA0125) near the 3′ side ofVH64 in the reverse orientation toVH6, which has an extremely short coding region but extensive 5′ and 3′ untranslated regions and shows lymphoid-restricted expression,62 with as-yet-unidentified function.
VH pseudogenes
We were interested in analyzing how often pseudogenes, which retained both their abilities to transcribe and rearrange with intact promoters and RS sequences (13 of 81; 16%) but failed to yield productive proteins, were used. Although pseudogenes make up a sizable proportion of the total rearrangeable segments (13 of 55; 24%), only 2 (3%) of such pseudogenes were found rearranged in our study. From an examination of their locations in the VH locus (Figures 5and 6), it can be seen that the majority of the rearrangeable pseudogenes (9 of 13; 69%) lie at 500 kb or more from JH1. This area corresponds to the plateau of the exponential-decay usage curve; therefore, a low rate of usage can be expected. Even so, pseudogene usage appeared to be lower than expected. Although all VH segments with intact promoters and RS sequences can rearrange, restriction selection of wasteful VH-(D)-JH recombinations may occur.
JH segment usage
We found that JH4, JH5, and JH6 were greatly overrepresented, in keeping with most previous reports in childhood ALL63-65 and adult PB,66 with the exception that they all report a predominance order of JH4 being greater than or equal to JH6, which is greater thanJH5, whereas we find an order of JH4 greater than JH5, which is greater thanJH6. Preimmune cells from fetal liver use JH3 and JH4, with littleJH5 and very little JH6and JH1.29,30 Cuisiner and colleagues24 revealed that although the overall order of preference in fetal liver wasJH3 = JH4, which is greater than(JH1 = JH2 = JH5 = JH6),on close inspection, JH6 usage, which was otherwise rare, strongly associated with VH6usage. On the contrary, none of the JH6-positive clones in adult B-lineage ALL were VH6 positive (Table 5). This is good evidence that the increasedVH6 usage observed in adult preimmune cells is unrelated to that seen in fetal B cells and that separate mechanisms give rise to the observed repertoires.
In summary, we have shown that the profiles of VH gene family usage in normal BM and PB are in broad agreement with theoretical values calculated under the assumption that all rearrangeable VH segments have an equal probability of being selected. Furthermore, B-lineage cells from clonally expanded populations from B-cell lymphoma and CLL conform to this pattern. However, clones from early B-cell progenitors of precursor populations (from ALLs) do not conform to this pattern, so the assumption made does not hold true. Detailed analyses have revealed that rate of VH usage is proportional to proximity to the JHlocus. It is plausible that most B cells first undergo rearrangement preferentially using JH-proximal VH, and that further rounds of VH recombining with (D)-JHoccur later on, with the effect of normalizing the observed VH repertoire. The final repertoire would appear unbiased with all VH segments represented in their correct proportions, excluding a role for this phenomenon in leukemogenesis.
We thank Richard Morris (Population Sciences and Epidemiology, Royal Free Hospital, London, United Kingdom) for his kind help with statistical analyses; Julie Burrett (Clinical Trial Service Unit, Oxford, United Kingdom) for clinical information on patients; Dr Paul Travers (Anthony Nolan Centre, London, United Kingdom) and Professor Lucio Luzzatto (Istituto Scientifico Tumori, Genoa, Italy) for helpful discussions; and all of the United Kingdom clinicians involved in the Adult UKALL XII study who supplied bone marrow samples for this study.
Supported by the Kay Kendall Leukaemia Fund and the Leukaemia Research Fund, United Kingdom.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Letizia Foroni, Department of Haematology, Royal Free and University College of London (Royal Free Campus), Pond St, London NW3 2QG, United Kingdom; e-mail: letizia@rfc.ucl.ac.uk.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal