Abstract
The members of tumor necrosis factor receptor (TNFR) superfamily have been designated as the “guardians of the immune system” due to their roles in immune cell proliferation, differentiation, activation, and death (apoptosis). This study reports the cloning of a new member of the TNFR superfamily, RELT (ReceptorExpressed in Lymphoid Tissues). RELT is a type I transmembrane glycoprotein with a cysteine-rich extracellular domain, possessing significant homology to other members of the TNFR superfamily, especially TNFRSF19, DR3, OX40, and LTβ receptor. The messenger RNA of RELT is especially abundant in hematologic tissues such as spleen, lymph node, and peripheral blood leukocytes as well as in leukemias and lymphomas. RELT is able to activate the NF-κB pathway and selectively binds tumor necrosis factor receptor-associated factor 1. Although the soluble form of RELT fusion protein does not inhibit the one-way mixed lymphocyte reaction, immobilized RELT is capable of costimulating T-cell proliferation in the presence of CD3 signaling. These results define a new member of the TNFR superfamily that may be a potential regulator of immune responses.
Introduction
The tumor necrosis factor receptor (TNFR) superfamily includes at least 20 members, such as TNFR type I (p55), TNFR type II (p75), Fas (CD95), CD40, CD27, CD30, 4-1BB (CDw137), and OX40 (CD134), that are recognized as key immunomodulatory molecules. Their functions include the induction of differentiation, proliferation, activation, and death of specific immune cells.1 This family of proteins is characterized by the presence of cysteine-rich extracellular domains that have been shown to be important for ligand binding.2 The canonical cysteine-rich extracellular domains usually consist of 6 cysteines with considerable variability in the domain length, depending on the particular receptor.1 The overall homology between different receptors is low; the highest homology, approaching 30%, is observed in the extracellular domains.2,3 The highest level of sequence divergence exists in the cytoplasmic domain, with the exception of death domains that are found in the death-inducing receptors such as Fas (CD95) and Death Receptors (TRAIL-R).1
In addition to the presence of extracellular cysteine-rich domains, many members of the TNFR superfamily have been shown to activate the NF-κB pathway, either by ligand-mediated cross-linking of the receptor or by overexpression of the receptor.4-7Activation of NF-κB by TNFR superfamily members is believed to be mediated primarily by TNF receptor-associated factors (TRAFs), although not all members of the TNFR superfamily utilize the TRAF family of proteins to signal.8,9 The TRAF family of proteins is currently comprised of 6 members, TRAFs 1-6, with different members of the TNFR superfamily binding specific TRAF family members.8
We have searched a human expressed sequence tagged (EST) database for novel molecules homologous to TNFR superfamily members that are important for activation and/or differentiation of T lymphocytes. By searching the EST database with the cysteine-rich extracellular domain of OX40, we have identified a partial EST sequence corresponding to a new member of the TNFR superfamily that we designated RELT (Receptor Expressed in LymphoidTissues). Here we report the complementary DNA (cDNA) cloning, amino acid sequence analysis, tissue and cell distribution of RELT messenger RNA (mRNA), and initial biological characterization of RELT.
Materials and methods
Cloning of RELT cDNA
A partial cDNA sequence encoding the 3′ end of RELT was identified from an EST database, based on its homology to OX40. The 5′ end of RELT was isolated by 5′RACE (rapid amplification of cDNA ends; Life Technologies, Gaithersburg, MD), using HL60 poly A RNA isolated with TRI reagent (Sigma, St Louis, MO), and poly A tract III system (Promega, Madison, WI). Full-length RELT was amplified from random primed Raji Burkitt lymphoma cell line cDNA, using the proofreading DNA polymerase PFU (Stratagene, La Jolla, CA), cloned into pTeasy vector (Promega) after addition of overhanging adenosines with TAQ polymerase (PE Biosystems, Foster City, CA), and sequenced. All subsequent constructs were derived from this plasmid, using the DNA polymerase PFU.
Northern blots
Dot blots and multiple tissue blots (Clontech, Palo Alto, CA) were probed according to the manufacturer's instructions, using a polymerase chain reaction (PCR) fragment of the 3′ end of the RELT that had been 32P labeled by the random primer method (Roche Molecular Biochemicals, IN).
Media, cell lines, transfections, binding assays, and reporter assays
HEK 293 cell lines were purchased from American Tissue Culture Center (ATCC, Rockville, MD) and grown in Dulbecco modified Eagle medium (Life Technologies) supplemented with glutamine, penicillin, streptomycin, and 10% fetal bovine serum (Hyclone, Logan, UT). For luciferase assays, 2 × 105 HEK 293 cells were transfected in 6-well plates by the GenePorter transfection reagent (Gene Therapy Systems, San Diego, CA) according to the manufacturer's instructions. Cells were lysed 24 to 48 hours after transfection with reporter lysis buffer (Promega). For TRAF2 dominant-negative (TRAF2DN) experiments, 293T cells (gift of Dr C. Paya, Mayo Clinic, Rochester, MN) were transfected with a mixture of the indicated plasmids in Fugene 6 (Roche Molecular Biochemicals), and cell lysates were harvested 36 hours after transfection for luciferase assay. The TRAF2DN plasmid contains a truncated form of TRAF2, corresponding to amino acid 87-501. The β-galactosidase constitutively active reporter plasmid (TK-βgal) and NF-κB reporter plasmid (κBluc) were gifts of Dr Carlos Paya.
TRAF binding assays and Western blot analysis
For TRAF binding assays, 5 × 105 293 cells were transfected with 5 μg hemagglutinin (HA)-tagged RELT plasmid and 5 μg FLAG-tagged TRAF 1, 2, 3, 5, or 6 plasmids (generous gifts from Dr D. Goeddel, Tularik, San Francisco, CA) by the calcium phosphate method. The transfected cells were harvested 36 to 48 hours after transfection in lysis buffer (1% NP-40, 150 mM NaCl, 50 mM Tris pH 7.5, 0.5% bovine serum albumin [BSA] [wt/vol], 2 μg/mL aprotinin, 2 μg/mL leupeptin, 1 μg/mL pepstain A, 100 μg/mL phenylmethylsulfonyl fluoride). Whole cell lysates were immunoprecipitated with anti-FLAG M2 beads (Sigma) for 4 hours at 4°C with rotation, washed 5 times with lysis buffer (without BSA), boiled 5 minutes, separated by 10% sodium dodecylsulfate–polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membrane, probed with anti-HA horseradish peroxidase (HRP; Roche), and developed with enhanced chemiluminescence (ECL; Amersham, Piscataway, NJ). For FLAG protein expression controls, blots were stripped and probed with biotinylated anti-FLAG M2 (Sigma), followed by streptavidin HRP (Biosource International, Camarillo, CA), and developed with ECL.
RELT and Fc fusion protein
The RELT Fc fusion protein was constructed by ligating a PCR fragment corresponding to the extracellular domain of RELT to human immunoglobulin G1 (IgG1) Fc domain.10 The 293 cells were transfected by the calcium phosphate method with the fusion protein construct. Fusion protein was purified by a protein G column (Pierce, Rockford, IL), and dialyzed against 3 1-L changes of phosphate-buffered saline (PBS). Presence of human Fc fusion protein was confirmed by a sandwich enzyme-linked immunosorbent assay specific for human Fc. RELT-hFc fusion protein or control hIgG (Sigma) were biotinylated, using the EZ-Link NHS-LC-LC-Biotin kit (Pierce). Total protein concentration in protein preparations was quantitated with Coomassie Plus Protein Assay Reagent (Pierce) according to the manufacturer's instructions.
T-cell costimulation and mixed lymphocyte reaction
Primary T cells were isolated from buffy coats of healthy donors as described previously.11 Briefly, peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Paque Plus (Pharmacia, Piscataway, NJ) and incubated 2 hours at 37°C to remove plastic adherent cells. Nonadherent cells were further purified by passing through nylon wool column and were more than 80% pure for CD3+ cells. T-cell costimulation assays were performed as described previously.12 Briefly, 96-well flat-bottomed plates were coated with antihuman CD3 (clone HIT3a, Pharmingen, San Diego, CA) at either 37°C for 2 hours or 4°C overnight in 50 μL PBS. The wells were washed 3 times in 100 μL PBS, coated with recombinant protein by incubating at 37°C for 2 hours, and then washed another 3 times in PBS. T cells were plated at a density of 1 × 106 cells/mL in triplicate. T cells were allowed to proliferate for a total of 72 hours and were pulsed with 1 μCi of3H thymidine during the last 12 to 18 hours. For mixed lymphocyte reaction, responder nylon wool–purified T cells and stimulator PBMCs (irradiated for 4000 rads) were mixed at varying ratios in round-bottom 96-well plates with a total of 3 × 105 cells/well. The number of nylon wool–purified cells was kept constant, and the number of stimulator PBMCs was varied to reflect the indicated ratios. All wells contained either 10 μg/mL of hIgG (Sigma) or RELT-hFc. Cells were incubated for 5 days and pulsed with 1 μCi of 3H thymidine for the last 18 hours. All points are in triplicate.
Results and discussion
In an attempt to find new members of the TNFR superfamily, we searched the Human Genome Sciences EST database with cysteine-rich domains characteristic of TNFR superfamily members and found an EST that was homologous to OX40. Isolation and sequencing of the full-length cDNA by 5′-RACE revealed a novel member of the TNFR superfamily when searched against the National Center for Biotechnology Information (NCBI) public database (Figure1A). We named the new molecule RELT, based on its limited mRNA expression profile.
Sequence analysis of RELT.
(A) Amino acid sequence of RELT. Single underline is signal peptide, double underline is location of transmembrane region as determined by hydrophilicity analysis. Arrow indicates potential proteolytic cleavage site. Asterisk indicates potential N-glycosylation site. Sequence analysis was performed with MacVector 6.5. (B) Amino acid alignment of the cysteine-rich extracellular domains of RELT with the cysteine-rich domain of other members of the TNFR superfamily. Dark gray indicates identical amino acid, and light gray indicates similar amino acid homologies. Amino acid alignment was performed with ClustaW. (C) Alignment of human RELT with the putative rat RELT derived from a partial EST sequence and full-length macaque sequence. GenBank Accession numbers AI071067, AB046039, and AF319553 are for rat, macaque, and human sequences, respectively.
Sequence analysis of RELT.
(A) Amino acid sequence of RELT. Single underline is signal peptide, double underline is location of transmembrane region as determined by hydrophilicity analysis. Arrow indicates potential proteolytic cleavage site. Asterisk indicates potential N-glycosylation site. Sequence analysis was performed with MacVector 6.5. (B) Amino acid alignment of the cysteine-rich extracellular domains of RELT with the cysteine-rich domain of other members of the TNFR superfamily. Dark gray indicates identical amino acid, and light gray indicates similar amino acid homologies. Amino acid alignment was performed with ClustaW. (C) Alignment of human RELT with the putative rat RELT derived from a partial EST sequence and full-length macaque sequence. GenBank Accession numbers AI071067, AB046039, and AF319553 are for rat, macaque, and human sequences, respectively.
TNFR superfamily members are evolutionarily conserved, and much of the amino acid conservation is observed in the extracellular domain, which can approach 70% amino acid identity, between species.13When we searched the nonhuman NCBI EST databases for the presence of an orthologue of RELT, a partial rat EST sequence and a full-length macaque EST sequence were found. Both the rat and macaque amino acid sequences are highly identical to the human sequence, with the macaque sequence having 96% amino acid identity (Figure 1C). The extremely high identity shared between the human, macaque, and rat proteins indicates that RELT exhibits evolutionary conservation.
BLAST search of NCBI databases demonstrates that the extracellular region of RELT has 2 cysteine-rich domains, one complete and one incomplete, that are homologous to TNFR superfamily members, such as TNFRSF19,14 DR3/TR6,15 OX40,16and LTβ receptor.17 The RELT cysteine residues are highly conserved when compared to other TNFR superfamily members (Figure 1B). The intracellular domain does not have significant homology to any known protein sequence in the database. The sequence contains several putative protein kinase C sites and a tyrosine phosphorylation site but no death domains.
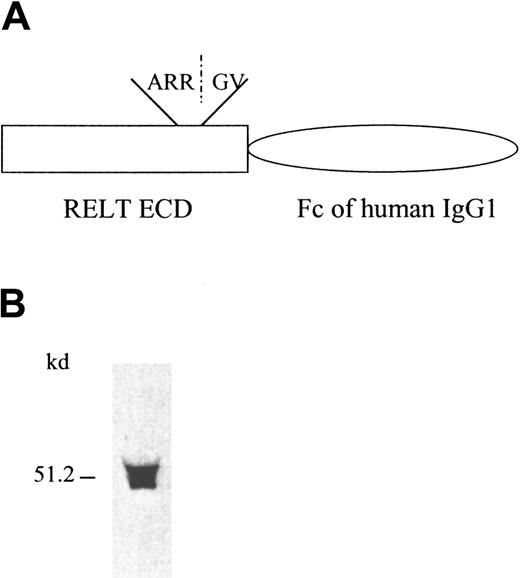
The nucleotide context of the most 5′ ATG of the RELT cDNA sequence conforms to the Kozak consensus sequence and is preceded by a GC-rich region. Hydrophilicity analysis of the amino acid sequence reveals that the methionine coded by the 5′ most start codon lies within a span of hydrophobic amino acids (start methionine to amino acid 25) that is most likely the signal peptide (Figure 1A). These characteristics indicate that this ATG is the start translation point for RELT and that we have isolated the full open reading frame of RELT. The proposed start codon was indeed able to initiate translation because 2 forms of the protein, a full-length protein that was HA epitope tagged in the cytoplasmic domain and a soluble extracellular domain human Fc (hIgG1) fusion protein were detectable from transfected cells (Figures2B and 4B). Surprisingly, Western blot analysis of the soluble receptor fusion protein revealed the presence of 2 different bands (Figure 2B) at approximately the predicted molecular weight. We obtained 2 major amino acid sequences on N-terminal amino acid sequencing of the soluble receptor fusion protein. One major sequence matched the predicted N-terminus of the protein after the signal peptide had been cleaved. The other major sequence, beginning with amino acid 131 (GVEV…), corresponded to an internal amino acid sequence of RELT, indicating that the smaller molecular weight band that was detected by Western blot may in fact be a truncated form of RELT. This smaller molecular weight form of RELT was detected in Western blot analysis of both the RELT-hFc fusion protein supernatants (Figure 2B) as well as the RELT-HA immunoprecipitates (data not shown). Further studies will be required to ascertain whether this smaller molecular weight form of RELT occurs naturally or is an artifact of the expression systems used. Truncated forms of TNFR superfamily are known to exist and occur either through proteolytic cleavage of the extracellular domain, as is the case for p55 and p75 TNF receptors,18 or through alternative splicing of the mRNA, as is the case for 4-1BB and Fas.19 20 The proposed translation initiation codon has an in-frame stop codon that ultimately encodes an open reading frame of 430 amino acids, which is also the longest open reading frame of the isolated sequence (Figure 1A). The isolated nucleotide sequence also contains a canonical polyadenylation signal that is approximately 1100 nucleotides from the proposed stop site. The full-length protein has a predicted molecular weight of 46 kDa, pI of 9.6, one N-glycosylation site in the extracellular domain, and a large hydrophobic region from amino acid 156-191, which is the transmembrane domain. The predicted molecular weight corresponds well to the approximate weight observed in a HA epitope-tagged full-length protein that was expressed in 293 cells (Figure 4B). These observations indicate that we have isolated a full-length cDNA clone of RELT that encodes a protein that can be expressed in an eukaryotic expression system.
RELT-hFc fusion protein and its expression.
(A) Diagram of RELT-hFc fusion protein construct. ECD is the extracellular domain of RELT that is linked to the Fc portion of hIgG1. ARR/GV is the amino acid sequence of the proposed cleavage site for the truncated form of the soluble protein. (B) Western blot analysis of recombinant RELT-hFc fusion protein from transfected 293 supernatant. After SDS-PAGE and transfer to nitrocellulose filter paper, fusion protein was detected with a goat antihuman Fc specific HRP-conjugated polyclonal antibody.
RELT-hFc fusion protein and its expression.
(A) Diagram of RELT-hFc fusion protein construct. ECD is the extracellular domain of RELT that is linked to the Fc portion of hIgG1. ARR/GV is the amino acid sequence of the proposed cleavage site for the truncated form of the soluble protein. (B) Western blot analysis of recombinant RELT-hFc fusion protein from transfected 293 supernatant. After SDS-PAGE and transfer to nitrocellulose filter paper, fusion protein was detected with a goat antihuman Fc specific HRP-conjugated polyclonal antibody.
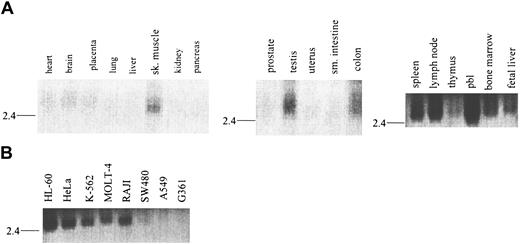
Expression of RELT in normal human tissues (Figure3A) and tumor lines (Figure 3B) was analyzed by Northern blot and dot blot (data not shown). RELT expression is limited primarily to hematologically important tissues and immune cell–derived lines, as determined by Northern blot. RELT mRNA is approximately a 2.6-kilobase (kb) transcript that is most abundant in peripheral blood leukocytes, lymph node, spleen, and bone marrow. RELT message is also present at low or barely detectable levels in other tissues, such as skeletal muscle, testis, and colon (Figure 3A and data not shown), but it is completely absent in other tissues such as brain, kidney, and pancreas. In addition, RELT mRNA is also found in fetal hematopoietic tissues such as liver, spleen, thymus, and lung (Figure 3A, data not shown), indicating a possible role of RELT expression in development of the immune system. Recapitulating the tissue Northern blot data, RELT mRNA is expressed in the majority of hematopoietic cell lines tested, including T cell (MOLT 4), B cell (RAJI), and myeloid (HL-60, K-562) cell lines, and at very low levels in the colorectal cell line (SW480). Conversely, RELT mRNA is not expressed in cell lines that are non-hematopoietically derived such as a melanoma (G361) and lung carcinoma (A549) (Figure 3B). These results indicate that the pattern of RELT mRNA expression is not unique to one specific hematologic cell subset(s) but has a ubiquitous distribution pattern among all hematologic cell subsets. In addition, the tissues that are known to have high levels of these cell types also have the highest level of RELT message. This distribution pattern indicates that RELT may have a function that is vital for hematologic cells and tissues.
Tissue distribution of RELT mRNA.
Northern blot analysis of RELT mRNA distribution in (A) normal human tissues and (B) transformed human cell lines.
Tissue distribution of RELT mRNA.
Northern blot analysis of RELT mRNA distribution in (A) normal human tissues and (B) transformed human cell lines.
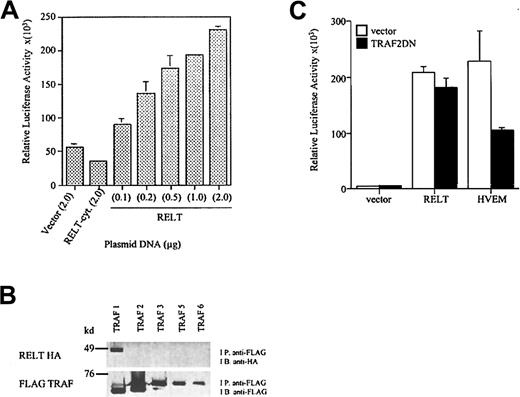
To investigate whether RELT is functionally homologous to the other members of the TNFR superfamily, we examined the potential of RELT to activate NF-κB. We transiently cotransfected HEK 293 cells with the full-length cDNA of RELT and a κB luciferase reporter plasmid. 293 cells that had been transfected with the RELT reporter plasmid were able to significantly induce NF-κB activation when compared to vector control or an expression plasmid containing a cytoplasmic deletion mutant of RELT (Figure 4A). The ability of RELT to activate NF-κB is dependent on the amount of RELT expression plasmid used to transfect the cells, with increasing concentration of RELT expression plasmid leading to increased reporter activity (Figure 4A). Activation and nuclear translocation of NF-κB is associated with the up-regulation of a variety of immunoregulatory molecules involved in lymphocyte and antigen-presenting cell activation.21
RELT is able to activate NF-κB and binds TRAF1.
(A) Activation of NF-κB by RELT. The 293 cells were transfected with increasing doses of RELT, starting at 0.1 μg, 0.2 μg, 0.5 μg, 1.0 μg, and 2.0 μg of DNA. Vector and RELT cytoplasmic domain deletion mutant (RELT-cyt) were used at a concentration of 2.0 μg. All wells were transfected with 0.5 μg κB-Luc and 0.5 μg TK-βgal. The total amount of DNA transfected in all wells was 3 μg. All values are normalized to β-gal activity. The experiment is representative of 3 experiments. (B) RELT binds TRAF1. The 293 cells were transfected with 5 μg RELT-HA and 5 μg of either FLAG TRAF 1, 2, 3, 5, or 6. Cell lysates were immunoprecipitated (I.P.) with anti-FLAG beads, separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted (I.B.) with anti-HA HRP. Blots were then stripped and re-probed with anti-FLAG M2 to detect the presence of tagged proteins. (C) Activation of NF-κB by RELT is not inhibited by a dominant-negative form of TRAF2. The 293T cells were transfected with a total of 3 μg DNA. All wells were transfected with 0.25 μg κB-luc and 0.25 μg TK-βgal. Cells were also transfected with 2 μg of either vector, RELT, or HVEM and 0.5 μg TRAF2DN or vector. Transfections were performed in duplicates, and this experiment is representative of 2 experiments.
RELT is able to activate NF-κB and binds TRAF1.
(A) Activation of NF-κB by RELT. The 293 cells were transfected with increasing doses of RELT, starting at 0.1 μg, 0.2 μg, 0.5 μg, 1.0 μg, and 2.0 μg of DNA. Vector and RELT cytoplasmic domain deletion mutant (RELT-cyt) were used at a concentration of 2.0 μg. All wells were transfected with 0.5 μg κB-Luc and 0.5 μg TK-βgal. The total amount of DNA transfected in all wells was 3 μg. All values are normalized to β-gal activity. The experiment is representative of 3 experiments. (B) RELT binds TRAF1. The 293 cells were transfected with 5 μg RELT-HA and 5 μg of either FLAG TRAF 1, 2, 3, 5, or 6. Cell lysates were immunoprecipitated (I.P.) with anti-FLAG beads, separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted (I.B.) with anti-HA HRP. Blots were then stripped and re-probed with anti-FLAG M2 to detect the presence of tagged proteins. (C) Activation of NF-κB by RELT is not inhibited by a dominant-negative form of TRAF2. The 293T cells were transfected with a total of 3 μg DNA. All wells were transfected with 0.25 μg κB-luc and 0.25 μg TK-βgal. Cells were also transfected with 2 μg of either vector, RELT, or HVEM and 0.5 μg TRAF2DN or vector. Transfections were performed in duplicates, and this experiment is representative of 2 experiments.
Because NF-κB activation by TNFR superfamily members is mediated primarily by the TRAF family of signaling molecules, we studied whether RELT can bind any of the TRAF molecules. 293 cells were cotransfected with a FLAG-tagged TRAF expression plasmid and a HA-tagged RELT expression plasmid. Immunoprecipitation of transfected cell lysates with anti-FLAG Sepharose beads followed by immunoblotting with anti-HA demonstrated that RELT coprecipitates only with TRAF1 but not with TRAFs 2, 3, 5, or 6 (Figure 4B, upper panel). Lack of binding of RELT to other TRAFs is not due to insufficient expression of TRAF proteins since high-level expression of TRAF 2, 3, 5, and 6 were detected in the immunoprecipitates (Figure 4B, lower panel). Therefore, RELT is unique among the TNFR superfamily members in its ability to exclusively bind TRAF1. RELT and TRAF1 share similar mRNA expression patterns, as both are primarily limited to hematologic tissues.22 RELT probably signals NF-κB activation independently of the known TRAF molecules in 293 cells. In addition, activation of NF-κB does not occur by TRAF1-mediated recruitment of NF-κB inducing TRAF molecules to the RELT receptor complex since 293 cells do not express TRAF1 even after tumor necrosis factor α (TNF-α) stimulation.23 The inability of RELT to activate NF-κB via TRAF molecules is further supported by the finding that, when RELT and TRAF2DN are coexpressed, there is no inhibition of RELT-mediated NF-κB activation. In the same assay, herpes virus entry mediator (HVEM), a TNFR superfamily member that is known to signal NF-κB activation by TRAF2, is inhibited by the TRAF2DN construct (Figure 4C).
It has been reported that TRAF1 expression is up-regulated by NF-κB activation and that TRAF1 can in turn modulate NF-κB activation.7 We are currently investigating the possibility that TRAF1 may modulate the NF-κB activity induced by RELT. It is of interest that TRAF1 overexpression was shown to protect CD8+ T cells from activation-induced cell death24 and potentiates the antiapoptotic activities of inhibitor of apoptosis 1 and 2.25 Furthermore, TRAF1 is up-regulated in certain types of lymphomas, most notably Hodgkin Reed-Sternberg cells that are known to express a wide spectrum of TNFR superfamily receptors and are believed to be in a constitutively activated state that leads to the survival of the cells.26 27 Whether RELT plays a role in any of these TRAF1-related functions will be the subject of further investigation.
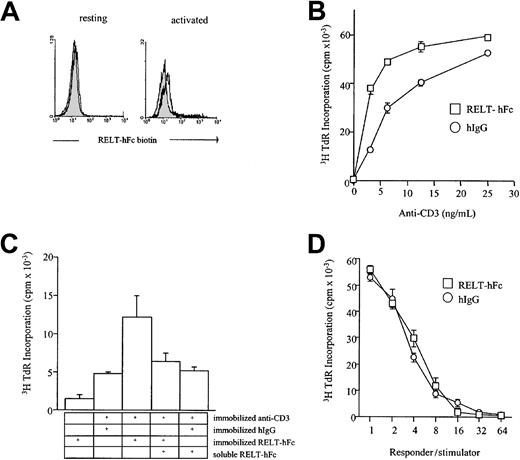
On the basis of the limited tissue distribution of RELT mRNA to hematologic tissues and peripheral leukocytes, we investigated the possible role of RELT as an immunomodulatory molecule. We created an extracellular domain fusion protein linked to the human IgG1 Fc and expressed the protein in 293 cells (Figure 2A,B). Inclusion of RELT-hFc up to 20 μg/mL in the cultures of allogeneic mixed lymphocyte reaction (MLR) neither decreased nor increased the proliferation of T cells in a wide range of responder-to-stimulator ratios (Figure 5D and data not shown). This result suggests that the interaction between RELT and its putative ligand is not required for the induction of MLR. When the purified RELT-hFc protein was immobilized to plastic plates with varying concentrations of anti-CD3 antibody, there was a dose-dependent increase in T-cell proliferation that was increased when compared to the wells that had been coated with a control hIgG (Figure 5B), indicating that a ligand of RELT can costimulate T-cell proliferation. Inhibition, by a soluble form of RELT-hFc, of the costimulatory activity of the immobilized RELT-hFc indicates that this costimulatory activity is RELT-hFc specific (Figure 5C). Several studies have shown that ligands of TNFR superfamily, including 4-1BBL and FasL,28-30 are able to signal T-cell proliferation in vitro. Therefore, it is likely that RELT binds a putative ligand on T cells and can transmit a T-cell proliferative signal. Indeed, a biotinylated form of RELT-hFc is able to bind phorbol 12-myristate 13-acetate (PMA)– and ionomycin– stimulated CD3+ cells by flow cytometry. In summary, our data identify a new member of the TNFR superfamily that is selectively expressed in hematopoietic tissues and potentially participates in the activation of hematopoietic cells.
Effect of RELT-hFc fusion protein in T-cell responses.
(A) RELT-hFc binds PMA and ionomycin-activated T cells. Nylon wool–purified T cells were activated for 18 hours in 200 ng/mL ionomycin and 20 ng/mL PMA and then double stained with RELT-hFc biotin and anti-CD3 phycoerythrin. Histogram shows CD3 gated cells. Filled histogram is hIgG biotin control, and open histogram is RELT-hFc biotin. (B) RELT-hFc costimulates T-cell proliferation. For proliferation assays, 96-well plates were coated with the indicated concentration of anti-CD3, washed with PBS, and coated with 10 μg/mL of the indicated protein. After coating with fusion protein, wells were washed again with PBS, and T cells were added. T cells were incubated for 72 hours and pulsed with 1 μCi 3H thymidine for the last 18 hours before being harvested. All CPM values are from triplicate wells. This experiment is representative of 3 experiments. (C) Addition of soluble RELT-hFc selectively inhibits RELT-hFc costimulation of T-cell proliferation. Proliferation assays were performed as in (A). Soluble RELT-hFc was added at a dose of 10 μg/mL, and the dose of anti-CD3 is 6.25 ng/mL. (D) RELT-hFc does not inhibit the one-way mixed lymphocyte reaction. Nylon wool–purified T cells and stimulator PBMCs that had been irradiated for 3000 rads were incubated at various responder-to-stimulator ratios with 10 μg/mL hIgG or RELT-hFc. The mixed cells were incubated for 5 days and pulsed with 1 μCi 3H thymidine for the last 18 hours. All CPM values are from triplicate wells. This graph is representative of 3 experiments.
Effect of RELT-hFc fusion protein in T-cell responses.
(A) RELT-hFc binds PMA and ionomycin-activated T cells. Nylon wool–purified T cells were activated for 18 hours in 200 ng/mL ionomycin and 20 ng/mL PMA and then double stained with RELT-hFc biotin and anti-CD3 phycoerythrin. Histogram shows CD3 gated cells. Filled histogram is hIgG biotin control, and open histogram is RELT-hFc biotin. (B) RELT-hFc costimulates T-cell proliferation. For proliferation assays, 96-well plates were coated with the indicated concentration of anti-CD3, washed with PBS, and coated with 10 μg/mL of the indicated protein. After coating with fusion protein, wells were washed again with PBS, and T cells were added. T cells were incubated for 72 hours and pulsed with 1 μCi 3H thymidine for the last 18 hours before being harvested. All CPM values are from triplicate wells. This experiment is representative of 3 experiments. (C) Addition of soluble RELT-hFc selectively inhibits RELT-hFc costimulation of T-cell proliferation. Proliferation assays were performed as in (A). Soluble RELT-hFc was added at a dose of 10 μg/mL, and the dose of anti-CD3 is 6.25 ng/mL. (D) RELT-hFc does not inhibit the one-way mixed lymphocyte reaction. Nylon wool–purified T cells and stimulator PBMCs that had been irradiated for 3000 rads were incubated at various responder-to-stimulator ratios with 10 μg/mL hIgG or RELT-hFc. The mixed cells were incubated for 5 days and pulsed with 1 μCi 3H thymidine for the last 18 hours. All CPM values are from triplicate wells. This graph is representative of 3 experiments.
We thank Drs D. Goeddel and C. Paya for generous gifts of TRAF constructs and NF-κB reporter plasmid, respectively; S. Hu for helpful discussion and suggestions; and K. Jensen and V. Ramnath for editing the manuscript.
Supported in part by grants from the Mayo Foundation and from the National Institutes of Health (NIH). G.Z. and K.T. are supported by NIH postdoctoral training grant CA09127 and by U.S. Army breast cancer research fellowship, respectively.
J.N. and L.C. share senior authorship.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Lieping Chen, Department of Immunology, Mayo Clinic, 200 First St SW, Rochester, MN 55905; e-mail:chen.lieping@mayo.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal