Abstract
A pivotal role has been assigned to Myb in the control of myeloid cell growth. Although Myb is a target of retinoic acid, little is known about the mechanisms by which it may contribute to induced growth arrest in leukemia cells. Indeed, few Myb target genes are known to be linked to proliferation. Myeloblastin is involved in the control of proliferation in myeloid leukemia cells. It is expressed early during hematopoiesis and is a granulocyte colony-stimulating factor–responsive gene. Myeloblastin can confer factor-independent growth to hematopoietic cells, an early step in leukemia transformation. The myeloblastin promoter contains PU.1, C/EBP, and Myb binding sites, each of which are critical for constitutive expression in myeloid cells. Inhibition of myeloblastin expression in leukemia cells growth-arrested by retinoic acid is demonstrated to depend on Myb down-regulation. Myb is shown to induce myeloblastin expression and abolish its down-regulation by retinoic acid. Altogether, the data offer a clue as to how a myeloid-specific transcriptional machinery can be accessible to regulation by retinoic acid and point to myeloblastin as a novel target of Myb. This link between Myb and myeloblastin suggests a previously nonidentified Myb pathway through which growth arrest is induced by retinoic acid in myeloid leukemia cells.
Introduction
Retinoic acid (RA) plays a major role in inducing growth arrest and differentiation of myeloid leukemia cells both in vitro and in vivo.1-5 However, little is known about those myeloid genes that respond to RA and are involved in growth control. Myeloblastin (MBN)6 is a serine protease with a broad spectrum of proteolytic activity. Inside myeloblastic cells, a main enzymatic target appears to be the Sp1 transcription factor, which is physiologically truncated by MBN and is known to contribute to growth regulation.7 An involvement of MBN in the control of leukemia cell growth was initially suggested by the fact that its expression was serum-dependent and down-regulated by growth-inhibiting inducers such as all-trans retinoic acid (ATRA).6 Furthermore, down-regulation of MBN expression by antisense oligodeoxynucleotides inhibited proliferation of promyelocytic-like leukemia cells.6 MBN is overexpressed in myeloid leukemia cells,8 and several reports suggest that T-cell responses to MBN peptides can be used for adaptative T-cell therapy in acute and chronic myeloid leukemias.9,10 MBN is a granulocyte colony-stimulating factor (G-CSF) target gene and can confer factor-independent growth when ectopically overexpressed in an early hematopoietic cell line.11 This has strongly reinforced the view that MBN is a key protease involved in the control of proliferation in normal hematopoietic progenitors and in contributing to early steps in their transformation.
The Myb family of transcription factors is strongly implicated in the regulation of cell growth and differentiation. In the murine system, inappropriate expression of c-Myb clearly contributes to leukemia transformation.12 The c-Myb promotes proliferation and blocks differentiation of hematopoietic cells in several experimental models.13,14 Targeting of c-Myb with antisense oligodeoxynucleotides inhibited hematopoiesis as well as proliferation of myeloid leukemia cells.15,16 Mice homozygous for the inactivated c-Myb gene had impaired definitive hematopoiesis with a drastic decrease in the number of progenitors likely to reflect a proliferation abnormality.17 Expression of c-Myb declines in RA-treated myelomonocytic leukemia cells, suggesting that the retinoic acid receptor (RAR) acts in part by down-regulating c-Myb expression. This may account for the observation that transcriptional activation of a Myb-responsive gene can be inhibited by RA.18 Furthermore, introduction of an exogenous RARα into v-Myb–transformed monoblasts permitted RA-dependent differentiation, indicating that transformation by v-Myb is recessive to RARα.18 Conversely, Myb functions as a potent inhibitor of RA-induced biologic responses.19 Although little is known about the mechanisms involved in these processes, a physical interaction exists between c-Myb and RAR.19However, despite a pivotal role assigned to Myb in the control of cell growth, few Myb-target genes are known to be linked to proliferation, and its relevance to ATRA-induced growth arrest in leukemic cells remains unresolved.
Because MBN is associated with growth control in myeloid leukemia cells, we have analyzed its transcriptional regulation by RA. For this, we have cloned the human and murine MBN promoters, which both harbor binding sites for the PU.1, C/EBP, and Myb transcription factors. Our data show that these factors are all critical for constitutive MBN expression and establish MBN as a novel target of Myb, mediating its regulation by ATRA in leukemia cells. This, together with the fact that MBN is involved in growth control and can provide factor-independent growth to hematopoietic cells, suggests that its association with Myb may play a role in mediating ATRA-induced growth arrest in myeloid leukemia cells.
Materials and methods
Leukemia cell lines culture conditions
Myeloblastic PLB-98520 and promyelocytic NB421 cells were cultured in RPMI 1640 medium with 10% fetal bovine serum and 2 mM L-glutamine (Gibco, Life Technologies, Paisley, UK). COS-7 cells were grown on 9-cm Petri dishes in Dulbecco modified Eagle medium containing 5% fetal bovine serum. Cells were grown in a humidified atmosphere of 5% CO2. Cell viability was assessed by standard trypan blue dye exclusion assay. Exponentially growing PLB-985 or NB4 cells (1 × 106/mL) were used to start suspension cultures. PLB-985 and NB4 cells (2 × 105/mL) were seeded 16 hours prior ATRA treatment. ATRA (Sigma-Aldrich, St Louis, MO) dissolved in ethanol was used at a final concentration of 10−6 M. Differentiation was assessed by the percentage of cells with cell-associated nitroblue tetrazolium (Sigma) and cell morphology under light microscopy on May-Grünwald-Giemsa–stained cytospin slides.
Northern blot analysis
Total RNA extracted using an RNeasy Mini Kit (Qiagen, Hilden, Germany) was electrophoresed and transferred onto Hybond-N nylon filters (Amersham Pharmacia Biotech, Uppsala, Sweden). Filters were prehybridized for 2 hours at 42°C in 50% formamide, 5 × SSC, 0.5% sodium dodecyl sulfate (SDS), 0.2% polyvinylpyrrolidone, 0.2% Ficoll, 50 mM sodium pyrophosphate (pH 6.5), 1% glycine, and 500 μg/mL single-stranded DNA. Hybridization was conducted for 15 hours at 42°C in 50% formamide, 5 × SSC, 0.5% SDS, 0.04% polyvinylpyrrolidone, 0.04% Ficoll, 20 mM sodium pyrophosphate (pH 6.5), 10% dextran sulfate, and 100 μg/mL single-stranded DNA. Filters were washed 30 minutes in 2 × SSC and 0.1% SDS at room temperature, followed by 60 minutes in 0.1 × SSC and 0.1% SDS at 60°C. The MBN probe was the full-length complementary DNA (cDNA). The c-Myb probe was a polymerase chain reaction (PCR) product obtained using 5′-AATTAAATACGGTCCCCTGAAGATG-3′ sense and 5′-CAGGTACTGCTACAAGGCTGCAAGG-3′ antisense primers. The glyceraldehydes-3-phosphate dehydrogenase (GAPDH) probe was a PCR product obtained using 5′-ATCACCATCTTCCAGGA-3′ sense and 5′-CCTGCTTCACCACCTTCTTG-3′ antisense primers. Probes were labeled with [α-32P]deoxycytidine triphosphate 110 TBq/mM (3000 Ci/mM) using a random primer labeling kit (Amersham Pharmacia Biotech).
Recombinant plasmids
Expression vectors for C/EBPα (MSV-C/EBPα), C/EBPβ (MSV-C/EBPβ), and C/EBPδ (MSV-C/EBPδ); for c-Myb (CMV-c-Myb)22; for PU.1 (pECE-PU.1)23; and for E1A 13S and E1A 12S Ntdl814 24 were obtained from A. Friedman, B. Lüscher, R. Maki, and P. Whyte, respectively.
A genomic library of SV129 mouse embryonic stem cell DNA was a gift from P. Chambon. A total of 1 ×106 recombinants were screened using a 5′ probe (85-base pair [bp]NotI-PvuII fragment) of the mouse MBN cDNA kindly provided by L. Hellman.25BamHI digestion of a positive clone and hybridization with the 5′-TCTGGAAGCTACCCATCC-3′ oligonucleotide yielded a 1.1-kilobase fragment that was subcloned and submitted to automated sequencing (Eurogentec, Seraing, Belgium). A 658-bp human MBN promoter region was subcloned into the pBLCAT6 vector,26 generating the pMBN-658 reporter plasmid. For constructing the pMBN-194, pMBN-137, and pMBN-91 deletion reporter plasmids, 194-bp, 137-bp, and 91-bp fragments, respectively, were generated using PCR and cloned into BamHI/XhoI sites of the pBLCAT6 vector. The 5′ primers were 5′-ATGGATCCGACTTGGGTGGGTGA-3′ from positions −194 to −180 (194-bp fragment), 5′-ATGGATCCAAAGCCCCCACCT-3′ from positions −137 to −124 (137-bp fragment), and 5′-ATGGATCCAAGGCAAAAGGAGGAAGT-3′ from positions −91 to −72 (91-bp fragment); the 3′ primer was 5′-ATCTCGAGGATATCGAATTCCTGCAG-3′. Constructions of pMBN-658PU.1mut, pMBN-658C/EBPmut, and pMBN-658Mybmut constructs were obtained by mutagenesis of the PU.1, c/EBP, and Myb binding sites using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The oligonucleotide sequences used were (mutated base pairs are in bold): PU.1, 5′-CAAGGCAAAAGGATTAAGTGGGGACCCAG-3′; C/EBP, 5′-CCAGCCTGGGCGTAGTGGGACTCAACGGCC-3′; c-Myb, 5′-GGGCAACTCATGGGCCTCTGGC-3′. Promoter constructs were verified by DNA sequencing.
In vivo expression assays
COS-7 cells were transfected using calcium phosphate coprecipitation27 of DNA vectors (adjusted to 14 μg per 9-cm Petri dish with pBluescript carrier DNA). Medium was changed after 16 hours. Cells were harvested after an additional 20-hour culture. Exponentially growing PLB-985 cells were washed twice in serum-free medium, once in Optimem medium, resuspended in the same medium at 15 × 106 cells in 0.5 mL, and electroporated (Gene Pulser, Biorad, Hercules, CA) at 300 V, 960 μF with 20 μg of (1) pBabe Puro28 alone or together with CMV–c-Myb (cells were then cultured for 48 hours prior to selection with 1 μg/mL puromycin [Sigma]); (2) reporter plasmid. Cells transfected with reporter plasmid were cultured in 10 mL of medium for 40 hours. Plasmid (5 μg) containing the luciferase cDNA driven by the cytomegalovirus promoter was an internal control for transfection efficiency. All transfections were performed at least in triplicate with independent template preparations. Assays to measure luciferase29 and chloramphenicol acetylase30were conducted as described.
Electrophoretic mobility shift assays
For whole-cell extracts, COS-7 cells were harvested in ice-cold phosphate-buffered saline, pelleted, washed twice in the same buffer, and resuspended in extraction buffer (0.4 M KCl, 20 mM Tris-HCl [pH 7.9], 20% glycerol, 5 mM dithiothreitol, 0.4 mM phenylmethylsulfonyl fluoride, and 2.5 ng each of leupeptin, pepstatin, aprotinin, antipain, and chymostatin [Sigma] per milliliter). After 2 freeze-thaw cycles with liquid nitrogen, the resulting cell lysate was cleared by centrifugation 15 minutes (4°C) at 10 000g and used for electrophoretic mobility shift assays (EMSAs). PLB-985 nuclear extracts were prepared essentially as described31; 2.5 ng each of leupeptin, pepstatin, aprotinin, antipain, and chymostatin (Sigma) per milliliter were added in both lysis and extraction buffers. EMSAs were carried out using PU.1 (human MBN promoter position −84 to −62), 5′-AGCTTAGGCAAAAGGAGGAAGTGGGGACG-3; mut PU.1, 5′-AGCTTAGGCAAAAGGATTAAGTGGGGACG-3′; C/EBP (human MBN promoter position −51 to −36), 5′-AGCTTGGGCATTGGGCAACTCG-3′; mut C/EBP, 5′-AGCTTGGGCGTAGTGGGACTCG-3′; Myb (human MBN promoter position −42 to −22), 5′-ATGGATCCGCAACTCAACGGCCTCTGGCAT-3′; and mut Myb, 5′-ATGGATCCGCAACTCATGGGCCTCTGGCAT-3′ oligonucleotides. Approximatively 0.25 ng (15 000 cpm) of a 32P-5′ end-labeled synthetic double-stranded oligonucleotide was incubated with 5 μg COS-7 cell lysate or PLB-985 nuclear extracts in the presence of poly(dIdC) as a nonspecific competitor in 10 mM Tris-HCl (pH 7.9), 50 mM KCl, 2 mM ethylenediaminetetraacetic acid (EDTA), 0.05% Nonidet P-40, and 0.3 mg/mL bovine serum albumin. After 15 minutes at room temperature, DNA-protein complexes were separated on a 5% polyacrylamide gel (acrylamide:bisacrylamide ratio, 37.5:1) in TBE (25 mM Tris base, 25 mM boric acid, 0.5 mM EDTA) at 4°C. For competition experiments, extracts were preincubated with poly(dIdC) and unlabeled competitor oligonucleotides for 15 minutes before addition of the probe and further incubation. For antibody supershift assays, affinity-purified polyclonal anti-PU.1, anti-C/EBPα, anti-C/EBPβ, anti-C/EBPδ, or anti–c-Myb antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) (1 μL) were added to the mixture and incubated for an extra 30 minutes before loading into the gel.
Results
PU.1, C/EBP, and Myb binding sites are all critical for constitutive MBN promoter activity
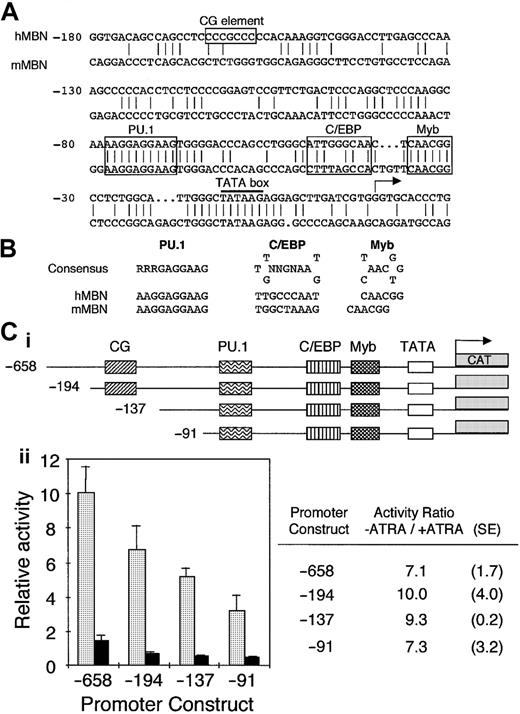
To assess critical MBN regulatory sequences, we have cloned human and mouse MBN promoter regions (GenBank accession numbers M96628 andAJ007030, respectively). Both promoter sequences had a TATA box and potential binding sites for Myb, C/EBP, and PU.1 (Figure1A). These sites exhibited consensus sequences in both the mouse and human promoters (Figure 1B). In addition, the human sequence harbored a CG element (Figure 1A). These elements were in agreement with putative binding sites indicated by Sturrock et al,32 except for C/EBP, which was not conserved between human and mouse and fit poorly with the C/EBP consensus sequence (data not shown). Mutation of this site did not modify the MBN promoter activity in transfected myeloid cells (data not shown). In contrast, we have identified another putative C/EBP binding site that differs from the C/EBP consensus sequence only by a G-to-C transition at the fifth position. A similar C/EBP site is present in the granulocyte-macrophage colony-stimulating factor receptor α promoter, harbors the same G-to-C transition, and binds C/EBPs.33 The Myb site is identical to a functional Myb binding site present in the neutrophil elastase promoter, which binds c-Myb with much lower affinity than, for example, the mim-1 Myb site.34
The human MBN promoter harbors conserved PU.1, C/EBP, and Myb binding sites: importance for MBN expression and down-regulation in ATRA-induced PLB-985 cells.
(A) Sequence homology between human and murine MBN promoters. Both strands were sequenced. The numbering is relative to the human transcription initiation site. The TATA box is underlined; the CG element as well as the PU.1, C/EBP, and Myb binding sites are boxed. (B) Comparison of the human and murine putative PU.1, C/EBP, and Myb binding sites with their consensus binding sites. The C/EBP site is in the reverse orientation. (C) ATRA affects MBN promoter activity. The pMBN 5′ deletion promoter mutants were fused to the CAT gene (i). PLB-985 cells were transfected with 20 μg each of the indicated MBN 5′ deletion mutants and cultured with or without ATRA (10−6 M). CAT activity was measured after normalization to the luciferase internal control (ii). The graph on the left shows untreated (−ATRA, ░) and ATRA-treated (+ATRA, ▪) cells. CAT activities presented for each pMBN construct were calculated relative to the pMBN-658. On the right are calculated −ATRA/+ATRA ratios as well as standard errors for each construct.
The human MBN promoter harbors conserved PU.1, C/EBP, and Myb binding sites: importance for MBN expression and down-regulation in ATRA-induced PLB-985 cells.
(A) Sequence homology between human and murine MBN promoters. Both strands were sequenced. The numbering is relative to the human transcription initiation site. The TATA box is underlined; the CG element as well as the PU.1, C/EBP, and Myb binding sites are boxed. (B) Comparison of the human and murine putative PU.1, C/EBP, and Myb binding sites with their consensus binding sites. The C/EBP site is in the reverse orientation. (C) ATRA affects MBN promoter activity. The pMBN 5′ deletion promoter mutants were fused to the CAT gene (i). PLB-985 cells were transfected with 20 μg each of the indicated MBN 5′ deletion mutants and cultured with or without ATRA (10−6 M). CAT activity was measured after normalization to the luciferase internal control (ii). The graph on the left shows untreated (−ATRA, ░) and ATRA-treated (+ATRA, ▪) cells. CAT activities presented for each pMBN construct were calculated relative to the pMBN-658. On the right are calculated −ATRA/+ATRA ratios as well as standard errors for each construct.
To investigate the mechanisms involved in MBN down-regulation in ATRA-treated growth-arrested leukemia cells, deletion constructs of the human MBN promoter (Figure 1Ci) cloned into the pBLCAT6 reporter plasmid were transfected into myeloblastic PLB-985 cells. Deletion from position −658 to −91 had minimal effect on MBN promoter constitutive activity, indicating that the remaining proximal promoter region retained the major elements (Figure 1Cii). The activity of the pMBN-658 reporter plasmid was reduced by 7-fold in ATRA-induced cells (Figure1C). The −91 deletion construct was still responsive to ATRA, indicating that this part of the promoter harbored responsive elements involved in the inhibition of MBN expression in myeloid leukemia cells growth-arrested by ATRA (Figure 1Cii).
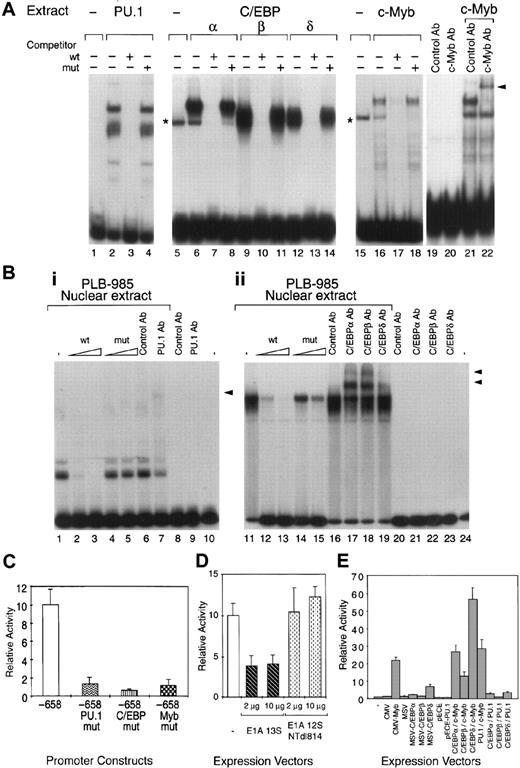
EMSAs were first conducted using total extracts from COS-7 cells transfected with vectors expressing recombinant PU.1, C/EBPα, C/EBPβ, C/EBPδ, and c-Myb. Total extracts from cells expressing the PU.1, C/EBPα, C/EBPβ, C/EBPδ, or c-Myb recombinants specifically bound to their respective PU.1, C/EBP, and c-Myb probes (Figure2A) as confirmed by competition experiments. Addition of anti–c-Myb antibodies specifically supershifted the c-Myb complex (Figure 2A). Although, similarly to other investigators in the field, we were unable to assess c-Myb DNA binding in myeloid cell nuclear extracts, likely because of high levels of protease activity, our results indicate that c-Myb binds specifically to its site in the MBN promoter. The PU.1 and C/EBP binding sites in the MBN promoter were able to bind their respective nuclear factors present in proliferating myeloid cells as confirmed by EMSA with nuclear extracts from untransfected PLB-985 cells. Indeed, PU.1- and C/EBP-specific complexes were detected with the PU.1- and C/EBP-specific probe (Figure 2B), respectively. Addition of the anti-PU.1 antibody specifically supershifted this complex (Figure2B). Similarly, C/EBP complexes were specifically supershifted with C/EBPα, C/EBPβ, and C/EBPδ antibodies (Figure 2B). Altogether, our results demonstrate that the PU.1, C/EBP, and Myb binding sites in the MBN promoter were able to bind their respective factors.
Functional PU.1, C/EBP, and c-Myb binding sites are present in the human MBN promoter.
(A) Recombinant human PU.1, C/EBPs, and c-Myb specifically bind functional sites in the MBN promoter. Five micrograms of whole-cell extract from COS-7 cells either untransfected (lanes 1, 5, 15) or transfected with PU.1 (lanes 2-4), C/EBPα (lanes 6-8), C/EBPβ (lanes 9-11), C/EBPδ (lanes 12-14), or c-Myb (lanes 16-18) were preincubated with 2 μg poly(dIdC) and a 400-fold molar excess of the wild-type (wt) or mutated (mut) competitors as indicated, before addition of the specific probe (PU.1, lanes 1-4; C/EBP, lanes 5-14; and c-Myb, lanes 15-18). Binding reaction mixtures were assayed by EMSA. The asterisks mark unspecific complexes. Supershift experiments were performed with 1 μL c-Myb antibody (Ab) and unrelated control Ab (lanes 19-22). Arrowhead indicates the specific supershifted complex. (B) Myeloid nuclear factors bind to the PU.1 and C/EBP sites. Nuclear extracts (1 μg) from exponentially growing PLB-985 cells were subjected to EMSA (lanes 1-7 and 11-19). DNA binding assays were performed with the PU.1 (lanes 1-10) or the C/EBP (lanes 11-24) probe in the presence of 5 μg and 0.2 μg poly(dIdC), respectively. Competition assays were carried out with a 40- and 200-fold molar excess of the wt or mut oligonucleotides. Arrows indicate the specific PU.1 or C/EBP complexes. Supershift experiments were performed with 1 μL anti-PU.1 (PU.1 Ab) (lanes 7-9), anti-C/EBPα, anti-C/EBPβ, anti-C/EBPδ (C/EBPα, C/EBPβ, C/EBPδ Abs) (lanes 17-19) and unrelated (control Ab) Abs (lanes 6, 8, 16, 20). Arrowheads indicate specific supershifted complexes. (C) PU.1, C/EBP, and Myb binding sites are all critical for MBN promoter activity in PLB-985 cells. Wt or indicated point mutation constructs (PU.1mut, C/EBPmut, and Mybmut) were transfected into cells. Transfection efficiencies were normalized by cotransfection of a CMV-luciferase vector. CAT activities relative to the wt (−658) are presented for PU.1mut, C/EBPmut, and Mybmut constructs. Error bars indicate SD from at least 3 different experiments. (D) CBP/p300 or related molecules are involved in MBN expression. PLB-985 cells were transfected with 20 μg pMBN-658 alone (−) or together with 2 and 10 μg of expression vector encoding E1A (13S) or the E1A 12S NTdl814 mutant. The average of 3 experiments are shown as relative CAT activity with SD as indicated. (E) Both c-Myb and C/EBPδ can cooperate to activate the MBN promoter. Five micrograms of pMBN-194 was transfected into COS-7 cells alone (−) or together with 1 μg of the CMV, CMV-Myb, MSV, MSV-C/EBPα, MSV-C/EBPβ, MSV-C/EBPδ, pECE, or pECE-PU.1 expression vectors or the indicated combinations of these vectors. CAT activities were assayed after 40 hours. The mean activations and standard errors observed in 3 determinations are shown.
Functional PU.1, C/EBP, and c-Myb binding sites are present in the human MBN promoter.
(A) Recombinant human PU.1, C/EBPs, and c-Myb specifically bind functional sites in the MBN promoter. Five micrograms of whole-cell extract from COS-7 cells either untransfected (lanes 1, 5, 15) or transfected with PU.1 (lanes 2-4), C/EBPα (lanes 6-8), C/EBPβ (lanes 9-11), C/EBPδ (lanes 12-14), or c-Myb (lanes 16-18) were preincubated with 2 μg poly(dIdC) and a 400-fold molar excess of the wild-type (wt) or mutated (mut) competitors as indicated, before addition of the specific probe (PU.1, lanes 1-4; C/EBP, lanes 5-14; and c-Myb, lanes 15-18). Binding reaction mixtures were assayed by EMSA. The asterisks mark unspecific complexes. Supershift experiments were performed with 1 μL c-Myb antibody (Ab) and unrelated control Ab (lanes 19-22). Arrowhead indicates the specific supershifted complex. (B) Myeloid nuclear factors bind to the PU.1 and C/EBP sites. Nuclear extracts (1 μg) from exponentially growing PLB-985 cells were subjected to EMSA (lanes 1-7 and 11-19). DNA binding assays were performed with the PU.1 (lanes 1-10) or the C/EBP (lanes 11-24) probe in the presence of 5 μg and 0.2 μg poly(dIdC), respectively. Competition assays were carried out with a 40- and 200-fold molar excess of the wt or mut oligonucleotides. Arrows indicate the specific PU.1 or C/EBP complexes. Supershift experiments were performed with 1 μL anti-PU.1 (PU.1 Ab) (lanes 7-9), anti-C/EBPα, anti-C/EBPβ, anti-C/EBPδ (C/EBPα, C/EBPβ, C/EBPδ Abs) (lanes 17-19) and unrelated (control Ab) Abs (lanes 6, 8, 16, 20). Arrowheads indicate specific supershifted complexes. (C) PU.1, C/EBP, and Myb binding sites are all critical for MBN promoter activity in PLB-985 cells. Wt or indicated point mutation constructs (PU.1mut, C/EBPmut, and Mybmut) were transfected into cells. Transfection efficiencies were normalized by cotransfection of a CMV-luciferase vector. CAT activities relative to the wt (−658) are presented for PU.1mut, C/EBPmut, and Mybmut constructs. Error bars indicate SD from at least 3 different experiments. (D) CBP/p300 or related molecules are involved in MBN expression. PLB-985 cells were transfected with 20 μg pMBN-658 alone (−) or together with 2 and 10 μg of expression vector encoding E1A (13S) or the E1A 12S NTdl814 mutant. The average of 3 experiments are shown as relative CAT activity with SD as indicated. (E) Both c-Myb and C/EBPδ can cooperate to activate the MBN promoter. Five micrograms of pMBN-194 was transfected into COS-7 cells alone (−) or together with 1 μg of the CMV, CMV-Myb, MSV, MSV-C/EBPα, MSV-C/EBPβ, MSV-C/EBPδ, pECE, or pECE-PU.1 expression vectors or the indicated combinations of these vectors. CAT activities were assayed after 40 hours. The mean activations and standard errors observed in 3 determinations are shown.
Deletion fragments of the MBN promoter were not adequate for studying the respective importance of the C/EBPs and Myb elements because removal of the PU.1 site resulted in a drastic diminution of the MBN promoter activity (data not shown). We therefore introduced point mutations into these sites. Mutation of PU.1, C/EBP, or Myb sites resulted in a drastic decrease of the promoter activity, indicating that all 3 sites were critical for the constitutive activity of the MBN promoter (Figure 2C) and suggesting that their action could be combinatorial. Indeed, it has been shown that c-Myb requires direct and specific interaction with the cyclic-AMP response element-binding protein (CREB)-binding protein (CBP) for its transactivation function.35,36 Similar observations were made with PU.137 and C/EBPβ,38 which also have physical and functional interaction with CBP/p300. We therefore investigated whether CBP/p300 could be required for full MBN expression and therefore be a potential candidate to support a combinatorial transcriptional effect of the 3 transcriptional factors on the MBN promoter. As previously described to demonstrate c-Myb35and C/EBP38 interactions with CBP/p300, we used the E1A 13S protein as a competitor that specifically interacts with CBP/p300 and blocks their activities. Transfection of an E1A 13S expression vector in PLB-985 cells reduced MBN promoter activity by 2.5-fold (Figure 2D). In contrast, no inhibition was detected using an E1A deletion mutant unable to interact with CBP/p300 (Figure 2D). Although E1A 13S can potentially interact with a variety of different target molecules, these results suggest that, in the context of the MBN promoter, CBP/p300 or related molecules are required for full MBN expression. To assess a potential cooperativity between c-Myb, PU.1, and C/EBP, the pMBN-194 reporter plasmid was transfected into COS-7 cells alone or together with the CMV, CMV-Myb, MSV, MSV-C/EBPα, MSV-C/EBPβ, MSV-C/EBPδ, pECE, or pECE-PU.1 expression vectors or combinations of these vectors. Compared with the empty vectors, c-Myb–, C/EBPδ-, and C/EBPα-containing vectors stimulated CAT expression by 20-, 6-, and 2-fold, respectively (Figure 2E), while C/EBPβ and PU.1 alone did not activate the MBN promoter in COS-7 cells. Cooperative activation was observed when c-Myb was cotransfected with C/EBPδ (Figure 2E). Cooperative DNA binding by c-Myb and C/EBP could not be demonstrated by EMSA analysis (data not shown). Altogether, these results suggest that (1) combinatorial action of Myb, PU.1, and C/EBP supports MBN promoter activity and (2) restricted positive cooperativity exists between C/EBPδ and Myb. Furthermore, together with Figure 2C, these results indicate that the binding of each factor to its specific site was necessary for constitutive activity of the MBN promoter but not sufficient individually for its full transactivation.
Inhibition of MBN expression in growth-arrested ATRA-treated leukemia cells is mediated by Myb down-regulation
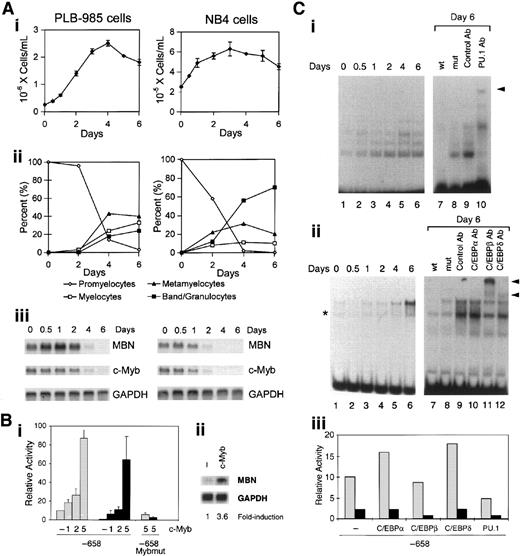
ATRA-induced growth inhibition in differentiating PLB-985 and NB4 leukemia cells (Figure 3Ai,ii, respectively) was accompanied by progressive down-regulation of the MBN messenger RNA (mRNA), which was barely detected at days 4 and 2, respectively (Figure 3Aiii). During these processes, the time at which MBN mRNA expression was inhibited closely paralleled that of c-Myb (Figure 3Aiii). We therefore overexpressed c-Myb in PLB-985 cells to test whether its down-regulation was required for ATRA to down-regulate the MBN promoter. In contrast to the expected ATRA-induced inhibition of MBN promoter activity in PLB-985 cells, overexpression of c-Myb (5 μg plasmid) resulted in an 8-fold induction of the MBN promoter activity (Figure 3Bi), which could no longer be decreased following treatment of the cells with ATRA (Figure 3Bi). To verify that potential ATRA-induced effects on MBN expression were not blunted by saturated levels of MBN as a result of c-Myb overexpression in PLB-985 cells, we used lower levels of engineered c-Myb expression (2 μg), which, following ATRA treatment, resulted in an MBN promoter activity similar to that found in untreated cells transfected with the empty plasmid (Figure 3Bi). This suggested that transfection of 2 μg c-Myb plasmid allowed for a quasi “normal” levels of c-Myb after ATRA treatment. Under these conditions, resistance to ATRA was still detectable. Indeed, while ATRA treatment resulted in a 10.6-fold decrease of the MBN promoter in control cells, a 2.9-fold and 2.4-fold decrease was observed with the same treatment in cells transfected with 1 and 2 μg c-Myb plasmid, respectively (Figure 3Bi). Resistance to ATRA was therefore likely due to exogenous c-Myb expression, which resulted in increased MBN promoter activity. In both untreated and ATRA-treated cells, induction of the MBN promoter activity by c-Myb overexpression and resistance to ATRA were dependent upon the integrity of the Myb binding site (Figure 3Bi). This strongly suggests that the binding of Myb to the MBN promoter is required for its regulation by ATRA. Notably, overexpression of c-Myb resulted in a 3.6-fold induction of the endogenous MBN mRNA (Figure 3Bii).
Transcriptional down-regulation of MBN by ATRA requires Myb down-regulation.
(A) ATRA-induced down-regulation of MBN mRNA in PLB-985 and NB4 cells correlates with growth arrest and down-regulation of c-Myb. Growth (i) and morphologic (ii) assessments of PLB-985 (left) and NB4 (right) at 2, 4, and 6 days of treatment with 10−6 M ATRA. (iii) Regulation of MBN and c-Myb mRNAs in PLB-985 cells untreated or treated for 0.5, 1, 2, 4, and 6 days with 10−6 M ATRA. For RNA blots, 3 μg total RNA was loaded in each lane. The lower part is a control hybridization of the same blots with GAPDH probe for assessment of RNA quantities. Panels Ai-iii are for PLB-985 and NB4 cells on left and right column, respectively. (B) Overexpression of c-Myb in PLB-985 cells abolished transcriptional down-regulation of MBN by ATRA. (i) Twenty micrograms of the pMBN-658 (−658) or pMBN-658Mybmut (−658 Mybmut) constructs were individually transfected (−) or cotransfected with 1, 2, or 5 μg CMV-Myb (c-Myb) into PLB-985 cells and cultured with (▪) or without (░) ATRA (10−6 M) for 40 hours. Transfection efficiencies were normalized by cotransfection of a CMV-luciferase vector. CAT activities were calculated relative to the wt (−658). The means and standard errors of 3 determinations are shown. (i) Autoradiogram of MBN mRNA expression in PLB-985 cells transfected with 5 μg of either the empty (−) or CMV-Myb (c-Myb) vector. The lower part is an autoradiogram of GAPDH mRNA for assessment of RNA quantities. Fold increase is indicated. (C) Overexpression of C/EBP and PU.1 in PLB-985 cells did not affect transcriptional down-regulation of MBN by ATRA. (i,ii) Induced PLB-985 growth arrest correlates with increased PU.1- and C/EBP-binding activity. Nuclear extracts from PLB-985 cells untreated or treated with 10−6M ATRA for the indicated days were assayed for PU.1 binding and C/EBP binding by EMSA performed with the PU.1 and C/EBP probes in the presence of 5 μg and 0.2 μg poly(dIdC), respectively (left panels). Competition experiments were carried out with a 400-fold molar excess of the indicated wt or mut competitor, and supershift experiments were performed with 1 μL PU.1 Ab; C/EBPα, C/EBPβ, C/EBPδ Abs; and unrelated control Abs (right). Arrows indicate the specific PU.1 or C/EBP complexes. Arrowheads indicate specific supershifted complexes. (iii) Twenty micrograms of the pMBN-658 (−658) was transfected (−) or cotransfected with 5 μg of the expression vectors for C/EBPα (MSV-C/EBPα), C/EBPβ (MSV-C/EBPβ), C/EBPδ (MSV-C/EBPδ), and PU.1 (pECE-PU.1) into PLB-985 cells and cultured with (▪) or without (░) ATRA (10−6 M) for 40 hours. Transfection efficiencies were normalized by cotransfection of a CMV-luciferase vector. CAT activities calculated relative to the wt (−658) as a reference are presented. This is representative of 3 independently performed experiments.
Transcriptional down-regulation of MBN by ATRA requires Myb down-regulation.
(A) ATRA-induced down-regulation of MBN mRNA in PLB-985 and NB4 cells correlates with growth arrest and down-regulation of c-Myb. Growth (i) and morphologic (ii) assessments of PLB-985 (left) and NB4 (right) at 2, 4, and 6 days of treatment with 10−6 M ATRA. (iii) Regulation of MBN and c-Myb mRNAs in PLB-985 cells untreated or treated for 0.5, 1, 2, 4, and 6 days with 10−6 M ATRA. For RNA blots, 3 μg total RNA was loaded in each lane. The lower part is a control hybridization of the same blots with GAPDH probe for assessment of RNA quantities. Panels Ai-iii are for PLB-985 and NB4 cells on left and right column, respectively. (B) Overexpression of c-Myb in PLB-985 cells abolished transcriptional down-regulation of MBN by ATRA. (i) Twenty micrograms of the pMBN-658 (−658) or pMBN-658Mybmut (−658 Mybmut) constructs were individually transfected (−) or cotransfected with 1, 2, or 5 μg CMV-Myb (c-Myb) into PLB-985 cells and cultured with (▪) or without (░) ATRA (10−6 M) for 40 hours. Transfection efficiencies were normalized by cotransfection of a CMV-luciferase vector. CAT activities were calculated relative to the wt (−658). The means and standard errors of 3 determinations are shown. (i) Autoradiogram of MBN mRNA expression in PLB-985 cells transfected with 5 μg of either the empty (−) or CMV-Myb (c-Myb) vector. The lower part is an autoradiogram of GAPDH mRNA for assessment of RNA quantities. Fold increase is indicated. (C) Overexpression of C/EBP and PU.1 in PLB-985 cells did not affect transcriptional down-regulation of MBN by ATRA. (i,ii) Induced PLB-985 growth arrest correlates with increased PU.1- and C/EBP-binding activity. Nuclear extracts from PLB-985 cells untreated or treated with 10−6M ATRA for the indicated days were assayed for PU.1 binding and C/EBP binding by EMSA performed with the PU.1 and C/EBP probes in the presence of 5 μg and 0.2 μg poly(dIdC), respectively (left panels). Competition experiments were carried out with a 400-fold molar excess of the indicated wt or mut competitor, and supershift experiments were performed with 1 μL PU.1 Ab; C/EBPα, C/EBPβ, C/EBPδ Abs; and unrelated control Abs (right). Arrows indicate the specific PU.1 or C/EBP complexes. Arrowheads indicate specific supershifted complexes. (iii) Twenty micrograms of the pMBN-658 (−658) was transfected (−) or cotransfected with 5 μg of the expression vectors for C/EBPα (MSV-C/EBPα), C/EBPβ (MSV-C/EBPβ), C/EBPδ (MSV-C/EBPδ), and PU.1 (pECE-PU.1) into PLB-985 cells and cultured with (▪) or without (░) ATRA (10−6 M) for 40 hours. Transfection efficiencies were normalized by cotransfection of a CMV-luciferase vector. CAT activities calculated relative to the wt (−658) as a reference are presented. This is representative of 3 independently performed experiments.
We then analyzed whether changes in PU.1 and C/EBP binding to their sites could be responsible for ATRA-induced MBN down-regulation. For this, EMSAs were conducted out with PLB-985 cell nuclear extracts using the PU.1 or the C/EBP probe (Figure 3Ci,ii, respectively). ATRA-induced growth arrest and differentiation of PLB-985 cells were accompanied by a progressive increased binding to both the PU.1 (Figure 3Ci, left) and C/EBP (Figure 3Cii, left) probes. Competition assays attested the specificity of the resulting DNA-protein complexes (Figure 3Ci,ii, right). The PU.1-retarded complex was specifically supershifted by PU.1 antibodies (Figure 3Ci, right). Incubation with anti-C/EBPβ and anti-C/EBPδ antibodies resulted in supershift of the C/EBP complexes and to a lesser extent of C/EBPδ complexes, respectively; no supershift was observed with anti-C/EBPα antibodies (Figure 3Cii, right). Overexpression of PU.1 in PLB-985 cells resulted in a slight decrease (about 2-fold) of the MBN promoter activity (Figure 3Ciii). In contrast to our observations with c-Myb and PU.1, overexpression of C/EBPα, C/EBPβ, and C/EBPδ had no effect on MBN transcriptional expression in ATRA-treated PLB-985 cells (Figure 3Ciii). This indicated that, despite increased binding to their specific sites in ATRA-treated PLB-985 cells, C/EBPs had no inhibitory effect on the ATRA response. These results indicate that MBN is a Myb target gene and that its down-regulation in growth-arrested ATRA-treated leukemia cells is dependent upon Myb down-regulation. Altogether, our results establish that (1) down-regulation of c-Myb mRNA correlated with that of MBN; (2) ectopic expression of c-Myb induced MBN promoter activity and enabled it to resist down-regulation by ATRA; (3) these 2 effects required the integrity of the specific Myb binding site; (4) the inhibitory effect of ATRA on the MBN promoter is not dependent upon C/EBPs; and (5) PU.1 may act as a transrepressor of MBN promoter activity via transcriptional repression of the c-Myb promoter.
Discussion
MBN is a myeloid growth-related gene.6 11 Although PU.1, C/EBP, and Myb transcription factors were all critical for constitutive expression of MBN, its down-regulation in leukemia cells growth-arrested by ATRA was dependent upon Myb. This and the fact that overexpression of c-Myb in leukemia cells inhibited down-regulation of MBN by ATRA point to MBN as a novel target of Myb in the context of proliferation.
The mechanism by which myeloid-specific genes can be expressed in immature myeloid cells has been exemplified by the study of neutrophil elastase, which is regulated as a result of a combinatorial activation by at least 3 factors, c-Myb, PU.1, and C/EBP, none of which by itself is myeloid-specific.34 PU.1 and C/EBP are involved in myeloid-specific gene expression.23,39 The fact that introduction of mutations in the PU.1 and C/EBP sites of the MBN promoter resulted in a drastic decrease in its constitutive transcriptional activity is consistent with the observation that MBN mRNA was markedly reduced in vivo in fetal liver of PU.1 and C/EBPα knockout mice.40 Furthermore, Zhang et al41demonstrated that loss of both the interleukin-6 and G-CSF receptors contributed to the absolute block in granulocyte maturation observed in C/EBPα-deficient hematopoietic cells and suggested that additional C/EBPα target genes are also important for the block in granulocyte differentiation observed in vivo in C/EBPα-deficient mice. Our data show that c-Myb can positively regulate MBN expression in myeloid leukemia cells. This is consistent with the fact that c-Myb is involved in the control of proliferation of definitive hematopoietic stem cells or multipotential progenitors as well as proliferation of myeloid leukemia cells.15-17 Expression of c-Myb decreases as normal hematopoietic stem or progenitor cells go into growth arrest and differentiation.42 It is also known that MBN mRNA is barely expressed in differentiated myeloid cells.6Therefore, the fact that in leukemia cells inhibition of both MBN and c-Myb mRNA expression is obtained through treatment by ATRA draws a parallel with their down-regulation during normal hematopoiesis. This is consistent with the view that, in leukemia cells, ATRA may mimic a process occurring during normal hematopoiesis. To this extent, it should be noted that, although G-CSF can induce MBN expression,11 a potential ATRA-induced increase in G-CSF receptor expression43 in PLB-985 cells did not affect the capacity of ATRA to induce MBN down-regulation as well as growth arrest and differentiation. Unlike c-Myb, B-Myb is ubiquitously expressed; A-Myb is expressed in a subpopulation of normal activated B lymphocytes and is not detected significantly in other hematopoietic cells. Although they have different expression patterns, a common feature of the Myb proteins is their expression in proliferating cells.44 Because anti–B-Myb antibodies suitable for EMSA are not available, our data do not indicate whether B-Myb can bind to the MBN Myb site. However, this is likely because Myb proteins display a high degree of homology within their DNA-binding domains recognizing a PyAACG/TG consensus sequence. The c-Myb and B-Myb can activate transcription of the same reporter genes,45,46 suggesting that they can regulate similar set(s) of growth control genes.47 Furthermore, expression of B-Myb closely parallels that of c-Myb in myeloid cells, particularly in ATRA-induced HL-60 cells.48
Studies of few genes regulated by Myb have indicated that it frequently acts in cooperation with other transcription factors. Indeed, our finding that functional PU.1, C/EBP, and Myb binding sites exist in the MBN promoter is reminiscent of previous observations with the neutrophil elastase34 and myeloperoxidase promoters. Our data are in agreement with previous work32 that established the importance of PU.1 for MBN promoter activity. However, our results contrast with previous indications32 that C/EBP and c-Myb were unlikely to play a significant role in the expression of MBN. Furthermore, it is of interest that PU.1 appeared to be involved in the regulation of MBN by phorbol myristate acetate (PMA), which decreased PU.1 binding to its site.32Although there was no evidence that overexpression or change in phosphorylation status of PU.1 would affect PMA-induced MBN transcriptional inhibition, these results are consistent with our observation that the PU.1 site is critical to MBN constitutive activity. Our results indicate that Myb specifically regulates MBN transcriptional inhibition by ATRA. We also found that, in contrast to PMA,32 ATRA up-regulates PU.1 binding to its site though overexpression of PU.1 could not abolish the negative effect of ATRA on MBN promoter activity. Altogether, previous work conducted with PMA32 and our present data suggest that PMA and ATRA down-regulate MBN promoter activity by affecting specific bindings to 2 different sites that are each critical to their constitutive transcriptional expression. Notably, overexpression of PU.1 resulted in a slight decrease in the MBN promoter activity. In PLB-985 cells, we have observed an increased PU.1 binding to its site upon ATRA treatment. In these cells, overexpression of PU.1 appeared to transrepress MBN promoter activity. Therefore, a transrepressor role for PU.1 may contribute to the down-regulation of MBN expression via inhibition of c-Myb transcription during myeloid differentiation.49
Cooperation between myeloid transcription factors for maximal promoter transactivation has been demonstrated.34,50,51 Recruitment of CBP, which has a strong histone acetyltransferase activity, by c-Myb and NF-M potentiated their transcriptional activities by bridging these 2 proteins.35 Our data show that C/EBPδ and Myb cooperatively activated the MBN promoter, reinforcing the view that cooperation between these factors is relevant to the activation of early myeloid genes. We have shown that CBP/p300 is likely to be required for full MBN expression. In view of the fact that the Myb, C/EBP, and PU.1 sites were all critical for MBN promoter activity, there is a possibility that CBP/p300 may support a combinatorial transcriptional effect of the 3 factors. Another possibility would be that, after recruitment to the MBN promoter by C/EBP and c-Myb, CBP/p300 may render the chromatin more accessible to the transcriptional machinery, allowing MBN transactivation by C/EBP and c-Myb.
It is assumed that leukemic cells descend from a small pool of progenitors with high proliferative activity.52This suggests that genes that are involved in controlling limited factor-dependent growth in normal hematopoietic progenitors might be continuously expressed, providing factor-independent growth to preleukemic cells. Such genes might be inhibited de novo when leukemia cells are forced into growth arrest by inducers such as ATRA. Our data provide a way by which ATRA can access a myeloid transcriptional machinery in which Myb is instrumental. Myb is known for its pivotal role in controlling cell growth and may therefore provide a way for ATRA to control proliferation in myeloid leukemia cells. Few Myb target genes are known to be linked to proliferation, and little is known about Myb involvement in growth inhibition induced by ATRA in leukemia cells. The fact that MBN, which can confer factor-independent growth to hematopoietic cells,11 is a target of Myb suggests a mechanism by which growth arrest may be induced by ATRA in myeloid leukemia cells.
We thank Drs A. Friedman, B. Lüscher, R. Maki, and P. Whyte for generous gifts of plasmids.
Supported by INSERM and grants from the Association Pour la Recherche sur le Cancer, the Lady Tata Memorial Trust, and the Ligue Nationale Contre le Cancer.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Yvon E. Cayre, Unité INSERM U417, Hôpital Saint Antoine, 184 Rue du Faubourg Saint Antoine, 75012 Paris, France; e-mail: cayre@st-antoine.inserm.fr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal