The pp65495-503 cytotoxic T-lymphocyte (CTL) epitope from cytomegalovirus (CMV) is universally recognized among CMV+ individuals who express an allele of the human leukocyte antigen A (HLA-A*0201). The relative binding affinity of the epitope to HLA-A*0201 is moderate, and its increased activity might prove beneficial in its use as a CTL epitope vaccine. A new approach to enhance the activity of T-cell epitopes is the use of positional scanning synthetic combinatorial libraries (PS-SCLs). Using a nonamer PS-SCL, the pp65495-503 epitope was modified after screening a CMV-specific T-cell clone (TCC) (3-3F4) from which the native peptide sequence was derived. Two peptides with amino acid substitutions at P1, P3, P7, and P8 are between 103 and 104 more active than the native epitope. Although the native CTL epitope terminates as a free acid, both tetrasubstituted peptides only function as CTL epitopes when the carboxyl terminus is amidated. Selective substitution of the native sequence based on PS-SCL screening results identified 3 amidated monosubstituted and disubstituted peptides that are better recognized than the native epitope by TCCs from a cohort expressing HLA-A*0201. In vitro stimulation of peripheral blood mononuclear cells with each of the peptide epitope analogs stimulated memory CTLs, which recognized CMV-infected targets among a high percentage of CMV+ individuals. Binding studies of peptide analogs with HLA-Ig (immunoglobulin) dimers and 2 different TCCs correlated with in vitro lysis results. These data suggest that increasing the activity of CTL epitopes while maintaining broad recognition is possible, which holds promise for vaccine development in infectious disease and cancer.
Introduction
Investigators in the last decade have sought to enhance the binding affinity of T-cell epitopes by manipulating residues involved in either TCR and/or major histocompatability complex (MHC) binding.1 The complexity of the molecular interactions between the trimolecular complex of TCR, peptide, and MHC has thwarted most attempts at significantly enhancing the binding affinity and activity of peptide MHC ligands.2 An approach first described in 1992 makes use of a positional scanning synthetic peptide combinatorial library (PS-SCL) for the identification of high-affinity ligands.3-5 The PS-SCLs are composed of peptide mixtures of uniform length, systematically arranged and having defined and mixture positions.6 A typical PS-SCL comprises “n” positional sublibraries (n = length of peptide). For example a nonamer PS-SCL is composed of 9 independent positional libraries, and it can be represented as O1XXXXXXXX, XO2XXXXXXX, XXO3XXXXXX, XXXO4XXXXX, XXXXO5XXXX, XXXXXO6XXX, XXXXXXO7XX, XXXXXXXO8X, or XXXXXXXXO9. The O positions represent 20 individually defined amino acids, and the X positions are composed of mixtures containing 19 of all 20 amino acids (excluding cysteine). Each positional library is composed of 20 different mixtures, which are serially repeated for each of the 9 positions that comprise a nonamer PS-SCL, for a total of 180 mixtures (9 × 20 mixtures). The screening of this PS-SCL provides information about the most effective residues at each position of the cytotoxic T-lymphocyte (CTL) epitope recognized by a T-cell clone (TCC).
Cytomegalovirus (CMV) is an important human pathogen affecting immunosuppressed patients including bone marrow transplantation recipients, solid organ donors, AIDS patients, and gestational fetuses.7-9 Previous studies10-12 and this study have shown the viral tegument protein pp65 to be recognized as immunodominant by most CMV+ individuals. We and others defined an immunodominant CTL epitope from pp65 (pp65495-503) recognized by a majority of CMV+patients in the context of an allele of human leukocyte antigen A (HLA-A*0201). Strategies using strong TH stimulators and lipids provided evidence that the pp65495-503 epitope functioned as a vaccine in HLA-A*0201 transgenic mice.13,14 The rationale for using combinatorial libraries is to improve the activity of peptide ligands, such as pp65495-503, which only activate CTLs in the nanomolar range,13 in order to reduce the amount of peptide vaccine needed to obtain a clinical response in an immunization strategy.15,16 Nonetheless, to be of potential clinical value, a CTL epitope analog would need to still maintain its ability to be recognized, as is pp65495-503, by most or all HLA-A*0201 expressors.17
We have screened the CMV-specific TCC, 3-3F4, with a nonamer PS-SCL, and described a series of peptide analogs that are recognized better than the native peptide sequence. Substitution peptides defined from the PS-SCL screen are biologically active only when amidated at the carboxyl (C) terminus, which is consistent with the fact that an amidated PS-SCL showed activity versus a C-terminal free-acid library. The object of this report is to define the properties of these amidated peptides. This includes studies of the molecular basis for their enhanced activity as defined by measurements of binding to either MHC or TCR components of the trimolecular recognition complex.
Materials and methods
Virus and cell lines
AD169 strain CMV (J. Zaia, City of Hope Medical Center, Duarte, CA) was used. Virus stocks of 5-10 × 106 pfu/mL were prepared from the supernatant of infected MRCvhbm5 fibroblasts as previously described13 and used to infect dermal fibroblasts. Epstein-Barr virus (EBV)–transformed B cell lines (EBVLCLs) were generated following standard methodology13,18 and were grown in medium (LCLM) consisting of RPMI 1640 medium (Gibco-BRL Life Technologies, Rockville, MD) supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES (4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid) buffer solution (Irvine Scientific, Santa Ana, CA), penicillin, streptomycin, and L-glutamine as described for FBM. T2 cells, the TAP− cell line,19 were maintained as described.13
Peptide synthesis and characterization
Alanine-scan peptides (Table2) were either synthesized on the multiple peptide synthesizer unit accessory (C terminus) or the main column (CONH2 terminus) of the Pioneer Peptide Synthesizer (Applied Biosystems Inc, Foster City, CA) using either Fmoc-L-VAL PEG-PS or Fmoc PAL-PEG-PS resin with HATU/DIPEA as activators. The peptide 44/46 sublibrary (Table4) was synthesized either using the Fmoc-protected VAL-Rink Amide-MBHA resin (CONH2 terminus) or Fmoc-VAL Wang resin (C terminus) (Anaspec Inc, San Jose, CA) using standard Fmoc protocols on the ABI 432A (Applied Biosystems). All peptides were cleaved from the resin and purified as described.6 This was followed by Matrix Assisted Laser Desorption Ionization Mass Spectrometry (MALDI) on the Kompact Probe (Kratos Analytical, Manchester, England) using alpha-cyano matrix and on high-pressure liquid chromatography (HPLC, Shimadzu SCL 10AVP) using a C18 column (Vydac Separations, Hesperia, CA) of 4.6 × 250-mm dimension composed of 5 μ × 300 particles. Sample purity was assessed at 70% to 80% based on the area percent of the sample peak in the chromatogram.
Libraries
L-amino acid synthetic combinatorial nonapeptide libraries—either C-amide (H-A1-Y9-NH2) or C-carboxyl (H-A1-Y9-COOH)—arranged in a positional scanning format were prepared as described previously using the simultaneous multiple peptide synthesis method.3 Each OX8 mixture consists of 1.7 × 1010 (198) different nonamer peptides in approximate equimolar concentration, and the total X9 library consists of 3.1 × 1012(9 × 20 × 198) different peptides. Assuming an average molecular weight of 1080 and a concentration of 100 μg/mL (93 μM) for nonapeptides in a mixture, the average concentration of individual nonapeptides in a mixture is approximately 5.5 × fM (10−15).
In vitro stimulation protocol
To prepare peptide-loaded antigen-presenting cells (APCs), 16-24 million of the Ficoll-separated peripheral blood mononuclear cells (PBMNCs) were incubated with 100 μM CMV pp65495-503peptide in a volume of 100 μL T-cell culture medium (TCM) and γ-irradiated (2400 rads) using the Isomedix Model 19 Gammator (Nuclear Canada, Parsippany, NJ). TCM consists of RPMI-1640 supplemented with 20% heat-inactivated HAB serum, 10 IU/mL recombinant interleukin-2 (rIL-2) (Chiron, Emeryville, CA), 25 mM HEPES buffer solution, 50 U/mL penicillin/50 μg/mL streptomycin, 2 mM L-glutamine, and 0.5 mM sodium pyruvate (Gibco). An additional aliquot of 10-20 million cells was incubated with saturating concentration of purified mouse antihuman CD4, CD16, and CD56 monoclonal antibody (mAb) (PharMingen, San Diego, CA). M450 Dynabead goat antimouse immunoglobulin G (IgG) (Dynal AS, Oslo, Norway) was then added to the mAb-labeled PBMNCs, and they were depleted of CD4+, CD16+, and CD56+ cells using a magnet.20 The resulting population was more than 80% CD8+ as determined using the fluorescence-activated cell sorter FACSCalibur (Becton Dickinson Immunocytometry Systems, Palo Alto, CA). We mixed 0.2 million depleted PBMNCs (effectors) with 5 million APCs in TCM and plated the cells in a 24-well plate at 2 mL per well for 2 weeks. They were fed with 10 IU/mL rIL-2 on days 5 and 10 and fresh medium when necessary. On day 14, pooled cells were assessed in a chromium release assay (CRA).
CRAs
CRAs were performed as previously described.13 For CMV-infected targets, 0.5 × 106 fibroblasts were pretreated with 800 U/106 cells of recombinant interferon (rIFN)-γ (Preprotech, Rocky Hill, NJ).21 CMV was added at a multiplicity of infection of 4, and virus infection occurred for 2 hours at 37°C in a 5% carbon dioxide (CO2) incubator. Infected fibroblasts were used at an effector-to-target (E/T) ratio of 50:10. EBVLCLs used as targets were pulsed with 50 μM pp65495-503 peptide and used at an E/T ratio of 25:5. For each target cell, spontaneous release was determined in the presence of medium only, and maximum release was determined after treatment with 2% sodium dodecyl sulfate (SDS) (Baker Chemicals, Phillipsburg, NJ). Specific cytotoxicity is defined as: 100 × [(Re-Rs)/Max-Rs)], where Re is the experimental release, and Rs is the spontaneous release.
Measurement of HLA dimer binding to TCC using
2-color flow cytometry
Soluble HLA-A2–Ig complexes (HLA dimers) were peptide-loaded as described.22 For each analog to be tested, 5 μg HLA dimers in 10 μL phosphate-buffered saline (PBS)/0.02% NaN3 (Baker Chemical) were incubated for 3 days at 4°C with 2 μL of a 2 mg/mL peptide solution. Each peptide-loaded dimer complex was tested at least twice with 2 different TCCs, 3-3F4 and VB57, and the fluorescence indices indicated are an average of both experiments (Figure 6).
T2 HLA class I assembly assay
For each analog to be tested, 250 000 T2 cells19cultured in Iscove modified Dulbecco medium (IMDM) (Gibco) were resuspended in 100 μL and incubated overnight with 100 μM peptide together with 15 μg/mL human β2-microglobulin23 (Sigma Chemical Co, St Louis, MO). Peptide-loaded cells were washed once at 4°C with cold PBS/0.05% bovine serum albumin (Sigma)/0.02% NaN3 (PBSA) followed by incubation with 1 μg per sample of the murine mAb BB7.2 with specificity for cell-surface HLA-A*0201 (ATCC) for 30 minutes at 4°C. After washing twice with cold PBSA, fluorescein isothiocyanate (FITC)–conjugated rat antimouse IgG2a/2b (PharMingen) at 1:40 dilution was added as second antibody, and the cells were incubated for another 30 minutes at 4°C. The cells were washed twice in cold PBSA, and mean fluorescence intensity (MFI) of 104 gated cells was measured on the FACSCalibur.
TCRAV gene usage of TCC derived from CMV+volunteers after IVS
Polymerase chain reaction (PCR) methods established in our laboratory were used to define the TCRAV gene segment usage by individual TCCs.24,25 RNA was prepared from 10-100 million TCCs by the Trizol (Promega Company, Madison, WI) method, and complementary (c)DNA was prepared using AMV reverse transcriptase as described in the package insert.25 We separated 30 μL of each reaction on a 1.3% agarose gel. PCR products were sequenced using the dideoxy method with IRD800-labeled primers on a Licor automated sequencing apparatus (Licor Inc, Lincoln, NE) (details upon request).
Results
Universal usage of the pp65495-503 CTL epitope in CMV+
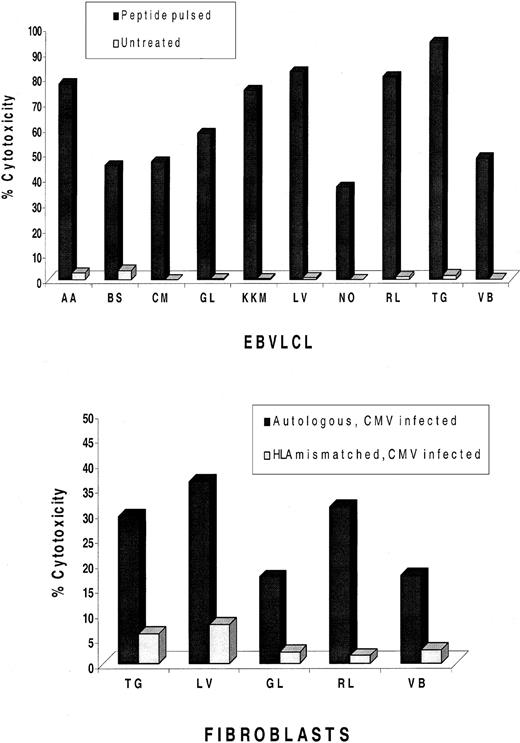
Whether the pp65495-503 CTL epitope is universally recognized in CMV+ and HLA-A*0201+ subjects was examined in 10 individuals with random haplotypes (Table1). A one-step IVS procedure that is a modification of a previous published method26 was carried out using the pp65495-503 CTL epitope as the immunogen.17 In 10 of 10 cases, it was possible to derive a pp65495-503–specific CTL response from HLA-A*0201 donors, and in 5 of 5 cases, it was possible to derive the same response against CMV-infected fibroblasts (Figure1). The results confirm and extend previous findings that CMV infection in HLA-A*0201+ healthy donors stimulates a pp65495-503–specific immune response that is independent of haplotype and is likely to be universal in its expression.13,20 27
Haplotypes of cytomegalovirus-positive donors and T-cell receptor structure of T-cell clones isolated from peripheral blood mononuclear cells evaluated by in vitro stimulation
| Donor . | HLA-A1 . | HLA-A2 . | HLA-B1 . | HLA-B2 . | TCC . | TCRA . | TCRB . |
|---|---|---|---|---|---|---|---|
| AA | A2 | A2 | B44 | B60 | |||
| BS | A2 | A26 | B51 | B62 | BS25 | AV2S1A2-AJ45-AC | BV7S2A1-BJ2.1-BC2 |
| CM | A2 | A3 | B7 | B14 | |||
| GL | A2 | A68 | B51 | B60 | 3-3F4 | AV18S2-AJ17.3-AC | BV13S1-BJ1.2-BC1 |
| JE | A2 | A68 | B7 | B17 | |||
| KKM | A2 | A1 | B27 | B44 | |||
| LV | A2 | A2 | B51 | B55 | LVB | ND | ND |
| NO | A2 | A3 | B47 | B51 | — | — | — |
| RB | A2 | A2 | B52 | B60 | — | — | — |
| TG | A2 | A68 | B62 | B62 | TGA2 | AV3S1-AJ9.12-AC | BV13S2A1-BJ2.1-BC2 |
| VB | A2 | A3 | B44 | B62 | VB57 | AV18S1-AJ20-AC | BV20S1A3-BJ1.1-BC1 |
| Donor . | HLA-A1 . | HLA-A2 . | HLA-B1 . | HLA-B2 . | TCC . | TCRA . | TCRB . |
|---|---|---|---|---|---|---|---|
| AA | A2 | A2 | B44 | B60 | |||
| BS | A2 | A26 | B51 | B62 | BS25 | AV2S1A2-AJ45-AC | BV7S2A1-BJ2.1-BC2 |
| CM | A2 | A3 | B7 | B14 | |||
| GL | A2 | A68 | B51 | B60 | 3-3F4 | AV18S2-AJ17.3-AC | BV13S1-BJ1.2-BC1 |
| JE | A2 | A68 | B7 | B17 | |||
| KKM | A2 | A1 | B27 | B44 | |||
| LV | A2 | A2 | B51 | B55 | LVB | ND | ND |
| NO | A2 | A3 | B47 | B51 | — | — | — |
| RB | A2 | A2 | B52 | B60 | — | — | — |
| TG | A2 | A68 | B62 | B62 | TGA2 | AV3S1-AJ9.12-AC | BV13S2A1-BJ2.1-BC2 |
| VB | A2 | A3 | B44 | B62 | VB57 | AV18S1-AJ20-AC | BV20S1A3-BJ1.1-BC1 |
Peripheral blood and skin biopsy specimens were obtained after individual consent and according to the provisions of the City of Hope Institutional Review Board. Individuals were first screened by known HLA type (per the City of Hope or National Marrow Donor program) and were checked for CMV seropositivity by indirect immunofluorescence (Hemagen Diagnostic, Columbia, MD) at the City of Hope Clinical Microbiology Laboratory. Both HLA alleles are shown for each individual as determined by serologic analysis using microcytotoxicity assay. TCCs were isolated from IVS cultures of selected donors, and theTCR gene structure was identified by PCR and DNA sequencing as described in “Materials and methods.”
HLA indicates human leukocyte antigen; TCC, T-cell clone; TCR, T-cell receptor; CMV, cytomegalovirus; ND not determined; PCR, polymerase chain reaction.
IVS of PBMNCs from HLA-A*0201 and CMV+volunteers.
Peripheral blood was magnetically depleted of CD4, CD16, and CD56 T lymphocytes and incubated for 12-14 days with autologous APCs pulsed with pp65495-503 peptide as described in “Materials and methods.” At the top of the figure, CRAs were conducted with IVS effectors, together with autologous and mismatched EBVLCLs at an E/T of 25:1 or 5:1 (data not shown), that were either peptide-pulsed or untreated as indicated in the legend. Percentages shown in the histogram reflect specific cytotoxicity after subtraction of values with mismatched EBVLCLs. At the bottom of the figure, an additional aliquot of IVS effectors was incubated with autologous or mismatched CMV-infected fibroblasts at an E/T of 50:1 or 10:1 (data not shown). Not all IVS cultures generated enough effector cells to conduct both EBVLCL and fibroblast experiments.
IVS of PBMNCs from HLA-A*0201 and CMV+volunteers.
Peripheral blood was magnetically depleted of CD4, CD16, and CD56 T lymphocytes and incubated for 12-14 days with autologous APCs pulsed with pp65495-503 peptide as described in “Materials and methods.” At the top of the figure, CRAs were conducted with IVS effectors, together with autologous and mismatched EBVLCLs at an E/T of 25:1 or 5:1 (data not shown), that were either peptide-pulsed or untreated as indicated in the legend. Percentages shown in the histogram reflect specific cytotoxicity after subtraction of values with mismatched EBVLCLs. At the bottom of the figure, an additional aliquot of IVS effectors was incubated with autologous or mismatched CMV-infected fibroblasts at an E/T of 50:1 or 10:1 (data not shown). Not all IVS cultures generated enough effector cells to conduct both EBVLCL and fibroblast experiments.
Alanine scanning mutagenesis identification of TCR and MHC contact residues
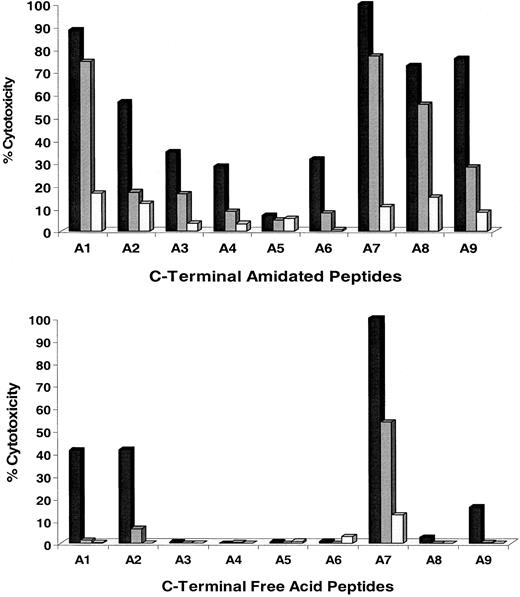
Experiments were conducted to determine which residues of the CMV-pp65495-503 T-cell epitope are able to tolerate amino acid side-chain modification and still maintain activity.28 Alanine substitution of the native residues in each position of CMV-pp65495-503(asparagine-leucine-valine-proline-methionine-alanine-threonine-valine; N-L-V-P-M-V-A-T-V) was carried out.29 Two different forms of each alanine-substituted peptide were synthesized, ending in either an amidated or free-acid C terminus (Table 2). The amidated forms were evaluated because the corresponding C-terminal amide form of the PS-SCL was active (see below). Results of CRAs for both series of alanine-substituted peptide ligands are shown (Figure2). Three different concentrations of peptides were used to sensitize APCs; this was followed by a standard CRA using TCC 3-3F4. A7 represents the native sequence of pp65495-503 that is an alanine in the corresponding position (P7) of the nonamer peptide.
Alanine scanning library of pp65495-503cytotoxic T-lymphocyte epitope
| Peptide name . | Position . | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| P1 . | P2 . | P3 . | P4 . | P5 . | P6 . | P7 . | P8 . | P9 . | |
| A1 | A | L | V | P | M | V | A | T | V |
| A2 | N | A | V | P | M | V | A | T | V |
| A3 | N | L | A | P | M | V | A | T | V |
| A4 | N | L | V | A | M | V | A | T | V |
| A5 | N | L | V | P | A | V | A | T | V |
| A6 | N | L | V | P | M | A | A | T | V |
| A7 (native) | N | L | V | P | M | V | A | T | V |
| A8 | N | L | V | P | M | V | A | A | V |
| A9 | N | L | V | P | M | V | A | T | A |
| Peptide name . | Position . | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| P1 . | P2 . | P3 . | P4 . | P5 . | P6 . | P7 . | P8 . | P9 . | |
| A1 | A | L | V | P | M | V | A | T | V |
| A2 | N | A | V | P | M | V | A | T | V |
| A3 | N | L | A | P | M | V | A | T | V |
| A4 | N | L | V | A | M | V | A | T | V |
| A5 | N | L | V | P | A | V | A | T | V |
| A6 | N | L | V | P | M | A | A | T | V |
| A7 (native) | N | L | V | P | M | V | A | T | V |
| A8 | N | L | V | P | M | V | A | A | V |
| A9 | N | L | V | P | M | V | A | T | A |
Eight peptide sequences were synthesized with either an amidated or free-acid terminus (either −NH2 or −COOH) as described in “Materials and methods.” The substitution of a native amino acid for an alanine is shown in bold for each position for the nonamer peptide pp65495-503. The native sequence corresponds to peptide A7.
Analysis of the recognition of alanine-scan substitutions of pp65495-503 peptide by CRA.
Alanine-substituted peptides (Table 2) were pulsed onto T2 cells (▪, 5 nM; , 0.5 nM; ■, 0.05 nM) as described in “Materials and methods.” C-terminal amidated (top) and free-acid (bottom) forms of all substitution peptides were evaluated. Experiments were repeated twice on different days with similar results.
, 0.5 nM; ■, 0.05 nM) as described in “Materials and methods.” C-terminal amidated (top) and free-acid (bottom) forms of all substitution peptides were evaluated. Experiments were repeated twice on different days with similar results.
Analysis of the recognition of alanine-scan substitutions of pp65495-503 peptide by CRA.
Alanine-substituted peptides (Table 2) were pulsed onto T2 cells (▪, 5 nM; , 0.5 nM; ■, 0.05 nM) as described in “Materials and methods.” C-terminal amidated (top) and free-acid (bottom) forms of all substitution peptides were evaluated. Experiments were repeated twice on different days with similar results.
, 0.5 nM; ■, 0.05 nM) as described in “Materials and methods.” C-terminal amidated (top) and free-acid (bottom) forms of all substitution peptides were evaluated. Experiments were repeated twice on different days with similar results.
Analysis of individual substitutions
The amidated forms of the peptide analogs have a greater tolerance for alanine substitutions than the free-acid forms, which is apparent by the difference in recognition of peptides A2-A6 and A8-A9, depending on their terminus structure. Substitution of alanine for asparagine at P1 is well tolerated by both the C-terminal amidated and free-acid forms of the epitope. In every other position, except P8 and P9 for the amidated form, the most precipitous decline in recognition occurs for peptides A3-A6 of the amidated forms and A3-A6 and A8 of the free-acid analogs. Interestingly, the anchor at P9, substituted by an alanine, does not show a substantial effect on recognition of the amidated form, whereas the equivalent free-acid form is severely impacted. These results suggest that the addition of the amide at the C terminus changes the conformation or binding characteristics of the CTL epitope to either MHC or TCR.30 31
Scanning of TCC 3-3F4 using PS-SCL
Using PS-SCL, the goal was to identify from the library individual nonapeptides that have greater activity than the native peptide. Two different nonapeptide PS-SCLs that differed in their C terminus were tested; one was amidated, and the other had a carboxylated C terminus. The amidated nonapeptide PS-SCL gave significantly better lysis results than the free-acid library, and only those results are presented. The 180 mixtures making up the PS-SCL were screened at 50 μg/mL in duplicate wells on different days with similar results (Figure 3). The responses varied between 0% and 90% specific cytotoxicity with a background of 5.0% without peptide. The sensitivity of recognition of the pp65495-503epitope by the 3-3F4 CTL is several logs higher than the concentration of individual peptide sequences in each of the test pools (5.5 fM). These results suggest that one or more peptides of high potency are responsible for the activity of the mixture, a phenomenon which may be associated with degeneracy of recognition by the TCR (“Discussion”).32 33
CRA results of PS-SCL screen of TCC 3-3F4.
Separate aliquots of T2 cells were pulsed with one of 180 mixtures (50 μg/mL) that make up the PS-SCLs and incubated with 3-3F4 cells as described in “Materials and methods.” All experimental points were obtained at the same time, and results shown represent the average of 2 experiments done on different days. The native amino acid at each position of the nonamer pp65495-503 peptide is indicated by a cross-hatched bar for each positional sublibrary graph.
CRA results of PS-SCL screen of TCC 3-3F4.
Separate aliquots of T2 cells were pulsed with one of 180 mixtures (50 μg/mL) that make up the PS-SCLs and incubated with 3-3F4 cells as described in “Materials and methods.” All experimental points were obtained at the same time, and results shown represent the average of 2 experiments done on different days. The native amino acid at each position of the nonamer pp65495-503 peptide is indicated by a cross-hatched bar for each positional sublibrary graph.
Scanning of TCC 3-3F4 using PS-SCL: results for P2 and P9 anchor positions
Results for the nonapeptide PS-SCL show that one or more mixtures at each position have substantial specific lysis. Mixtures having the defined amino acid that correspond to the native peptide showed specific lysis higher than 20% (Figure 3, native amino acid represented by hatched bar). Notably, the results at anchors P2 and P9 show that mixtures defined with amino acid residues that correspond to known motifs are among the highest responders at those positions (eg, leucine at P2 and valine at P9 for HLA-A*0201).34Interestingly, other highly ranked mixtures for P9 were defined as neutrally charged or containing aliphatic side chains such as isoleucine, leucine (an alternative residue for the P9 anchor), or alanine (Figure 3).35 Whereas mixtures defined with polar amino acids were ranked lower, those residues were not preferred as anchors for HLA-A*0201 binding peptides.36 We found a similar ranking of preferred residues for the P2 anchor position based on published motifs.35
Scanning of TCC 3-3F4 using PS-SCL: results for P1 and P3-P5
For P1, the majority of mixtures show similar activities ranging from 15% to 30% specific lysis (Figure 3). The fact that all mixtures show similar activity suggests that a number of amino acids are acceptable at this position. For P3, mixtures defined with neutrally charged amino acids, leucine, methionine, and isoleucine showed higher activity than the valine mixture (corresponds to native sequence); the alanine mixture was only slightly less active (Figure 3). The substantial difference in recognition of the 3 highest ranked mixtures compared to all others suggests a preference of those residues at P3. The most active mixtures for P4 and P5, proline and methionine, respectively, correspond to the native sequence. There is little difference in activity for the 3 most active mixtures for P5, nor is the level of lysis as high as the surrounding positions. Interestingly, the methionine at P5 is negatively affected by any substitution, including alanine, among individually synthesized peptides (Figure 2 and C.L.R., R.K., and D.J.D., unpublished data, 2000). This methionine is likely oriented away from the MHC peptide-binding pocket, making it available for TCR contact according to our molecular modeling simulations and in agreement with previous modeling studies (data not shown).37
Scanning of TCC 3-3F4 using PS-SCL: results for P6-P8
For P6, the most active mixture is defined with valine, and the second- and third-ranked mixtures are defined with amino acids of similar structure. P6 likely requires a small neutrally charged amino acid for interaction with MHC and/or TCR that is independent of other substitutions. P7 and P8 provide the most dramatic difference between the highest ranked mixtures and those that follow (Figure 3). The recognition that the alanine and threonine mixtures at P8 are equivalent (Figure 3) is consistent with results from the alanine-substitution study (Figure 2). Yet the PS-SCL screen results show that the most active mixtures have defined either serine or proline at P8. The summation of all of the changes predicts the identification of active peptides that substantially differ in sequence from the native CTL epitope.
Selection of analog peptides and evaluation of activity using TCC 3-3F4
Peptides were synthesized based on the amino acids defined in the most active mixtures of the PS-SCL. P1-P3, P6, and P9 were substituted with the fixed amino acid corresponding to the top-ranked mixture, whereas the top 2 amino acids were selected for P4, P5, P7, and P8, when the mixtures were recognized equivalently (Figure3). These individual peptides were made as C-terminal amides, which is consistent with the structure of the peptides contained in the PS-SCL screening library. The activity of the 16 peptides is shown as the minimal peptide concentrations for each analog that result in lysis between 15% and 30% (Table 3). Two of the peptides were substantially more active than the native epitope sequence in CRAs. These peptides, 44 and 46, each have 4 amino acid changes from the native peptide. Also, they differed from each other only in P7, in which peptide 44 has a valine and peptide 46 has a threonine. Interestingly, peptides 45 and 47, which share many of the substitutions of peptides 44 and 46, were far less active than the native sequence. Both peptides 45 and 47 use proline in P8, which suggests the importance of serine at P8 for T-cell recognition by the 3-3F4 clone.
Sixteen nonamers predicted from the results of positional scanning synthetic combinatorial library screen
| Position . | . | P1 . | P2 . | P3 . | P4 . | P5 . | P6 . | P7 . | P8 . | P9 . | . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Amino acid . | . | Y . | L . | L . | P . | M . | V . | V . | S . | V . | . |
| . | . | . | . | . | W . | Y . | . | T . | P . | . | . |
| Peptide name . | Peptide sequence . | Concentration, nM . | |||||||||
| 44 | H- | Y | L | L | P | M | V | V | S | V | 0.00005 |
| 46 | H- | Y | L | L | P | M | V | T | S | V | 0.0005 |
| 54 | H- | Y | L | L | W | M | V | T | S | V | 0.5 |
| 56 | H- | Y | L | L | W | Y | V | V | S | V | 50 |
| 47 | H- | Y | L | L | P | M | V | T | P | V | 500 |
| 52 | H- | Y | L | L | W | M | V | V | S | V | 500 |
| 53 | H- | Y | L | L | W | M | V | V | P | V | 500 |
| 55 | H- | Y | L | L | W | M | V | T | P | V | 500 |
| 58 | H- | Y | L | L | W | Y | V | T | S | V | 500 |
| 45 | H- | Y | L | L | P | M | V | V | P | V | > 500 |
| 48 | H- | Y | L | L | P | Y | V | V | S | V | > 500 |
| 49 | H- | Y | L | L | P | Y | V | V | P | V | > 500 |
| 50 | H- | Y | L | L | P | Y | V | T | S | V | > 500 |
| 51 | H- | Y | L | L | P | Y | V | T | P | V | > 500 |
| 57 | H- | Y | L | L | W | Y | V | V | P | V | > 500 |
| 59 | H- | Y | L | L | W | Y | V | T | P | V | > 500 |
| pp65495-503 | H- | N | L | V | P | M | V | A | T | V | 0.5 |
| pp65495-503 | H- | N | L | V | P | M | V | A | T | V | 0.5 |
| Position . | . | P1 . | P2 . | P3 . | P4 . | P5 . | P6 . | P7 . | P8 . | P9 . | . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Amino acid . | . | Y . | L . | L . | P . | M . | V . | V . | S . | V . | . |
| . | . | . | . | . | W . | Y . | . | T . | P . | . | . |
| Peptide name . | Peptide sequence . | Concentration, nM . | |||||||||
| 44 | H- | Y | L | L | P | M | V | V | S | V | 0.00005 |
| 46 | H- | Y | L | L | P | M | V | T | S | V | 0.0005 |
| 54 | H- | Y | L | L | W | M | V | T | S | V | 0.5 |
| 56 | H- | Y | L | L | W | Y | V | V | S | V | 50 |
| 47 | H- | Y | L | L | P | M | V | T | P | V | 500 |
| 52 | H- | Y | L | L | W | M | V | V | S | V | 500 |
| 53 | H- | Y | L | L | W | M | V | V | P | V | 500 |
| 55 | H- | Y | L | L | W | M | V | T | P | V | 500 |
| 58 | H- | Y | L | L | W | Y | V | T | S | V | 500 |
| 45 | H- | Y | L | L | P | M | V | V | P | V | > 500 |
| 48 | H- | Y | L | L | P | Y | V | V | S | V | > 500 |
| 49 | H- | Y | L | L | P | Y | V | V | P | V | > 500 |
| 50 | H- | Y | L | L | P | Y | V | T | S | V | > 500 |
| 51 | H- | Y | L | L | P | Y | V | T | P | V | > 500 |
| 57 | H- | Y | L | L | W | Y | V | V | P | V | > 500 |
| 59 | H- | Y | L | L | W | Y | V | T | P | V | > 500 |
| pp65495-503 | H- | N | L | V | P | M | V | A | T | V | 0.5 |
| pp65495-503 | H- | N | L | V | P | M | V | A | T | V | 0.5 |
Nonamer peptides with the substitutions as shown at each position were synthesized as described in “Materials and methods.” Substitutions predicted by the library screen of TCC 3-3F4 are shown on top of the table, immediately below each residue P of the CTL epitope, and the native sequence of the pp65495-503 CTL epitope is shown at the bottom of the table. Results of CRA are shown using peptide-sensitized HLA-A
0201–restricted EBVLCLs as target cells and TCC 3-3F4 effectors at an E/T ratio of 5. The terminus for each peptide and for pp65495-503 is −NH2, and the terminus for pp65495-503 is −OH. The concentration of peptide, in which specific killing was between 15% and 30%, represents the average results from at least 3 independent experiments carried out on different days using different batches of TCC-3F4. Peptides were tested using 10-fold dilutions ranging from 500-0.00005 nM.
CTL indicates cytotoxic T-lymphocyte; CRA, chromium release assay; EBVLCLs, Epstein-Barr virus–transformed B cell lines; E/T, effector-to-target; for other abbreviations, see Table 1.
Amide-substituted C terminus stabilizes multiple substitutions in pp65495-503 nonamer peptides
Peptides 44 and 46 were synthesized as free acids, and a CRA was performed comparing the sensitivity of recognition of the amide versus free-acid form of both sequences (data not shown). The free-carboxyl moiety causes the complete elimination of recognition of both peptides by TCC 3-3F4 and all other TCCs evaluated (data not shown) in comparison to the amidated forms (Table 3). This result suggests a role in the binding of the peptide to TCR and/or MHC by the C-terminal amide. The native pp65495-503 sequence was synthesized with an amidated C terminus and evaluated in a CRA. The amidated peptide was recognized almost equivalently to the free-acid form (Table 3). The alanine substitution study suggests that greater flexibility of substitution is tolerated when the C terminus is amidated (Table 3, compare amidated to free-acid results). These results are consistent with the bias of the amidated library in predicting active amidated sequences, but the magnitude of activity difference in sequences without the amidated C terminus is striking.
Recognition of peptides 44 and 46 by different individuals
We attempted to verify whether the increased sensitivity of recognition is a general property of these new amidated analogs. Therefore a CRA was performed using 4 additional TCCs, each derived from a different CMV+ volunteer (Tables 1 and 5). The tetrasubstituted peptides (Table 3, peptides 44 and 46) were not recognized by any of the TCCs in the cohort. The TCR usage of 4 of 5 TCCs from the cohort was determined, and they were all found to use different TCRBV and TCRAV gene segments (Table1). Therefore, different TCCs expressing unique TCRs may not be able to recognize peptides 44 and 46. The search for recognition was expanded as the peripheral blood of 4 individuals was stimulated under IVS conditions using peptides 44 and 46, including GL, the individual from whom the original TCC 3-3F4 was derived (Table 1). Only one of 4 individuals had cross-reactive T cells that were amplified by peptides 44 and 46, and the single responder was GL, from whom the screening TCC 3-3F4 was derived (data not shown). In summary, the data suggest that the native sequence is recognized by a diverse group of TCRs, and its activity is a compromise between broadness of recognition and protective immunity against CMV. In contrast, peptides 44 and 46 are both highly active and recognized by a restricted number of TCRs, with the apparent loss of universal recognition.
Broadening recognition by substituting native residues from pp65495-503 into peptides 44 and 46
An alternative approach was developed to restore broad recognition while preserving sensitivity by partially substituting native amino acids with library-directed changes. Two series of 21 peptides were constructed as both C-terminal amidated and free-acid forms, with single, double, or triple substitutions of amino acids of the native sequence based upon the sequence of the tetrasubstituted peptides 44 and 46. These peptides were assayed using a CRA with the screening TCC 3-3F4 and a representative TCC (VB57) expressing a different TCR (Tables 1 and 4). TCC 3-3F4 recognized 15 of 21 amidated peptides at a lower concentration than the native epitope (Table 3), although only 5 were 100-fold or more active (Table 4). As expected, the results for the free-acid forms of the same peptide sequence were different than for the amidated series (Table 4). For instance, peptide 193N (N = amide) was approximately 100-fold more active than the native epitope, and the equivalently substituted peptide 193H (H = free acid) lacking the amide was approximately 100-fold less reactive. Seven of the amidated peptides were recognized with a specific cytotoxicity greater than 15% at a concentration of 0.05 nM, which is a greater sensitivity than the native epitope has for the same TCC. Only one of the free-acid peptides (111H) was active below 0.05 nM (Table 4). These results further confirm that amidation of the C terminus of the nonamer provides greater flexibility of recognition than the free-acid form.
Directed-substitution sequences developed from peptides 44 and 46
| . | P1 . | P2 . | P3 . | P4 . | P5 . | P6 . | P7 . | P8 . | P9 . | . | . | . | . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Substitutions4-150 . | . | . | . | . | . | . | V . | . | . | . | . | . | . |
| . | Y . | . | L . | . | . | . | T . | S . | . | T-cell clones . | |||
| . | N . | L . | V . | P . | M . | V . | A . | T . | V . | 3-3F4 . | VB57 . | ||
| Peptide name . | . | . | . | . | . | . | . | . | . | −NH2 . | −COOH . | −NH2 . | −COOH . |
| 193 | Y | L | V | P | M | V | A | T | V | 0.0054-151 | > 5 | 0.05 | > 5 |
| 194 | N | L | L | P | M | V | A | T | V | 0.05 | > 5 | 0.5 | > 5 |
| 110 | N | L | V | P | M | V | V | T | V | 0.05 | 0.5 | > 5 | ND |
| 111 | N | L | V | P | M | V | T | T | V | 0.05 | 0.05 | > 5 | 5 |
| 112 | N | L | V | P | M | V | A | S | V | 0.5 | 5 | 0.5 | 0.5 |
| 116 | N | L | L | P | M | V | V | T | V | 0.5 | > 5 | > 5 | ND |
| 117 | N | L | L | P | M | V | T | T | V | 0.05 | 0.5 | > 5 | 5 |
| 118 | N | L | L | P | M | V | A | S | V | 0.05 | > 5 | 0.05 | > 5 |
| 119 | N | L | V | P | M | V | V | S | V | 0.005 | > 5 | > 5 | > 5 |
| 120 | N | L | V | P | M | V | T | S | V | 0.05 | 5 | > 5 | > 5 |
| 195 | Y | L | L | P | M | V | A | T | V | 0.005 | > 5 | 0.5 | > 5 |
| 113 | Y | L | V | P | M | V | V | T | V | 0.5 | > 5 | > 5 | ND |
| 114 | Y | L | V | P | M | V | T | T | V | 0.05 | 0.5 | 5 | 5 |
| 115 | Y | L | V | P | M | V | A | S | V | 0.005 | > 5 | 0.05 | 5 |
| 126 | N | L | L | P | M | V | V | S | V | 0.005 | 5 | 0.5 | > 5 |
| 127 | N | L | L | P | M | V | T | S | V | 0.05 | 5 | > 5 | > 5 |
| 124 | Y | L | V | P | M | V | V | S | V | 0.5 | > 5 | 5 | > 5 |
| 125 | Y | L | V | P | M | V | T | S | V | 0.05 | > 5 | > 5 | > 5 |
| 121 | Y | L | L | P | M | V | V | T | V | 5 | > 5 | > 5 | ND |
| 122 | Y | L | L | P | M | V | T | T | V | 5 | > 5 | > 5 | > 5 |
| 123 | Y | L | L | P | M | V | A | S | V | 0.05 | > 5 | 0.5 | > 5 |
| . | P1 . | P2 . | P3 . | P4 . | P5 . | P6 . | P7 . | P8 . | P9 . | . | . | . | . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Substitutions4-150 . | . | . | . | . | . | . | V . | . | . | . | . | . | . |
| . | Y . | . | L . | . | . | . | T . | S . | . | T-cell clones . | |||
| . | N . | L . | V . | P . | M . | V . | A . | T . | V . | 3-3F4 . | VB57 . | ||
| Peptide name . | . | . | . | . | . | . | . | . | . | −NH2 . | −COOH . | −NH2 . | −COOH . |
| 193 | Y | L | V | P | M | V | A | T | V | 0.0054-151 | > 5 | 0.05 | > 5 |
| 194 | N | L | L | P | M | V | A | T | V | 0.05 | > 5 | 0.5 | > 5 |
| 110 | N | L | V | P | M | V | V | T | V | 0.05 | 0.5 | > 5 | ND |
| 111 | N | L | V | P | M | V | T | T | V | 0.05 | 0.05 | > 5 | 5 |
| 112 | N | L | V | P | M | V | A | S | V | 0.5 | 5 | 0.5 | 0.5 |
| 116 | N | L | L | P | M | V | V | T | V | 0.5 | > 5 | > 5 | ND |
| 117 | N | L | L | P | M | V | T | T | V | 0.05 | 0.5 | > 5 | 5 |
| 118 | N | L | L | P | M | V | A | S | V | 0.05 | > 5 | 0.05 | > 5 |
| 119 | N | L | V | P | M | V | V | S | V | 0.005 | > 5 | > 5 | > 5 |
| 120 | N | L | V | P | M | V | T | S | V | 0.05 | 5 | > 5 | > 5 |
| 195 | Y | L | L | P | M | V | A | T | V | 0.005 | > 5 | 0.5 | > 5 |
| 113 | Y | L | V | P | M | V | V | T | V | 0.5 | > 5 | > 5 | ND |
| 114 | Y | L | V | P | M | V | T | T | V | 0.05 | 0.5 | 5 | 5 |
| 115 | Y | L | V | P | M | V | A | S | V | 0.005 | > 5 | 0.05 | 5 |
| 126 | N | L | L | P | M | V | V | S | V | 0.005 | 5 | 0.5 | > 5 |
| 127 | N | L | L | P | M | V | T | S | V | 0.05 | 5 | > 5 | > 5 |
| 124 | Y | L | V | P | M | V | V | S | V | 0.5 | > 5 | 5 | > 5 |
| 125 | Y | L | V | P | M | V | T | S | V | 0.05 | > 5 | > 5 | > 5 |
| 121 | Y | L | L | P | M | V | V | T | V | 5 | > 5 | > 5 | ND |
| 122 | Y | L | L | P | M | V | T | T | V | 5 | > 5 | > 5 | > 5 |
| 123 | Y | L | L | P | M | V | A | S | V | 0.05 | > 5 | 0.5 | > 5 |
ND indicates not determined; TCC, T-cell clone.
The substitutions at each position are shown above the peptide sequences; this is given for positions that were varied in the library screen (Table 3) (ie, P1, P3, P7, and P8). Each peptide was made either with an amidated (NH2) or free-acid (COOH) terminus.
The concentration of peptide is that in which specific killing was between 15% and 30%. Peptides were tested using 10-fold dilutions between 500 and 0.00005 (3-3F4) or 0.005 (VB57) nM. The best peptides for each TCC and terminus are bolded.
Recognition profile of TCC 3-3F4 versus VB57
The amino acid substitutions that resulted in increased recognition by TCC 3-3F4 can be consolidated into a few categories. Most striking is the substitution of the tyrosine for the asparagine in P1, which is present in 3 of the 5 most active peptides. Even as a single substitution it provides a 100-fold increase in reactivity compared to the native epitope (Tables 3 and 4). The substitution of a serine in P8 is also present in 3 of the 5 highest ranking peptides recognized by the TCC 3-3F4. In summary, the 5 best peptides recognized by TCC 3-3F4 have between 1 and 3 substitutions. In contrast, TCC VB57, which expresses a different TCR (Table 1), evokes a different spectrum of responses (Table 4). There are 5 peptides that are recognized at a concentration of 0.005 nM using TCC 3-3F4, whereas there are no peptides recognized at 0.005 nM using TCC VB57. Nonetheless, TCC VB57 recognizes 8 peptides at 0.5 nM, and 3 peptides are still recognized at 0.05 nM, which is better than the recognition of the native peptide sequence (Table 3). Three peptide sequences, 115,118, and 193, are recognized with greater sensitivity by both TCCs and should be evaluated as candidate CTL epitope vaccines. These results demonstrate that with the assistance of a diverse PS-SCL, it is possible to engineer peptides with enhanced recognition to TCCs expressing different TCRs.
Cross-recognition of antigen analogs by TCCs specific for the native epitope sequence
Whether PS-SCL–directed substitution peptides can maintain an equivalent breadth of recognition as the native peptide sequence (ie, pp65495-503) will be an important criterion for their usefulness as vaccines. The best recognized of the antigen analogs (C-terminal amides 115, 118, and 193) by both TCC 3-3F4 and VB57 were evaluated for recognition by a cohort of 3 additional TCCs from 3 separate individuals (Table 5). Peptide 118 is better recognized than the native sequence in 3 of 5 TCCs, and peptides 115 and 193 are better in 2 of 5 TCCs. In the best cases, the sensitivity is at least 10 times greater than the native epitope, although there are cases when the recognition is equivalent between the analog and native sequences (Table 5). The TCC data confirm that it is possible to derive a sequence that is better recognized than the native CTL epitope by several TCCs with different TCRs.
Recognition of substitution peptides by a cohort of T-cell clones from human leukocyte antigen A5-1500201 and cytomegalovirus-seropositive individuals
TCCs were derived from individuals after in vitro stimulation using the native sequence of pp65495-503 as described in Figure 4. Limiting dilution cloning was conducted with a portion of the cells, and individual clones were evaluated using CRAs with the peptides shown above. The amount of peptide used to sensitize HLA-A
0201–expressing EBVLCLs is expressed as either 5.0 or 0.5 nM. Specific cytotoxicity is shown as either greater than 30%, ▪; greater than 15% and less than 30%, ░; or less than 15%, ■.
Three antigen analogs of pp65495-503 are recognized by multiple individuals
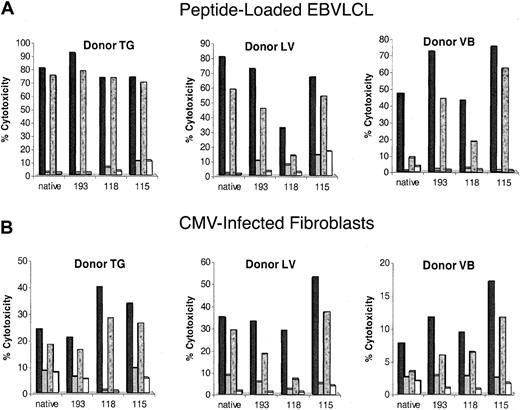
Whether a CMV-specific memory T-cell repertoire can be derived for each of the 3 peptides can be best addressed by conducting IVS experiments. Peripheral blood from 3 individuals was separately stimulated with amidated peptides 115, 118, and 193 and the native sequence. All 3 individuals vigorously responded to the IVS conducted with 3 of 3 analog peptides, as shown by the CRA using peptide-sensitized autologous and mismatched LCL (Figure4A). Effector cells from the cultures of 3 individuals specifically recognized CMV-infected fibroblasts after stimulation by all 3 analog peptides (Figure 4B). Peptide 115 stimulated greater CTL activity against CMV-infected targets than the native peptide for all 3 donors, and peptide 118 was more effective for donor TG. The results are consistent for both the high and low E/T ratio, especially in the case of CMV targets. The differences are not as profound for peptide-loaded targets, but their physiologic significance is not as great compared to the CMV targets. These results contrast with data showing that peptides 44 and 46 are only recognized by a single individual (data not shown). The success of these activity studies in a small cohort of individuals warrants further examination of the extent of recognition of peptides 115, 118, and 193 in a larger group of CMV+ individuals.
IVS of CMV+ volunteers with 3 analog peptides.
Aliquots of magnetically depleted lymphocytes were incubated with autologous APCs loaded with one of 3 amidated analog peptides or the native sequence under IVS conditions as described in the legend to Figure 1 and in “Materials and methods.” Either peptide-loaded autologous (black and stippled bars) or HLA-mismatched (gray and white bars) EBVLCLs (A) or autologous (black and stippled bars) or HLA-mismatched (gray and white bars) CMV-infected fibroblasts (B) were used as targets as described in the legend to Figure 1 and in “Materials and methods.” E/T ratios were 25 (black and gray bars) and 5 (stippled and white bars) for panel A and 50 (black and gray bars) and 10 (stippled and white bars) for panel B.
IVS of CMV+ volunteers with 3 analog peptides.
Aliquots of magnetically depleted lymphocytes were incubated with autologous APCs loaded with one of 3 amidated analog peptides or the native sequence under IVS conditions as described in the legend to Figure 1 and in “Materials and methods.” Either peptide-loaded autologous (black and stippled bars) or HLA-mismatched (gray and white bars) EBVLCLs (A) or autologous (black and stippled bars) or HLA-mismatched (gray and white bars) CMV-infected fibroblasts (B) were used as targets as described in the legend to Figure 1 and in “Materials and methods.” E/T ratios were 25 (black and gray bars) and 5 (stippled and white bars) for panel A and 50 (black and gray bars) and 10 (stippled and white bars) for panel B.
MHC binding affinity of antigen analogs: T2 assembly assay
Changes to MHC class I binding of CTL epitopes can be measured independently from changes in TCR binding affinity. The approach is to use the T2 assembly assay that measures the relative strength of peptide binding to HLA-A*0201.23 38 Five pp65495-503 analog peptides and the native sequence were evaluated as both C-terminal free acids and amides along with a known HLA-A*0201 binding control peptide. T2 cells were incubated with 100 μM of each test peptide overnight, and surface HLA-A*0201 was evaluated using mAb BB7.2 as described in “Materials and methods.” Results shown in Figure 5 for the amide peptides indicate only slight changes in MHC binding, which is consistent with retention of the native sequence at the major anchor positions, P2 and P9. Although peptides 44 and 46 have been shown to be strongly immunogenic when using TCC 3-3F4, their MHC affinity measured by this assay is not dramatically different than the native sequence. As expected, there is equivalent MHC binding of both forms of the native sequence that reflects the observed similarity in recognition as measured by lysis assays. The contrast between acid and amide forms in binding to HLA-A*0201 is not as pronounced for the 3 more universally recognized analogs, 115, 118, and 193, despite a large difference in ability to mediate lysis. In summary, heightened recognition of the amidated antigen analogs is likely the result of interactions with both MHC class I and the TCR components of the trimolecular complex.
T2-assembly assay with analog and control peptides.
C-terminal amidated (▪ and free-acid ( ) peptides, whose sequences are shown in Tables 3 and 4, and free-acid control peptides (native, pp65495-503; HBVc, HBV core antigen18-27 CTL epitope58) were incubated (100 μM) with T2 cells overnight. The cells were labeled with the mAb BB7.2, and fluorescence was measured by flow cytometry as described in “Materials and methods.” Shown are representative results from 3 separate measurements. The fluorescence ratio was calculated as follows: MFI of T2 cells with peptide/MFI of T2 cells without peptide. The T2 binding assay was repeated twice with similar results, and the ratios shown in the figure represent average values.
) peptides, whose sequences are shown in Tables 3 and 4, and free-acid control peptides (native, pp65495-503; HBVc, HBV core antigen18-27 CTL epitope58) were incubated (100 μM) with T2 cells overnight. The cells were labeled with the mAb BB7.2, and fluorescence was measured by flow cytometry as described in “Materials and methods.” Shown are representative results from 3 separate measurements. The fluorescence ratio was calculated as follows: MFI of T2 cells with peptide/MFI of T2 cells without peptide. The T2 binding assay was repeated twice with similar results, and the ratios shown in the figure represent average values.
T2-assembly assay with analog and control peptides.
C-terminal amidated (▪ and free-acid ( ) peptides, whose sequences are shown in Tables 3 and 4, and free-acid control peptides (native, pp65495-503; HBVc, HBV core antigen18-27 CTL epitope58) were incubated (100 μM) with T2 cells overnight. The cells were labeled with the mAb BB7.2, and fluorescence was measured by flow cytometry as described in “Materials and methods.” Shown are representative results from 3 separate measurements. The fluorescence ratio was calculated as follows: MFI of T2 cells with peptide/MFI of T2 cells without peptide. The T2 binding assay was repeated twice with similar results, and the ratios shown in the figure represent average values.
) peptides, whose sequences are shown in Tables 3 and 4, and free-acid control peptides (native, pp65495-503; HBVc, HBV core antigen18-27 CTL epitope58) were incubated (100 μM) with T2 cells overnight. The cells were labeled with the mAb BB7.2, and fluorescence was measured by flow cytometry as described in “Materials and methods.” Shown are representative results from 3 separate measurements. The fluorescence ratio was calculated as follows: MFI of T2 cells with peptide/MFI of T2 cells without peptide. The T2 binding assay was repeated twice with similar results, and the ratios shown in the figure represent average values.
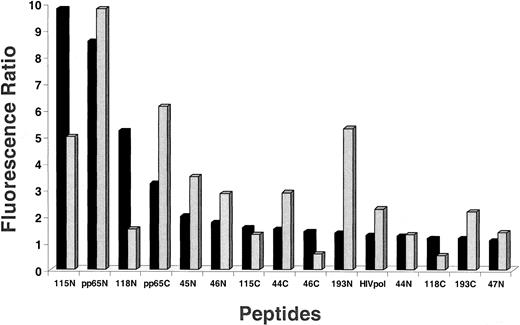
TCR binding affinity of antigen analogs using HLA dimers
An approach to distinguish how particular substitutions of TCR contact residues contribute to the activity of analog peptides of pp65495-503 is to use a soluble HLA reagent that only recognizes TCR after binding a specific peptide. The reagent used to evaluate peptide affinity to TCR is referred to as an HLA-Ig dimer.22,39 The dimer reagent is a chimera of the HLA class I α1 and α2 domains attached to the Fab portions of Ig molecules.40 41 HLA-A2–Ig dimer aliquots were individually loaded with a series of peptides and incubated with 2 different TCCs, 3-3F4 and VB57 (Figure6). Amidated peptides 115, pp65495-503, and 118 bind well to 3-3F4, although peptides 44, 46, and 193 are unexpectedly poor binders. VB57 has a similar binding profile, except that 118 is a poor binder, and 193 is among the better binders. Of great interest is the fact that the free-acid forms of binding peptides 115, 118, and 193 (VB57 only) fail to bind to either TCC, which is in contrast to their amidated counterparts. The free-acid peptides bind with similar low affinity to the TCC as a control C-terminal free-acid peptide derived from human immunodeficiency virus (HIV) pol protein. The binding data agree with the large difference in recognition between amidated and free-acid forms of the library peptides derived from the screening of PS-SCLs.
Flow-cytometric measurement of HLA-Ig dimer binding to TCCs mediated by antigen analogs and control peptides.
We incubated 10.0 μg HLA-Ig dimers with 660-fold molar excess peptide for 3 days at 4°C. Aliquots of 1.0 μg peptide-loaded dimers were mixed with 0.2 million of either TCC 3-3F4 (▪) or VB57 ( ) and incubated as described in “Materials and methods.” As controls, dimers were also loaded with the pp65495-503 native peptide, an irrelevant peptide (pp65 B*0702 epitope), or 2 μL PBS/0.02% NaN3 in all experiments (data not shown). Cells were washed in PBS/2% FBS/0.02% NaN3 (WB) and subsequently stained with 0.9 μL per sample of goat antimouse IgG1-PE (phycoerythrin) (Caltag, Burlingame, CA). After 2 additional washings in WB, cells were analyzed on the FACSCalibur. The fluorescence ratio between the mean fluorescence of the peptide-loaded dimers and the mean fluorescence of the PBS/0.02% NaN3–loaded control dimers was calculated. Peptide names ending in N are amidated, and those ending in C are free acids. Experiments were repeated twice with similar results. Measurements with both cell lines were normalized so they could be displayed on the same axis. HIVpol = HIVpol (18-27), which indicates aa position in HIV polymerase protein.
) and incubated as described in “Materials and methods.” As controls, dimers were also loaded with the pp65495-503 native peptide, an irrelevant peptide (pp65 B*0702 epitope), or 2 μL PBS/0.02% NaN3 in all experiments (data not shown). Cells were washed in PBS/2% FBS/0.02% NaN3 (WB) and subsequently stained with 0.9 μL per sample of goat antimouse IgG1-PE (phycoerythrin) (Caltag, Burlingame, CA). After 2 additional washings in WB, cells were analyzed on the FACSCalibur. The fluorescence ratio between the mean fluorescence of the peptide-loaded dimers and the mean fluorescence of the PBS/0.02% NaN3–loaded control dimers was calculated. Peptide names ending in N are amidated, and those ending in C are free acids. Experiments were repeated twice with similar results. Measurements with both cell lines were normalized so they could be displayed on the same axis. HIVpol = HIVpol (18-27), which indicates aa position in HIV polymerase protein.
Flow-cytometric measurement of HLA-Ig dimer binding to TCCs mediated by antigen analogs and control peptides.
We incubated 10.0 μg HLA-Ig dimers with 660-fold molar excess peptide for 3 days at 4°C. Aliquots of 1.0 μg peptide-loaded dimers were mixed with 0.2 million of either TCC 3-3F4 (▪) or VB57 ( ) and incubated as described in “Materials and methods.” As controls, dimers were also loaded with the pp65495-503 native peptide, an irrelevant peptide (pp65 B*0702 epitope), or 2 μL PBS/0.02% NaN3 in all experiments (data not shown). Cells were washed in PBS/2% FBS/0.02% NaN3 (WB) and subsequently stained with 0.9 μL per sample of goat antimouse IgG1-PE (phycoerythrin) (Caltag, Burlingame, CA). After 2 additional washings in WB, cells were analyzed on the FACSCalibur. The fluorescence ratio between the mean fluorescence of the peptide-loaded dimers and the mean fluorescence of the PBS/0.02% NaN3–loaded control dimers was calculated. Peptide names ending in N are amidated, and those ending in C are free acids. Experiments were repeated twice with similar results. Measurements with both cell lines were normalized so they could be displayed on the same axis. HIVpol = HIVpol (18-27), which indicates aa position in HIV polymerase protein.
) and incubated as described in “Materials and methods.” As controls, dimers were also loaded with the pp65495-503 native peptide, an irrelevant peptide (pp65 B*0702 epitope), or 2 μL PBS/0.02% NaN3 in all experiments (data not shown). Cells were washed in PBS/2% FBS/0.02% NaN3 (WB) and subsequently stained with 0.9 μL per sample of goat antimouse IgG1-PE (phycoerythrin) (Caltag, Burlingame, CA). After 2 additional washings in WB, cells were analyzed on the FACSCalibur. The fluorescence ratio between the mean fluorescence of the peptide-loaded dimers and the mean fluorescence of the PBS/0.02% NaN3–loaded control dimers was calculated. Peptide names ending in N are amidated, and those ending in C are free acids. Experiments were repeated twice with similar results. Measurements with both cell lines were normalized so they could be displayed on the same axis. HIVpol = HIVpol (18-27), which indicates aa position in HIV polymerase protein.
Discussion
Peptides derived from viral proteins that bind to cell surface MHC class I can mediate an adaptive immune response through interaction with CD8+ T lymphocytes.42,43 In many cases differentiation of these cells into mature CTLs leads to clearance or control of viral infection. Hepatitis B virus (HBV),44HCV,45 and HIV46-48 are among the best-characterized viruses in terms of CTL epitopes that are recognized by the human immune system. CTL epitopes between amino acids 8 and 11 specifically bind to MHC molecules as minimal-length peptides, and synthetic versions sensitize targets for lysis by CD8+CTLs.19,35,49,50 More importantly, several in vivo vaccination studies in animal models have shown these peptides to be capable of eliciting CTLs that clear viral infections.42 51 Although vaccines containing minimal-length epitopes have been shown to elicit CTLs, as yet there has been no successful reported attempt to control disease, either viral or cancer, in humans with these peptides. The object of this report is to develop strategies for the identification of more antigenic/immunogenic peptides using CTL epitopes or their analogs that can be generated using combinatorial chemistry methods.
APCs are exceptionally sensitive to the CTL epitope in its minimal form, and synthetic derivatives of naturally processed epitopes in minute amounts sensitize cells for in vitro lysis, often at picomolar concentration.52-54 The recognition of the pp65495-503 CTL epitope is only sensitive in the nanomolar range to promote lysis of EBVLCLs by human pp65-specific TCCs13 (this paper). Reducing the concentration of the pp65495-503 CTL epitope, which is necessary for recognition by T cells, might be a favorable property in developing a vaccine for clinical use.15 Analysis of the sequence of the pp65495-503 peptide did not reveal any deleterious amino acid or unacceptable anchor motif whose substitution would cause heightened sensitivity of recognition. CMV isolates have limited protein sequence variation, and separate studies have confirmed the conservation of amino acid sequence of the pp65495-503 epitope between most viral isolates derived from healthy donors.12 59
There are several reported cases in which peptide epitopes have been altered to enhance favorable binding interactions with either MHC class I or II as a means to increase activity. Peptides from the melanoma-specific antigen gp-100 could be predictably altered at the anchor positions for increased binding to HLA-A*0201, which resulted in the increased efficiency of CTL stimulation from melanoma patients.55 Recent clinical data suggested that the directed-substitution approach was inadequate to produce ligands that stimulated high-affinity CTLs that cause tumor regression.2 Studies using the positional scanning format have identified high-potency ligands in many cases that exceed the binding affinity of the natural ligand.33,56 57 The focus of these previous reports has been either to discover new epitopes or enhance existing epitope activity specific to individual inbred mouse strains or human TCCs. In contrast, our studies have a dual goal of enhancing affinity to TCR and/or MHC and simultaneously preserving the desirable property of universal recognition of the pp65495-503 CTL epitope; both are necessary properties of a vaccine for broad clinical use.
Results for P1, P3, P7, and P8 highlight the strength of the approach of simultaneously varying all amino acids at each position of the epitope. The favored defined amino acid sublibrary for each of those positions was nonintuitive, as based on published motif information. A consistent property of both alanine and PS-SCL scans is the advantage of C-terminal amidation to obtain high-level recognition of analogs. A rescreen of the same TCC 3-3F4 by a C-terminal free-acid library resulted in uniformly low recognition of all sublibraries, even those having fixed amino acids that correspond to the native sequence (data not shown). In summary, ranking of preferred sublibrary mixtures as a result of PS-SCL screening is more complex than a simple correspondence to native amino acid residues, and C-terminal amidation provides significant recognition enhancement to nonnative sequence analogs.
Sequences corresponding to combinations of the best-recognized defined amino acid library at each of 9 positions were selected, which resulted in 16 peptides that were synthesized as terminal amides. Only 2 of the 16 synthesized peptides were significantly more active than the native sequence. In 3 of 4 instances in which 2 amino acids were evaluated at one position, only one provided any benefit to activity, and in some cases the alternative proved to be deleterious for recognition (see P5 and P8) or equally tolerated, such as valine or threonine at P7. Significantly, the free-acid forms of all 16 peptides proved to be nonimmunogenic using either T2 cells or EBVLCLs (data not shown). In contrast, there was no difference in activity between the amidated and free-acid forms of the native epitope sequence. Collectively, these data show that the screening and deconvolution of the nonapeptide PS-SCL were effective at identifying superior immunogens and that they are stabilized by C-terminal amidation.
The critical test for evaluating the analog epitopes is whether they would be recognized equivalently by T cells from other subjects who recognized the native epitope sequence. Because the analogs were not cross-reactive with TCCs specific for the native sequence, IVS was employed to evaluate whether any distinct repertoire of memory TCCs specific for peptides 44 and 46 could be stimulated from peripheral blood. Although peripheral blood from 4 individuals who responded to the native epitope sequence was stimulated with peptides 44 and 46, only T cells from the individual from whom we derived the PS-SCL screen clone showed any positive response. These results suggest that peptide:TCR contacts were altered by insertion of new residues at P1, P3, P7, and P8.
A second set of individual peptide analogs, which was based on the unique residue modification of peptides 44 and 46, was generated. We evaluated 21 synthetic peptides with different combinations of altered residues using TCC 3-3F4, and several displayed significant activity, even when only a single residue was altered from the native sequence. As expected from the results of the previous screen of 16 analogs, 20 of 21 analogs displayed increased activity as amidated versus free-acid derivatives, but only one free-acid analog was more potent than its amidated counterpart. Results from both the alanine scan and library screen suggested that P1 could tolerate substitution without detriment to recognition. As an example, peptide 193N incorporates tyrosine in place of asparagine and is at least 10-fold more active than the native sequence. Recent work with new synthetic epitope analogs has shown that several amino acids, including serine, threonine, lysine, and even arginine, can be inserted at P1 in place of asparagine. For several residue combinations, a 10-fold greater activity than the native epitope was obtained using serine at P1 (C.L.R., R.K., and D.J.D., unpublished data, 2000).
A novel aspect of this study is that analogs were also evaluated using a different TCC with equivalent recognition of pp65495-503to 3-3F4, but expressing a unique TCR. In that case, 3 analogs still showed 10-fold to 30-fold greater activity than the native sequence (Tables 4 and 5). Expanding the repertoire by 3 new TCCs gave mixed results because one TCC recognized 118N better than the native sequence, whereas a different individual's TCC was negative. Although preliminary, the data suggested that a combination of 2 peptides (118N and 193N) might be more active and broadly recognized than the native sequence. The failure of all 3 peptides to be recognized as universally as the native epitope sequence by cross-reactive TCC was not unexpected. A more effective strategy of determining the breadth of recognition of analogs is to examine whether a separate repertoire exists that recognizes them in different individuals. IVS was used to examine the recognition of the 3 epitopes that were cross-reactive to TCC VB57, an approach that proved effective for evaluating the native sequence epitope and peptides 44 and 46. The results confirmed and expanded what was learned from individual TCCs, as peptides 118N and 193N seemed most broadly recognized, and 115N had more limited recognition. The IVS data suggest that a unique repertoire exists for peptide 193N because in 2 cases, T cells that recognized it were derived from individuals for which TCCs against the native epitope were not cross-reactive (Table 5). In 2 additional cases, TCCs specific for the native epitope either did not recognize peptides 115N and 118N (LV) or 115N and 193N (TG), yet IVS revealed a T-cell repertoire in both individuals for the seemingly unrecognized peptides. Furthermore, all 3 analogs expanded peripheral blood CTLs that not only recognized peptide-coated targets, but were able to independently lyse CMV-infected fibroblasts.
In summary, the IVS results are consistent with broad recognition of 2 of the 3 analogs. Two analogs have properties that qualify them as candidate vaccines because of their enhanced activity coupled with substantial recognition by populations that also recognize the native epitope. IVS data suggest that several of the most potent analogs are broadly recognized, but future studies defining alternative recognition patterns with different TCCs may yield more conclusive information about the best choices of residues to maximize CMV reactivity at a given position of the epitope. Studies in HLA-A*0201 transgenic mice might reveal the extent of their improved activity compared to the native sequence, but clinical trials in humans who are at risk for disease may be the only way to discern whether increased activity leads to efficacy against viral infection.
The authors acknowledge expert technical assistance from J. Papp, Agnes Gardner, and Joy Mead. We also would like to thank G. Hawkins for advice on the growth and maintenance of CMV strains, M. Carmen-Villacres for assistance with flow cytometry, X. Liu for TCR repertoire analysis, M. Sherman for assistance in molecular modeling, J. Pascal (Mixture Sciences Inc, San Diego, CA) for library peptide synthesis and characterization, and M. Wills (University of Cambridge, England) for providing details of IVS procedures. The personnel at the City of Hope General Clinical Research Center, including Judy Brent and Joanne Schifflett, are gratefully acknowledged for their professional and enthusiastic interaction with volunteers, whose specimens were critical to the success of this study. J. Ellenhorn is acknowledged for obtaining all volunteer skin biopsies and for suggestions to improve the manuscript. J. A. Zaia is acknowledged for scientific and clinical advice throughout this study and for the CMV vaccine initiative. The expert assistance of Carole Smith and Rose-Marie Imstepf is gratefully acknowledged in the organization of this manuscript.
Supported by grants 1RO1-CA77544 and 1PO1-CA30206 (Project IV) (D.J.D.) and grants 1RO1-AI29575 and AI44129 (J.S.) from the Public Health Service; grant CA33572 from the City of Hope Cancer Center; grant 5MO1-RR00043-38 (City of Hope Medical Center) from General Clinical Research Center; Translational Research Grant 6116-98 (D.J.D.) from the Leukemia Society of America; and a grant (J.S.) from the National Multiple Sclerosis Society.
Correspondence about the use of combinatorial peptide libraries should be addressed to C. Pinilla: pinilla@tpims.org.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Don J. Diamond, Laboratory of Vaccine Research, Fox South, Beckman Research Institute, the City of Hope Medical Center, Duarte, CA; e-mail: ddiamond@coh.org.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal