The membrane glycoprotein CD36 is involved in platelet aggregation, inhibition of angiogenesis, atherosclerosis, and sequestration of malaria-parasitized erythrocytes. In this study, immunoprecipitations with anti-CD36 antibodies were performed to identify proteins that associate with CD36 in the platelet membrane. Platelets were solubilized in 1% Triton X-100, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), Brij 96, or Brij 99, and the proteins that coprecipitated with CD36 were identified by peptide mass spectrometry and Western blotting. The tetraspanin protein CD9 and the integrins αIIbβ3 and α6β1 specifically coprecipitated with CD36 from platelets that were solubilized in CHAPS and Brij 99 but not from platelets that were solubilized in Triton X-100. Only CD9 is coprecipitated with CD36 from platelets that were solubilized in Brij 96. Reciprocal immunoprecipitations with antibodies to CD9, α6, αIIb, or β3 from Brij 99–solubilized platelets coprecipitated CD36. Coprecipitation of CD36, CD9, and α6β1 was also observed on platelets from a patient with Glanzmann thrombasthenia, indicating that αIIbβ3 is not required for the other proteins to associate. Colocalization of α6 and CD36, of CD9 and CD36, and of α6 and CD9 was observed on intact platelets prior to solubilization, using double immunofluorescence microscopy. These data indicate that CD36 associates with CD9 and integrins on human blood platelets. These associated proteins may mediate or participate in some of the diverse biological functions of CD36.
Introduction
CD36 is a transmembrane glycoprotein that has been shown to participate in multiple biological functions, including platelet aggregation, inhibition of angiogenesis, uptake of oxidized low-density lipoprotein and long-chain fatty acids, cell adhesion, and the sequestration of Plasmodium falciparum–infected erythrocytes.1 As a scavenger receptor, CD36 is involved in the uptake of oxidized low-density lipoprotein by macrophages and the formation of foam cells during arterial atherogenesis.2-4 As a thrombospondin 1 (TSP-1) receptor on platelets, CD36 is involved in reinforcing the molecular bridge that is formed between platelets by fibrinogen and the αIIbβ3 integrin during aggregation.5-7 This model is supported by observations that antibodies to TSP-1 inhibit platelet aggregation (for review see Lawler8). As a TSP-1 receptor on endothelial cells, CD36 reportedly mediates the antiangiogenic effect of TSP-1.9 The effect of TSP-1 on angiogenesis involves the inhibition of endothelial cell migration and the induction of apoptosis. The latter effect reportedly involves the activation of p38 mitogen-activated protein kinase and caspases.9
The original fluid mosaic model of the plasma membrane visualized membrane proteins as free-floating entities in a sea of lipids.10 Data indicate that the plasma membrane is far less homogeneous than this model implies.10-14 Membrane proteins and lipids associate to form functional domains in the plasma membrane. The function of the individual constituents of these domains can be modulated by the other protein or lipid components. Thus, the analysis of a membrane protein's function should be performed with a knowledge of the proteins that associate with it. CD36 has been shown to associate with the Src family protein tyrosine kinases Fyn, Lyn, and Yes in platelets and endothelial cells.15-17 The activity of CD36-associated Lyn has been shown to be regulated by the non–receptor-type tyrosine kinase Chk.18 In endothelial cells, signaling through Fyn reportedly mediates CD36-dependent inhibition of angiogenesis by TSP-1.9 The relatively short (6-9 amino acids) cytoplasmic domain of CD36 is similar to CD4 and CD8 (in that 2 cysteine residues are appropriately spaced for metal ion–dependent association with Lck).17,19 However, Fyn, Lyn, and Yes do not contain the C-X-X-C sequence that has been shown to be necessary for Lck to associate with CD4. This observation raises the possibility that other proteins may associate with CD36 to facilitate signal transduction. Cross-linking studies have demonstrated that CD36 is in close proximity to αIIbβ3 in blood platelets.20Thrombospondin-1, fibrinogen, and αIIbβ3 have also been shown to colocalize on activated platelets by immuno-electron microscopy.21 The potential role of CD36 as a signaling receptor and its association with αIIbβ3 may explain the activation of platelets by the CD36 ligand TSP-1 and by antibodies to CD36.22-24 Additional evidence for the association of CD36 with other membrane proteins has been provided by chemical cross-linking. A 113 000-d form of CD36 has been identified on cross-linked platelets.25 Because the cross-linking reagent that was used was not reversible, the authors could not distinguish between an altered conformational state and the cross-linking of a protein with an approximate mass of 25 000 d to the 88 000-d CD36 polypeptide.
Previous studies on CD36-associated proteins have used Triton X-100 to solubilize the platelet membrane. This detergent disrupts hydrophobic interactions that may occur in the plane of the membrane. To identify proteins that may be associated with CD36, we have solubilized platelets in 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), Brij 96, and Brij 99. Under these conditions, we have identified CD9 and the integrins αIIbβ3 and α6β1 as proteins that are specifically associated with CD36. Immunofluorescence studies show that CD36, CD9, and α6β1 colocalized on the platelet membrane prior to detergent solubilization.
Materials and methods
Antibodies
Mouse monoclonal antibodies to CD36 (FA6-152), the αIIb integrin subunit (SZ22), the β3 integrin subunit (SZ21), CD9 (ALB6), and platelet glycoprotein Ib (SZ2) were obtained from Immunotech (Westbrook, ME). Mouse monoclonal antibodies to CD36 (185-1G2, 1E8) were also obtained from Neomarkers (Fremont, CA). Mouse monoclonal antibodies to Fyn (clone 25), Yes (clone 1), Lyn (clone 42), and focal adhesion kinase (clone 77) were obtained from Transduction Labs (San Diego, CA). A mouse monoclonal antibody to integrin-associated protein (B6H12) was obtained from Pharmingen (San Diego, CA), and a mouse monoclonal antibody to the α6 integrin (4F10) was obtained from Biodesign International (Kennebunkport, ME). Polyclonal antiserum to the β3 integrin subunit (SC-6627), CD9 (SC-7639), and the α6 integrin subunit (SC-6597) were obtained from Santa Cruz (Santa Cruz, CA). A polyclonal antiserum to the β1 integrin subunit was obtained from Research Diagnostics (Flanders, NJ). The polyclonal αvβ3 antibody was obtained from Gibco/BRL (Gaithersburg, MD).
The rabbit anti-CD36 polyclonal antibody 1207 was provided by Dr Tandon (Otsuka America Pharmaceutical, Rockville, MD); the mouse monoclonal antibody to PECAM-1 (CD31) designated PECAM 1.3 was provided by Dr Peter J. Newman (Blood Research Institute, Milwaukee, WI); the rabbit antihuman CD31 antiserum was provided by Steven Albelda (University of Pennsylvania School of Medicine, Philadelphia, PA); anti-Chk monoclonal antibodies (13G2 and 18E12) were provided by Dr Naoto Yamaguchi (University of Shizuoka, Shizuoka, Japan); and the rabbit anti-α6 and rat anti-α6 (GoH3) antibodies were provided by Drs. Leslie Shaw and Arthur Mercurio (Beth Israel Deaconess Medical Center, Boston, MA).
Preparation of human blood platelets
Fifty milliliters of human blood was drawn from the donor, into a one-ninth volume of anticoagulant acid citrate dextrose (National Institutes of Health formula A). The blood from the patient with Glanzmann thrombasthenia was kindly provided by Dr Alan D. Michelson (University of Massachusetts Medical School, Worcester, MA). Indated and recently outdated platelet phoresis products were also used in some studies and gave equivalent results to freshly drawn platelets. The blood was centrifuged at 500 rpm for 15 minutes to remove erythrocytes. The platelet-rich plasma was transferred to a new tube and centrifuged at 2500 rpm for 20 minutes to sediment the platelets. The platelet pellet was washed twice in 10 to 15 volumes of pH 6.5 buffer that contained 0.102 M NaCl, 3.9 mM K2HPO4, 3.9 mM Na2HP04, 22 mM NaH2PO4, and 5.5 mM glucose and was finally suspended in 15 mM Tris-HC1 (pH 7.6), 0.14 M NaC1, and 5 mM glucose (tris-buffered saline with glucose [TBSG]) at 5 × 108/mL. The washed platelets were either used immediately for biotin labeling or directly lysed in lysis buffers (250 mM NaC1, 25 mM Tris-HCl [pH 7.5], 5 mM EDTA, 2 μg/mL aprotinin, 100 μg/mL phenylmethylsulfonyl fluoride [PMSF], and 1% of the various detergents). Brij 96 was purchased from Sigma (St Louis, MO), Brij 99 was purchased from Janssen Chimica (Ann Arbor, MI), Triton X-100 was purchased from Fisher Scientific (Pittsburgh, PA), and CHAPS was purchased from Boehringer Mannheim (Indianapolis, IN). Both labeled and unlabeled platelet lysates could be stored at −20°C for up to a month before use.
Biotin labeling and immunoprecipitation
The membrane surface proteins of the platelets were labeled with biotin, using the enhanced chemiluminescence (ECL) protein Biotinylation System according to protocols supplied by the manufacturer (Amersham Pharmacia Biotech, Piscataway, NJ). The cells were treated with lysis buffer, containing 1% of the various detergents for 20 minutes at 4°C, and the samples were centrifuged at 13 000 rpm for 15 minutes at 4°C in a microcentrifuge. The cell lysate was either used immediately for immunoprecipitation experiments or stored at −20°C. To determine the amount of CD36 that was not recovered in the supernatant, the pellet was rinsed twice in lysis buffer and dissolved in 500 μL sample buffer for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (see below). To preclear the samples, 1 mL cell lysate in a microcentrifuge tube, 5 μg nonimmune immunoglobulin G (IgG) and 20 μL (pellet volume) of protein A beads (Amersham Pharmacia Biotech) were mixed for 1 hour at 4°C. After removal of the protein A beads by centrifugation, 5 μg antibody and 20 μL (pellet volume) protein A beads were added, and the samples were incubated for 2 to 3 hours at 4°C. The beads were washed 4 times with lysis buffer, and the precipitated immunocomplex was eluted in 40 μL of 2 × SDS-PAGE loading buffer by boiling for 4 minutes. The eluted samples were separated by SDS-PAGE either in the presence or absence of 1% dithiothreitol as described previously.26
Detection of biotinylated proteins and immunoblotting
After SDS-PAGE, the proteins were transferred to nitrocellulose membrane (Bio-Rad, Hercules, CA) as described previously.26 The membrane was blocked in 5% blocking reagent (nonfat dry milk) in phosphate-buffered saline (PBS; pH 7.4) containing 0.1% Tween 20 (PBS-T) for 1 hour. The membrane was rinsed twice in PBS-T and incubated for 1 hour in streptavidin–horseradish peroxidase (HRP; Amersham Pharmacia Biotech) solution. After 3 washes in PBS-T, ECL detection was performed with the ECL Western Blotting Detection regents according to manufacturer's instructions (Amersham Pharmacia Biotech). For immunologic detection, the electrophoretic transfer membrane was incubated in 5% blocking reagent in PBS-T for 1 hour. The primary antibody (diluted in the blocking solution) was added, and the membrane was incubated for 1 hour at 22°C with mixing. After 3 washes in PBS-T, the HRP-conjugated secondary antibody was added, and the blot was incubated for 1 hour. The membrane was washed 3 times in PBS-T, and the bands were visualized by using ECL detection.
Mass spectrometric peptide sequencing
Anti-CD36 monoclonal antibody FA6-152 (1 mg) and 2 mL (pellet volume) of protein A Sepharose beads were added to 100 mL Brij 99 lysates of human blood platelets for the large-scale immunoprecipitation. The eluted immunocomplex was concentrated, using a microconcentrater (Amicon, Beverly, MA) and separated by SDS-PAGE. Coomassie blue–stained bands were subjected to in gel reduction, carboxyamidomethylation, and tryptic digestion (Promega, Madison, WI). Multiple peptide sequences were determined in a single run by microcapillary reverse-phase chromatography directly coupled to a Finnigan LCQ quadrupole ion trap mass spectrometer (MS) equipped with a custom nanoelectrospray source. The ion trap was programmed to acquire successive sets of 3 scan modes, consisting of full scan MS over alternating ranges of 395 to 800 m/z and 800 to 1300 m/z, followed by 2 data-dependent scans on the most abundant ion in the full scan. These dependent scans allowed the automatic acquisition of a high-resolution (zoom) scan to determine charge state and exact mass and tandem mass spectrometry (MS/MS) spectra for peptide sequence information. MS/MS spectra were acquired with a relative collision energy of 30%, an isolation width of 2.5 d, and recurring ions were dynamically excluded. Interpretation of the resulting MS/MS spectra of the peptides was facilitated by programs developed in the Harvard Microchemistry Facility and by database correlation with the algorithm SEQUEST.27 28
Sucrose density ultracentrifugation
Platelet extracts were separated on sucrose gradients, using the method of Dorahy et al.29 Biotinylated platelets (1 × 109) were solubilized in 1 mL ice-cold lysis buffer that contained 25 mM 2-[N-morpholino]ethanesulfonic acid (MES, pH 6.5), 0.15 M NaCl, and 2 mM PMSF that contained 1% Triton X-100 or Brij 99. The lysates were briefly sonicated and adjusted to 40% sucrose by addition of 1 mL 80% sucrose in 25 mM MES (pH 6.5) and 0.15 M NaCl. A step gradient was formed by layering 1.5 mL of 30% sucrose in 25 mM MES (pH 6.5) and 0.15 M NaCl over the platelet extract in 40% sucrose. A series of steps that decrease by 5% were formed by sequential overlaying with the appropriate level of sucrose in 25 mM MES (pH 6.5) and 0.15 M NaCl. The gradients were centrifuged at 200 000g at 4°C for 16 hours in a SW41 rotor. The gradients were fractionated into 11 equal (1 mL) fractions, and the pellet at the bottom of the tube was washed in lysis buffer and dissolved in 1 mL SDS-PAGE sample buffer.
Immunofluorescence localization of platelet membrane proteins
Human platelets isolated from fresh plasma were plated on poly-L lysine-coated microscopic slides in a humidified chamber and allowed to adhere for 3 to 5 minutes. The platelets were fixed with 4% formaldehyde in PBS supplemented with 1 mM CaC12 and 2 mM MgC12 for 30 minutes, then washed with PBS. The platelets were permeabilized with 0.1% Triton X-100 or with 0.1% Brij 98 in PBS that contained 4% cold fish skin gelatin at 4°C for 1 hour and then incubated with (1) rabbit anti-CD36 antibody (1207, serum dilution 1:100) and mouse anti-CD9 antibody (2 μg/mL), (2) mouse anti-CD36 antibody (FA6-152, 2 μg/mL) and rabbit anti-α6 antibody (serum dilution 1:100), (3) rabbit anti-CD31 antibody (2 μg/mL) and mouse anti-CD36 antibody (2 μg/mL), or (4) rabbit anti-CD31 antibody (2 μg/mL) and mouse anti-CD9 antibody (2 μg/mL) at 4°C for 16 hours. Goat antirabbit conjugated to Texas Red (Molecular Probes, Eugene, OR), donkey affinity purified antirabbit conjugated to FITC, donkey affinity purified antimouse conjugated to Texas Red, donkey affinity purified antimouse-FITC (Jackson Immunoresearch Laboratories, West Grove, PA), and goat affinity purified antirat (with no cross-reactivity to mouse) conjugated to FITC (Cappel-Organon-Teknika, Durham, NC) were used as reporter secondary antibodies. Controls included normal rabbit IgG and normal mouse IgG at the same concentrations as the primary antibodies and omitting the primary antibodies.30
Antibody-induced capping was used to further establish membrane protein association. The antibodies were diluted in TBSG that contained 1 mg/mL bovine serum albumin. Freshly isolated platelets were plated on 10 μg/mL fibronectin-coated coverslips for 5 minutes, briefly rinsed with TBSG, and then incubated with the following first antibody combinations: (1) mouse anti-CD36 (2 μg/mL) and rat anti-α6 integrin (dilution 1:100); (2) mouse anti-CD9 (2 μg/mL) and rat anti-α6 integrin (dilution 1:100); (3) normal mouse IgG (10 μg/mL) and normal rabbit IgG (10 μg/mL) for 1 hour at 20°C. Platelets were washed for 5 minutes with buffer, then incubated with goat antirat FITC (1:200) and donkey antimouse Texas Red (1:200) for 1 hour at 20°C. Controls included normal mouse IgG, normal rabbit IgG, and normal rat IgG at the same concentration as the primary antibodies or omitting the primary antibodies. The samples were rinsed with buffer for 5 minutes, then fixed in 4% formaldehyde, and mounted with Vectashield.
The samples were viewed with a Zeiss 100 ×, 1.3 N.A. immersion oil objective and with a Bio-Rad MRC-1024 confocal microscope equipped with an Argon Krypton laser. To exclude any bleeding through from FITC (represented by green channel) to Texas Red (represented by red channel), the images were acquired sequentially for each channel (fluorochrome). Each image was acquired below the saturation level, within the intensity linear range of the instrument. The degrees of colocalization of each pair of antibodies were analyzed with the Bio-Rad Laser 3.2 software. The colocalization coefficients were calculated after subtracting the background in each channel. The program calculated 2 values, the colocalization coefficients, which represented the proportion of colocalizing objects in each component of the dual-color (RG) image. Colocalization of molecule A and molecule B at the same point in the sample was represented in the image by a voxel with green intensity above the green background and a red intensity above the red background. The colocalization coefficients C-red and C-green are proportional to the amount of the colocalizing objects in each component of the image, relative to the total amount of fluorescence in that component. The calculation is based on Pearson correlation coefficient that describes the degree of overlap between patterns of images.31
Results
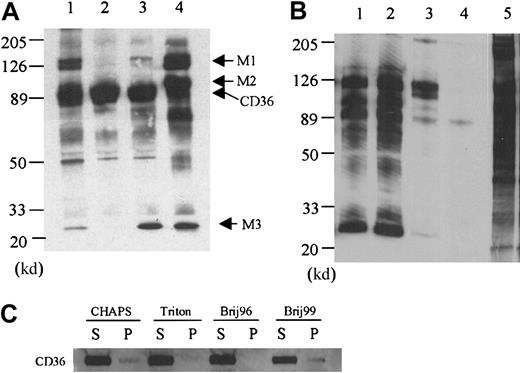
Immunoprecipitation of biotinylated platelet membrane proteins that have been solubilized in Triton X-100 with the anti-CD36 antibody FA6-152 yields a prominent band at 88 000 d (Figure1A, lane 2). A band at 220 000 d and several minor bands are variably observed in control immunoprecipitations with nonimmune IgG, indicating that these bands are not specific to the CD36 immunoprecipitation (data not shown). A comparable pattern is observed when the platelets are solubilized in Brij 96 except that a 25 000-d polypeptide is specifically coprecipitated (Figure 1A, lane 3). Because the biotinylation was performed with intact platelets, portions of the 25 000-d protein should be exposed on the extracellular side of the membrane. The platelets are washed twice after biotinylation and prior to solubilization to prevent labeling of cytoplasmic proteins. Whereas Fyn, Lyn, and Yes can be detected by Western blotting of the anti-CD36 immunoprecipitations (see below), biotinylation of these proteins is not observed, indicating that cytoplasmic proteins are not labeled with the protocol used in this study.
The effect of detergents on CD36-associated proteins.
(A) Platelet membrane proteins were labeled with biotin and lysed in 1% CHAPS (lane 1), Triton X-100 (lane 2), Brij 96 (lane 3), or Brij 99 (lane 4), and CD36 was immunoprecipitated with the monoclonal antibody FA6-152. In Triton X-100 platelet lysates (lane 2), only CD36 is precipitated, whereas in CHAPS (lane 1), Brij 96 (lane 3), and Brij 99 (lane 4) platelet lysates, additional protein bands can be coprecipitated. (B) Two other anti-CD36 monoclonal antibodies 1E8 (lane 1) and 185-1G2 (lane 2) also precipitate the complex in Brij 99 lysates, but a nonrelated antibody to CD31 (lane 3) and nonimmune mouse IgG (lane 4) do not. Lane 5 is the complete platelet lysate, showing that multiple membrane proteins are biotinylated and the bands that are immunoprecipitated with anti-CD36 antibody are not selectively labeled. (C) The distribution of CD36 in the supernatant (S) or pellet (P) after solubilization in the various detergents. Platelets (5 × 108) were solubilized in 1.0 mL lysis buffer that contained 1% of the various detergents. After 20 minutes at 4°C, the sample was centrifuged, and the supernatant was removed. The pellet was resuspended in one half the original volume of SDS-PAGE sample buffer and 10 μL supernatant or 5 μL solubilized pellet was subjected to Western blot analysis with an anti-CD36 antibody.
The effect of detergents on CD36-associated proteins.
(A) Platelet membrane proteins were labeled with biotin and lysed in 1% CHAPS (lane 1), Triton X-100 (lane 2), Brij 96 (lane 3), or Brij 99 (lane 4), and CD36 was immunoprecipitated with the monoclonal antibody FA6-152. In Triton X-100 platelet lysates (lane 2), only CD36 is precipitated, whereas in CHAPS (lane 1), Brij 96 (lane 3), and Brij 99 (lane 4) platelet lysates, additional protein bands can be coprecipitated. (B) Two other anti-CD36 monoclonal antibodies 1E8 (lane 1) and 185-1G2 (lane 2) also precipitate the complex in Brij 99 lysates, but a nonrelated antibody to CD31 (lane 3) and nonimmune mouse IgG (lane 4) do not. Lane 5 is the complete platelet lysate, showing that multiple membrane proteins are biotinylated and the bands that are immunoprecipitated with anti-CD36 antibody are not selectively labeled. (C) The distribution of CD36 in the supernatant (S) or pellet (P) after solubilization in the various detergents. Platelets (5 × 108) were solubilized in 1.0 mL lysis buffer that contained 1% of the various detergents. After 20 minutes at 4°C, the sample was centrifuged, and the supernatant was removed. The pellet was resuspended in one half the original volume of SDS-PAGE sample buffer and 10 μL supernatant or 5 μL solubilized pellet was subjected to Western blot analysis with an anti-CD36 antibody.
When the platelets are solubilized in lysis buffer that contains 1% CHAPS, the band at 25 000 d is again coprecipitated with CD36 (Figure1A, lane 1). In addition, bands at 140 000, 130 000, and 48 000 d are observed. The 48 000-d protein is observed in the anti-CD36 antibody immunoprecipitations from platelets that are solubilized in the other detergents, but it is most prominent in the CHAPS extracts. Solubilization in the detergent that is least likely to dissociate hydrophobic interactions, Brij 99, reveals the largest number of CD36-associated proteins. When the platelets are solubilized in Brij 99, bands with molecular weights of 140 000, 130 000, 100 000, 70 000, and 25 000 are observed (Figure 1A, lane 4). This complex is specific to CD36 in that the same bands are observed with other anti-CD36 monoclonal (185-1G2 and 1E8) and polyclonal (1207, not shown) antibodies, and they are not observed when the immunoprecipitations are performed with nonimmune IgG or with antibodies to CD31 (Figure 1B). Electrophoresis of the proteins that pelleted after solubilization revealed that the vast majority of CD36 is in the supernatant after solubilization with all of the detergents (Figure 1C).
To determine the composition of CD36-associated bands, Brij 99 extracts were scaled up by a factor of 100, and multiple lanes of the FA6-152 immunoprecipitations were electrophoresed. The gel was stained with Coomassie blue, and the 25 000- and 100 000-d bands were excised (data not shown). In addition, the 130 000- and 140 000-d bands were excised together as a single sample. In this study, we focused on the bands designated M1, M2, and M3 that are observed in both CHAPS and Brij 99 (Figure 1A). The identity of the proteins in each band was determined by microcapillary HPLC/ion trap MS. The results indicated that (1) the integrin subunits α6, β1, and αIIb comprise the M1 sample, (2) the β3 integrin subunit is present in the M2 band, and (3) the M3 band contains the light chains of the α6 and αIIb integrin subunits and the tetraspanin family (transmembrane 4 superfamily [TM4SF]) member CD9 (Table1).
Summary of mass spectrometric peptide sequencing
| Band . | Protein . | Accession no. . | No. of MS/MS Spectra . |
|---|---|---|---|
| M1 | αIIb | 107334 | 30 |
| α6 | 106765 | 24 | |
| β1 | 4504767 | 7 | |
| M2 | β3 | 124968 | 10 |
| CD36 | 115982 | 11 | |
| M3 | CD9 | 4502693 | 10 |
| α6 | 106765 | 2 | |
| αIIb | 107334 | 2 |
| Band . | Protein . | Accession no. . | No. of MS/MS Spectra . |
|---|---|---|---|
| M1 | αIIb | 107334 | 30 |
| α6 | 106765 | 24 | |
| β1 | 4504767 | 7 | |
| M2 | β3 | 124968 | 10 |
| CD36 | 115982 | 11 | |
| M3 | CD9 | 4502693 | 10 |
| α6 | 106765 | 2 | |
| αIIb | 107334 | 2 |
For each protein identified, the number of peptide sequences determined by tandem mass spectrometry on an ion trap mass spectrometer as described in “Materials and methods.” The accession number is the identifier for the protein in the National Center for Biotechnology Information nonredundant protein database.
MS/MS indicates tandem mass spectrometry.
We have conducted Western blot analysis on the anti-CD36 antibody immunoprecipitates to confirm the presence of the various proteins that have been identified by MS. The 25 000-d band (M3) does stain when the Western blots are probed with an anti-CD9 antibody (Figure2). The M1 sample that contains the 130 000- to 140 000-d proteins Western blots with antibodies to α6, αIIb, and β1 integrin subunits, and the 100 000-d band (M2) stains with an antibody to the β3 integrin subunit (Figure 2). Western blotting has also been used to probe for the presence of CD36-associated proteins that have been reported in the literature and that we did not observe by MS. The Src-family kinases, Fyn, Lyn, and Yes, are detected in Triton X-100, CHAPS, and Brij 99 (Figure 2). By contrast, we do not detect Chk. We also do not detect 2 other receptors for TSP-1, the integrin-associated protein (CD47) or the αv integrin subunit. In addition, we did not detect platelet glycoprotein Ibα or focal adhesion kinase.
Western blot analysis of CD36-associated proteins.
Three detergents, CHAPS (lanes 1,2), Triton X-100 (lanes 3 and 4), and Brij 99 (lanes 5 and 6), were used to lyse the platelets. The immunoprecipitations with the anti-CD36 antibody (lanes 1, 3, and 5) were compared with those with nonimmune mouse IgG (lanes 2,4,6). The results show that αIIb, β3, α6, β1, and CD9 were detected in CHAPS and Brij 99 platelet lysates but were not detected in Triton X-100 platelet lysates. CD36 and the Src gene family members, Lyn, Fyn, and Yes, were detected in all of the detergent lysates.
Western blot analysis of CD36-associated proteins.
Three detergents, CHAPS (lanes 1,2), Triton X-100 (lanes 3 and 4), and Brij 99 (lanes 5 and 6), were used to lyse the platelets. The immunoprecipitations with the anti-CD36 antibody (lanes 1, 3, and 5) were compared with those with nonimmune mouse IgG (lanes 2,4,6). The results show that αIIb, β3, α6, β1, and CD9 were detected in CHAPS and Brij 99 platelet lysates but were not detected in Triton X-100 platelet lysates. CD36 and the Src gene family members, Lyn, Fyn, and Yes, were detected in all of the detergent lysates.
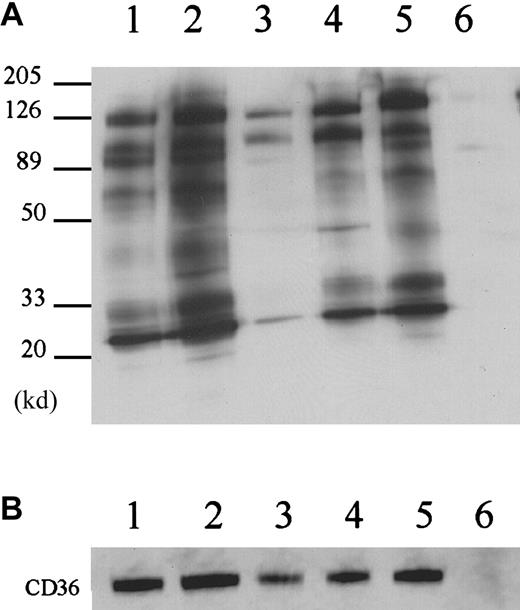
To confirm the specificity of the association of CD36 with CD9, αIIbβ3, and α6β1, we immunoprecipitated the Brij 99 platelet extracts with antibodies to CD9, αIIb, α6, and β3 and conducted Western blot analysis for CD36. Immunoprecipitation with the anti-CD9 antibody produces multiple bands from biotinylated platelets (Figure3, lane 2). The CD36 band is readily visible at 88 000 d. The protein composition of the anti-CD9 immunoprecipitates is very similar to that of the anti-CD36 immunoprecipitates (Figure 3, lanes 1,2). A maximum of 64% of the total CD36 was immunoprecipitated by multiple rounds of anti-CD9 immunoprecipitation (data not shown). This value may underestimate the degree of association because some CD36 and CD9 may have dissociated during the solubilization and subsequent manipulations. CD36 was also coprecipitated with anti-α6 antibody (Figure 3, lane 5). The anti-αIIb and anti-β3 antibodies precipitated a band at 88 000 d that is relatively weak but is readily detectable with the anti-CD36 antibody by Western blotting (Figure 3B).
Reciprocal immunoprecipitations.
(A) The reciprocal immunoprecipitations with anti-CD9 (lane 2), anti-αIIb (lane 3), anti-β3 (lane 4), and anti-α6 (lane5) were compared to nonimmune mouse IgG (lane 6) and anti-CD36 antibody FA6-152 (lane 1). Anti-CD9, β3 and α6 antibodies precipitated similar proteins to the anti-CD36 antibody FA6-152. (B) Immunoprecipitated samples were subjected to Western blot analysis with anti-CD36 antibody. Note that 10 μg anti-αIIb antibody was used in the immunoprecipitation to make the CD36 more readily detectable.
Reciprocal immunoprecipitations.
(A) The reciprocal immunoprecipitations with anti-CD9 (lane 2), anti-αIIb (lane 3), anti-β3 (lane 4), and anti-α6 (lane5) were compared to nonimmune mouse IgG (lane 6) and anti-CD36 antibody FA6-152 (lane 1). Anti-CD9, β3 and α6 antibodies precipitated similar proteins to the anti-CD36 antibody FA6-152. (B) Immunoprecipitated samples were subjected to Western blot analysis with anti-CD36 antibody. Note that 10 μg anti-αIIb antibody was used in the immunoprecipitation to make the CD36 more readily detectable.
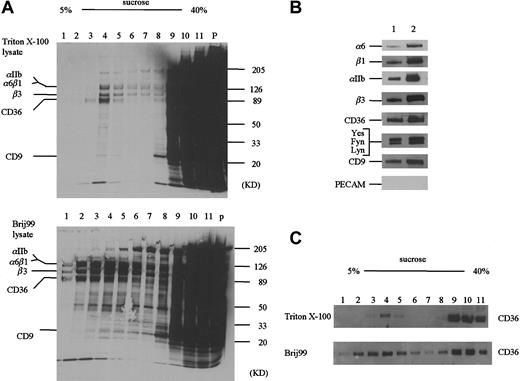
A portion of the CD36 molecules have been reported to associate with a Triton X-100 insoluble, low-density fraction that is rich in cholesterol and can be separated by sucrose-gradient ultracentrifugation.29 We have used this technique to fractionate Triton X-100 and Brij 99–solubilized platelets (Figure4). A considerably greater amount of protein is associated with the low-density factions in the Brij 99–solubilized platelets as compared to the Triton X-100 (Figure 4A). Western blotting reveals that CD36 partitions into both the low- and high-density fractions (Figure 4C). In both Triton X-100 and Brij 99–solubilized samples, we detect α6, αIIb, β3, and CD9 in the low-density fractions (Figure 4B). In addition, Fyn, Lyn, and Yes are present in the low-density fraction, but CD31 is not. These data show that the observation of the CD36 multiprotein complex is not unique to immunologic separations like immunoprecipitation.
Sucrose gradient ultracentrifugation of detergent extracts.
(A) Biotinylated blood platelets were solubilized in 1% Triton X-100 (A, top) or Brij 99 (A, bottom) and centrifuged on 5% to 40% sucrose step gradients (see “Materials and methods”). An equal volume (10 μL) of each fraction was subjected to SDS-PAGE. Whereas the majority of biotinylated proteins is associated with the high-density factions, a cholesterol-rich population centered on fraction 4 is observed in the lower-density fractions. (B) The presence of the various CD36-associated proteins in fraction 4 from the Triton X-100–solubilized platelets (lane 1) or the Brij 99–solubilized platelets (lane 2) was detected by Western blotting. (C) The distribution of CD36 within the sucrose gradients was determined by Western blotting with an anti-CD36 antibody.
Sucrose gradient ultracentrifugation of detergent extracts.
(A) Biotinylated blood platelets were solubilized in 1% Triton X-100 (A, top) or Brij 99 (A, bottom) and centrifuged on 5% to 40% sucrose step gradients (see “Materials and methods”). An equal volume (10 μL) of each fraction was subjected to SDS-PAGE. Whereas the majority of biotinylated proteins is associated with the high-density factions, a cholesterol-rich population centered on fraction 4 is observed in the lower-density fractions. (B) The presence of the various CD36-associated proteins in fraction 4 from the Triton X-100–solubilized platelets (lane 1) or the Brij 99–solubilized platelets (lane 2) was detected by Western blotting. (C) The distribution of CD36 within the sucrose gradients was determined by Western blotting with an anti-CD36 antibody.
Previous cross-linking studies have established that CD36 and αIIbβ3 are associated on the platelet membrane. The αIIbβ3 integrin has been reported to be associated with CD9 and other membrane proteins.32,33 We performed immunoprecipitations with the anti-CD36 antibody on platelets from a patient with Glanzmann thrombasthenia to establish that CD36 associates with the other proteins in the absence of αIIbβ3. The platelets from this patient contain about 3% of the normal levels of αIIbβ3 and contain normal levels of αvβ3.34 35 Immunoprecipitation of the thrombasthenic platelets with the anti-CD36 antibody revealed biotinylated bands at 140 000, 88 000, 70 000, 48 000, and 25 000 d when the platelets are solubilized in CHAPS (Figure5, lane 4) or Brij 99 (Figure 5, lane 1). A similar pattern of biotinylated bands was observed when these platelet extracts were immunoprecipitated with anti-CD9 antibody (Figure 5, lanes 2,5).
CD36-associated proteins on thrombasthenic platelets.
Platelet membrane proteins from a patient with Glanzmann thrombasthenia were labeled with biotin and lysed in Brij 99 (lanes 1, 2, and 3) or CHAPS (lanes 4, 5, and 6). The platelet lysates were immunoprecipitated with anti-CD36 antibody FA6-152 (lanes 1 and 4), anti-CD9 antibody (lanes 2 and 5) or nonimmune mouse IgG (lanes 3 and 6). The results show that CD36 was coprecipitated with CD9 and α6β1 in the absence of αIIbβ3. Lane 7 is the complete platelet lysate, showing that multiple platelet membrane proteins are biotinylated and the bands that are immunoprecipitated with anti-CD36 antibodies are not selectively labeled.
CD36-associated proteins on thrombasthenic platelets.
Platelet membrane proteins from a patient with Glanzmann thrombasthenia were labeled with biotin and lysed in Brij 99 (lanes 1, 2, and 3) or CHAPS (lanes 4, 5, and 6). The platelet lysates were immunoprecipitated with anti-CD36 antibody FA6-152 (lanes 1 and 4), anti-CD9 antibody (lanes 2 and 5) or nonimmune mouse IgG (lanes 3 and 6). The results show that CD36 was coprecipitated with CD9 and α6β1 in the absence of αIIbβ3. Lane 7 is the complete platelet lysate, showing that multiple platelet membrane proteins are biotinylated and the bands that are immunoprecipitated with anti-CD36 antibodies are not selectively labeled.
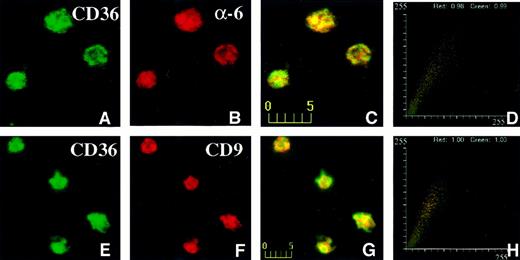
The distribution of CD36, CD9, and α6 on intact platelets was visualized by immunofluorescence confocal microscopy. The data shown in Figure 6 were taken from one experiment (of 4 experiments) and represent a typical distribution of CD36, CD9, and α6 on platelets. CD36, visualized in the green channel (Figure6A), did largely colocalize with α6 integrin, visualized in the red channel (Figure 6B), giving rise to yellow-orange color in the merged panel (Figure 6C). The fluorogram generated from the red and green intensities within the merged panel is shown in Figure 6D. The value for green (CD36) was 0.99 and for red was 0.98 (α6 integrin). For other fields of the same experiment, the lowest value for green (CD36) was 0.89, and the lowest value for red (α6 integrin) was 0.93. Thus, although the majority of CD36 and α6 integrin colocalize, distinct pools of each protein probably exist on the platelet membrane. CD36, visualized in the green channel (Figure 6E), also colocalized with CD9 protein, visualized in the red channel (Figure 6F), resulting in yellow-orange color in the merged panel (Figure 6G). The fluorogram generated from the red and green intensities within the merged panel is shown in Figure 6H. The values for red and green are both 1.00, indicating that all of the CD36 colocalized with CD9 in the platelet membrane. To establish that the values for the colocalization reflect the true association of the membrane glycoproteins and not coincident overlap in the fluorescent signals, we determined the colocalization of CD36 and CD9 with CD31, a membrane glycoprotein that we have not found to be associated with CD36 biochemically. The degree of colocalization of CD36 with CD31 ranged from 0.36 to 0.49 and that for CD9 and CD31 ranged from 0.27 to 0.51 (data not shown). These values are consistent with a low level of colocalization.31
Confocal immunofluorescence visualization of CD36 and α6 (A-D) or CD36 and CD9 (E-H) colocalization in human platelets.
Fixed human platelets were simultaneously incubated with mouse anti-CD36 and rabbit anti-α6 antibodies (A-D) or rabbit anti-CD36 and mouse anti-CD9 (E-H) antibodies, followed by incubation with multiple reporter antibodies. CD36 visualized in the green channel (A,E), colocalized with α6 integrin (B) or CD9 (F), visualized in the red channel giving rise to yellow-orange color in the merged panels (C,G). The fluorograms generated from the red and green intensities within the merged panel are shown in panels D and H. The axes represent red and green pixel intensities (0-255) in the merged panels (C and G).
Confocal immunofluorescence visualization of CD36 and α6 (A-D) or CD36 and CD9 (E-H) colocalization in human platelets.
Fixed human platelets were simultaneously incubated with mouse anti-CD36 and rabbit anti-α6 antibodies (A-D) or rabbit anti-CD36 and mouse anti-CD9 (E-H) antibodies, followed by incubation with multiple reporter antibodies. CD36 visualized in the green channel (A,E), colocalized with α6 integrin (B) or CD9 (F), visualized in the red channel giving rise to yellow-orange color in the merged panels (C,G). The fluorograms generated from the red and green intensities within the merged panel are shown in panels D and H. The axes represent red and green pixel intensities (0-255) in the merged panels (C and G).
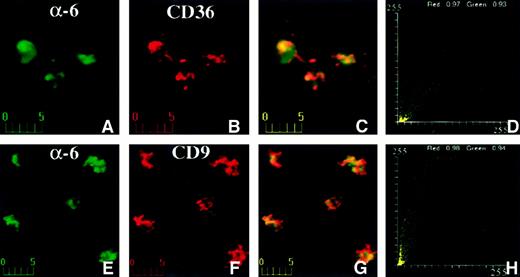
Transmembrane proteins like CD9, CD36, and α6 integrin have extracellular domains that can be laterally cross-linked with specific antibodies. If the proteins are associated with each other, this leads to redistribution of these proteins into patches on the cell membrane. We examined the patching behavior by incubating platelets with antimouse CD36 and antirat α6 integrin concurrently (Figure7) at 20°C for 1 hour, followed by incubation with reporter antibodies at 20°C for 1 hour. Analysis of confocal fluorescence images showed that both proteins redistributed in patches (Figure 7A-D). These patches extensively overlapped in closely coinciding yellow-orange areas of immunofluorescence (Figure 7C). The colocalization coefficients were 0.97 (CD36) and 0.93 (α6) for the red and green channels, respectively. These data indicate that CD36 and α6 integrin coclustered extensively and, thus, belong to the same membrane domain. Similar results were obtained when anti-CD9 and anti-α6 integrin antibodies were used (Figure 7E-H). The colocalization coefficients were 0.98 (CD9) and 0.94 (α6) for the red and green channels, respectively. No patching was observed for control antibodies (data not shown).
Confocal immunofluorescence visualization of CD36 and α6 (A-D) or CD9 and α6 (E-H) capping on human platelets.
Fresh platelets were plated on fibronectin-coated coverslips and incubated with mouse anti-CD36 and rat anti-α6 antibodies (A-D) or mouse anti-CD9 and rat anti-α6 antibodies (E-H) at 20°C for 1 hour. α6 was visualized in the green channel (A,E) and CD36 (B) or CD9 (F) was visualized in the red channel. Capping is apparent in the merged panels (C,G) as yellow-orange color and in the fluorograms (D,H). The platelets on fibronectin in this figure are approximately twice as large as those shown in Figure 6 on polylysine.
Confocal immunofluorescence visualization of CD36 and α6 (A-D) or CD9 and α6 (E-H) capping on human platelets.
Fresh platelets were plated on fibronectin-coated coverslips and incubated with mouse anti-CD36 and rat anti-α6 antibodies (A-D) or mouse anti-CD9 and rat anti-α6 antibodies (E-H) at 20°C for 1 hour. α6 was visualized in the green channel (A,E) and CD36 (B) or CD9 (F) was visualized in the red channel. Capping is apparent in the merged panels (C,G) as yellow-orange color and in the fluorograms (D,H). The platelets on fibronectin in this figure are approximately twice as large as those shown in Figure 6 on polylysine.
Discussion
We have shown that CD36 is a component of one or more multiprotein complexes on the platelet membrane. The association of CD36 with the other proteins is specific because (1) the constituents of these complexes are not observed in immunoprecipitates with nonimmune IgG or with an antibody to another membrane protein (PECAM-1); (2) large amounts of these complexes are not observed in all detergents, indicating that they are not generally associated with membrane solubilization; (3) chemical cross-linking is not required to observe the complexes; (4) similar complexes can be precipitated with antibodies to various components; and (5) double immunofluorescence studies indicate that CD36, α6, and CD9 colocalize on the surface of intact platelets prior to solubilization. Multiple specific interactions have been documented in the literature by using an equivalent experimental approach to the one used here.12 14 In addition, we have found that CD36 and CD9 also coimmunoprecipitated from human dermal microvessel endothelial cells (W-M.M., E.V., J.L., unpublished results, June 2000). The observation that the complexes contain membrane proteins and that the majority of these are disrupted by Triton X-100 suggests that the associations are hydrophobic and within the plane of the membrane. However, we cannot exclude the possibility that cytoplasmic proteins serve to connect CD36 with CD9 or integrins.
Associations between some of the protein components that are reported here have been shown directly or inferred indirectly in the literature. Platelet CD36 has been reported to associate with αIIbβ3 that has, in turn, been reported to associate with CD9.20,32,33Dorahy et al20 showed that CD36 and αIIbβ3 could be cross-linked to each other without evidence of inclusion of CD9. Rhinehart-Jones and Greenwalt25 identified a 113 000-d species that reacts with antibodies to CD36 after platelets are cross-linked with bis (sulfosuccinimidyl) suberate. The molecular weight of this species is equivalent to the sum of the molecular weights of CD36 and CD9. Our data show that CD36, CD9, αIIbβ3, and α6β1 coprecipitate in the absence of cross-linking reagents if mild detergents are used. Taken together, the data suggest that CD36, CD9, and αIIbβ3 are in close proximity to each other and may form a ternary complex.
Separation of Triton X-100–solubilized platelets on sucrose gradients produces a fraction that contains CD36 and the associated proteins. This appears to be a small percentage of the total CD36 because it is not detectable in the unfractionated extract and because Western blotting reveals higher levels of CD36 in the high-density fractions. The appearance of the CD36 distribution is different from that reported by Dorahy et al20 probably because we loaded equal volumes, whereas they loaded equal amounts of total protein onto the SDS gels. Because the majority of protein is found in the high-density fractions, the relative quantity of CD36 is decreased to levels that are not detectable. However, our data show that the majority of CD36 is in the high-density fractions after Triton X-100 solubilization. The quantity of protein associated with the low-density fractions is considerably greater in the Brij 99–solubilized platelets, suggesting that the cholesterol-rich microdomains are better preserved. Our data are consistent with those of Dorahy et al20 in that CD9, CD36, β3, and Src family kinases are present in the low-density fractions. These data indicate that the CD36-associated multiprotein complex is a constituent of the cholesterol-rich microdomains in the platelet membrane.
The double immunofluorescence studies reported here establish that CD36 and α6β1, as well as CD36 and CD9, colocalize on the platelet membrane prior to solubilization. The extent of colocalization of the probes indicates that the majority of CD36 is associated with CD9. This close association is consistent with the fact that CD9 is the only other major band observed in anti-CD36 immunoprecipitations from Brij 96–solubilized platelets. Our data on thrombasthenic platelets indicate that CD36, α6β1, and CD9 form complexes that do not require αIIbβ3. This conclusion is consistent with the observation that CD9 and CD36 coprecipitate from solubilized endothelial cells that lack αIIbβ3 (W-M.M., E.V., J.L., unpublished results, June 2000). The level of colocalization of CD36 and α6β1 is also high; however, it appears that distinct pools of each protein do exist. These data suggest that CD36, CD9, and α6β1 may also form a ternary complex. Because the TM4SF proteins self-associate to form networks, it is possible that the group of proteins that are coprecipitated from Brij 99– and CHAPS-solubilized platelets are clustered together as a single multiprotein complex on the platelet membrane. CD36 and the integrins may be incorporated into a network composed of multiple CD9 molecules. This type of relationship may explain the high level of colocalization of CD36 and CD9 even though there are approximately 2-fold to 3-fold more copies of CD9 than CD36 on platelets. The double immunofluorescence studies may overestimate the extent of colocalization. Because both CD9 and CD36 are present at relatively high copy numbers and because platelets are small in size, there is a possibility of some overlapping fluorescent signals that do not reflect specific association. We have performed capping experiments on platelets that are spread on fibronectin to further establish the close association of these membrane proteins. Concurrent incubation of platelets with anti-α6 and anti-CD36 or anti-CD9 antibodies cross-links the antigens into small areas of the membrane. The clustering coefficients indicate that the majority of CD36, CD9, and α6 are associated with each other. Because the antibodies used are IgGs, coclustering of the antigens into large patches should only occur if the antibodies are straddling protein complexes. The fact that CD36, CD9, and α6β1 form large patches in the presence of the antibodies indicates that they are associated on the platelet membrane.
The close apposition of CD36, α6β1, and αIIbβ3 raises the possibility that ligands may interact with more than one receptor. TSP-1 has been shown to bind to both CD36 and αIIbβ3.36,37 The binding of TSP-1 to both receptors concomitantly may significantly enhance the affinity of the interaction. Our data also raise the possibility that TSP-1 binding to CD36 may serve to strengthen the binding of laminin to α6β1 in much the same way that TSP-1 stabilizes fibrinogen binding to αIIbβ3.5 TSP-1 reportedly binds to both laminin and fibrinogen.8 38-40 The clustering of CD36, CD9, and integrins in the membrane may form specialized domains for platelet adhesion and aggregation.
The TM4SF proteins are widely expressed transmembrane proteins that are involved in cell migration, activation, proliferation, and differentiation.12,14 They appear to function by regulating the activity of other receptor systems, including integrins. TM4SF proteins reportedly recruit signaling proteins, including protein kinase C, to integrins, resulting in the phosphorylation of the cytoplasmic tails of α3 and α6.14 Our data raise the possibility that signal transduction by CD36 may involve CD9 and protein kinase C. Platelets exhibit similar responses to anti-CD36 and anti-CD9 antibodies.23,41 42 Both antibodies activate αIIbβ3 to bind fibrinogen and cause platelet aggregation. Platelet aggregation that is induced by an antibody to either antigen is inhibited by protein kinase C inhibitors. Because neither CD9 nor integrins have been reported to interact with Src family protein tyrosine kinases, the link between CD36 and these proteins remains to be determined. We are currently pursuing the identity of the proteins that coprecipitate with CD36 only in specific detergents in an effort to resolve this issue.
Ongoing studies seek to determine if CD36, CD9, and α6β1 form a complex on microvascular endothelial cells, C32 melanoma cells, and monocytes. The presence of this complex on these cells may help to explain the involvement of CD36 in various cellular processes. For example, CD36 reportedly mediates the effect of TSP-1 on endothelial cell migration and induction of apoptosis.9 Recently, an antibody to CD9 has been reported to inhibit endothelial cell migration.43 These data suggest that the antiangiogenic activity of TSP-1 may be mediated, in part, by CD9.
We thank Drs Martin Hemler, Richard Hynes, Chris Stipp, Keith R. Solomon, and Mary Herndon for helpful discussions and reagents, and Dan Kirby and Kerry Pierce for expertise in the HPLC, mass spectrometry, and peptide sequencing. We also wish to thank Mark Duquette for excellent technical support. We are grateful to Drs Narendra Tandon, Peter Newman, Steven Albelda, Naoto Yamaguchi, Leslie Shaw, and Art Mercurio for providing antibody preparations, and to Dr Alan Michelson for providing the blood from a patient with Glanzmann thrombasthenia. The manuscript was prepared by Regina Prout and Alexis Bywater.
Supported by grant HL28749 from the National Heart, Lung and Blood Institute of the National Institutes of Health.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jack Lawler, Department of Pathology, Beth Israel Deaconess Medical Center, 99 Brookline Ave, Boston, MA 02215; e-mail: lawler@mbcrr.harvard.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal