Using a variety of differentiation-inducible myeloid cell lines, we previously showed that the zinc-finger transcription factor early growth response gene 1 (Egr-1) is a positive modulator of macrophage differentiation and negatively regulates granulocytic differentiation. In this study, high-efficiency retroviral transduction was used to ectopically express Egr-1 in myeloid-enriched or stem cell–enriched bone marrow cultures to explore its effect on the development of hematopoietic progenitors in vitro and in lethally irradiated mice. It was found that ectopic Egr-1 expression in normal hematopoietic progenitors stimulates development along the macrophage lineage at the expense of development along the granulocyte or erythroid lineages, regardless of the cytokine used. Moreover, Egr-1 accelerated macrophage development by suppressing the proliferative phase of the growth-to-macrophage developmental program. The remarkable ability of Egr-1 to dictate macrophage development at the expense of development along other lineages resulted in failure of Egr-1–infected hematopoietic progenitors to repopulate the bone marrow and spleen, and thereby prevent death, in lethally irradiated mice. These observations further highlight the role Egr-1 plays in monocytic differentiation and growth suppression.
Introduction
Early growth response gene (Egr) 1 is a member of the Egr family of genes, which includes Egr-1,1,2 Egr-2,3 Egr-3,4 and Egr-4.5 These genes encode for zinc-finger transcription factors that have specificity to related but not identical guanine-cytosine–rich DNA binding motifs.6,7 Egr-1 was initially identified as an immediate-early growth response gene in cultured fibroblasts,1,6,8 but more recent studies have provided evidence that Egr-1 plays a role in the development, growth control, and survival of several cell types, including T cells and B cells,9 neuronal cells,10 and myeloid cells.11-13
We previously found evidence that Egr-1 plays a role in the development of hematopoietic cells along the macrophage lineage.11-13 We initially identified Egr-1 as a myeloid-differentiation primary response gene that is activated in the absence of de novo protein synthesis on 12-O-tetradecanoyl-phorbol-13-acetate (TPA)–induced macrophage differentiation of HL-60 cells.11 Using a variety of myeloid-differentiation–inducible cell lines, we showed that Egr-1 is a positive modulator of macrophage differentiation whose function varies according to the state of lineage commitment for differentiation and the hematopoietic cell type. In HL-60 cells, Egr-1 blocked granulocytic differentiation and restricted differentiation to the monocytic lineage. Egr-1 also blocked granulocyte colony-stimulating factor (G-CSF)–induced differentiation of interleukin (IL) 3–dependent 32Dcl3 hematopoietic precursor cells, endowing the cells with the ability to be induced by granulocyte-macrophage colony-stimulating factor (GM-CSF) for terminal differentiation along the macrophage lineage. Interestingly, Egr-1 was also found to at least partly mediate the ability of the homeobox gene Hox-B8 (Hox2.4) to endow 32Dcl3 cells with the potential to be induced by GM-CSF for terminal macrophage differentiation.14Furthermore, ectopic expression of Egr-1 in M1 myeloblastic leukemia cells activated the macrophage-differentiation program in the absence of the differentiation inducer IL-6.12
The complex process of blood cell formation, which is regulated throughout life, involves a hierarchy of hematopoietic progenitor cells in the bone marrow (BM) that proliferate and terminally differentiate along multiple distinct cell lineages, including the proliferation and differentiation of myeloid progenitor cells into a variety of mature myeloid cells.15-19 To understand how Egr-1 regulates normal hematopoietic cell development, we here used high-efficiency retroviral transduction to ectopically express Egr-1 in myeloid-enriched or stem cell–enriched BM cell cultures and explored its effects on the development of hematopoietic progenitors in vitro and in vivo. We found that regardless of the cytokine used, ectopic Egr-1 expression in normal hematopoietic progenitors stimulated their development to the macrophage lineage at the expense of development along other lineages, notably the granulocyte and erythroid lineages. Furthermore, Egr-1 accelerated macrophage development by suppressing the proliferative phase. Consequently, Egr-1–infected hematopoietic progenitors failed to repopulate the BM in lethally irradiated mice.
Materials and methods
Mice, BM, and cytokines
Femoral BM was prepared from 4- to 6-week-old inbred female Balb/c mice (Taconic, Germantown, NY) that had been given 3 mL 10% sodium caseinate (SC) (United States Biochemical Corp, Cleveland, OH) intraperitoneally 3 days earlier to enrich for myeloid progenitor cells or 150 mg/kg 5-fluorouracil (5-FU) (F8423; Sigma, St Louis, MO) intravenously 4 days earlier to enrich for proliferating stem cells.20 Both SC and 5-FU were prepared in sterile l × phosphate-buffered saline (PBS). Erythrolysis of BM was done by treating the cells with buffered ammonium chloride for 10 minutes (Stemcell Technologies, Vancouver, Canada). Cells were then washed 3 times with PBS to remove both ammonium chloride and lysed red blood cells.
For retroviral infection, erythrolysed nucleated BM cells were prestimulated to promote cell division with IL-3 (10% WEHI-3B–conditioned medium as a source of IL-3), IL-6 (10 ng/mL), and stem cell factor (SCF; 200 ng/mL) in α minimum essential medium (MEM; Gibco BRL, Grand Island, NY) supplemented with 20% heat-inactivated fetal calf serum (FCS; Stemcell Technologies) plus 1% penicillin and streptomycin (Cellgrow; Mediatech, Heydon, VA) for 48 hours in a humidified atmosphere with 10% carbon dioxide (CO2) at 37°C. The recombinant cytokines used in this study were human IL-6, human G-CSF, human GM-CSF, rat SCF, and human erythropoietin (Epo), which were generous gifts from Amgen Inc (Thousand Oaks, CA). WEHI-3B–conditioned medium as a source of IL-3, L-cell–conditioned medium as a source of macrophage colony-stimulating factor (M-CSF) and pokeweed mitogen–stimulated, spleen cell–conditioned medium (SCM) were prepared in our laboratory as described previously.21 22
Generation of retroviral particles and infection of BM cells
For transfection of packaging cell line Bosc23 (a gift from Dr Warren S. Pear,23 University of Pennsylvania), 1.5 × 106 Bosc23 cells were seeded in 75 cm2Falcon tissue-culture flasks containing 10 mL Dulbecco modified Eagle medium (Cellgrow; Mediatech) supplemented with 10% FCS plus 1% penicillin and streptomycin (Cellgrow; Mediatech). Cell were incubated in a humidified atmosphere with 5% CO2 at 37°C for 2 days. Control and Egr retroviral-expression vector murine stem cell virus (MSCV) EB neo Egr-1 were transfected into Bosc23 packaging cells by using the standard calcium phosphate DNA transfection method.24 Two days after transfection, the viral titer of culture supernatants was determined by infecting NIH3T3 cells and selecting for G418 (800 μg/mL)–resistant colonies.25For infection of BM cells, 2 days after transfection, Bosc23 transfectants were incubated with 10 μg/mL mitomycin C (Sigma) for 3 hours to prevent cell division of Bosc23 transfectants during cocultivation with BM cells. The flasks were washed 3 times with l × PBS to remove mitomycin C and cocultivated with 2 × 106 prestimulated BM cells in 10 mL α MEM supplemented with 20% FCS, l × penicillin and streptomycin, 10% IL-3 (WEHI-3B–conditioned medium), 10 ng/mL IL-6, 200 ng/mL SCF, and 8 μg/mL Polybrene for 4 days in a humidified atmosphere with 5% CO2 at 37°C. Under similar conditions, control BM cells were mock infected by cocultivating prestimulated BM cells with nontransfected, mitomycin C–treated normal Bosc23 cell cultures. After cocultivation, mock-infected (control) and infected BM cells were washed 3 times with PBS and used immediately in clonogenic progenitor assays. For all experiments, virus titers of greater than 5 × 106/mL were used, employing the same titer of control neo and Egr-1 virus for infection of BM cells.
For BM transplantation studies, washed cells were cultured for 24 hours in complete α MEM containing 10% IL-3, 200 ng/mL SCF, and G418 (750 μg/mL concentration). Cells were then washed 3 times and resuspended in PBS for injection into lethally irradiated mice.
In vitro clonogenic progenitor assay
Immediately after cocultivation, control and infected BM cells were assessed by in vitro progenitor cell colony-forming assays. Cells were washed and plated in 35-mm tissue-culture dishes (StemCell Technologies Inc) in 1.1 mL methylcellulose-based medium (Methocult HCC 3234; StemCell Technologies Inc) according to the manufacturer's instructions. Colonies were raised in the presence of various cytokines by supplementing methylcellulose-based medium with IL-3 (10% WEHI-3B–conditioned medium), SCM (10%), G-CSF (100 ng/mL), GM-CSF (100 ng/mL), or M-CSF (10% L-cell–conditioned medium). Cells were seeded at an initial density of 0.5 × 105 cells/dish with or without 650 μg/mL G418 (Gibco BRL) and were scored after 8 days.
Assays for differentiation-associated properties
For each sample, isolated BM colonies were pooled and cytospin smears prepared. Morphologic differentiation was determined by counting at least 300 cells on May-Grünwald-Giemsa–stained cytospin smears and scoring the proportion of blast cells, mature granulocytes, and macrophages.7,8 Immature blast cells are characterized by scant cytoplasm and round or oval nuclei and mature granulocyte-like cells by enlarged cytoplasm and lobulated nuclei. Mature macrophage-like cells are flattened, well spread out, and interspersed with numerous vacuoles in a greatly enlarged cytoplasm. Erythroid cells were identified by benzidine staining.26 To identify granulocytes and macrophages, nitroblue tetrazolium (NBT) staining and nonspecific esterase (NSE) staining, respectively, were done as described previously.11 Analysis of expression of macrophages and granulocyte-specific cell-surface markers on BM was done by using fluorescence-activated cell-sorting (FACS) analysis with fluorescein isothiocyanate (FITC)–conjugated F4/80, a rat monoclonal antibody to mouse macrophage antigen (Caltag Laboratories, Burlingame, CA), and FITC-conjugated Gr-1, an antimouse LyG6 (Pharmingen, San Diego, CA), respectively.
General recombinant DNA techniques, expression vectors, and reverse transcriptase–polymerase chain reaction
Plasmid preparations, restriction enzyme digestions, DNA fragment preparations, and agarose gel electrophoresis were done as described previously.12 MSCV EB neo, the retroviral plasmid expression vector used in this study, was a gift from Dr Robert G. Hawley (University of Toronto, Toronto, Canada).27 The 2.3-kb BamHI and SalI fragment of the full-length murine Egr-1 complementary DNA was cloned into the XhoI site of the MSCV EB neo retroviral vector by means of blunt-end ligation. To identify ectopic Egr-1 expression in Egr-1–infected BM cells, reverse transcription–polymerase chain reaction (RT-PCR) was done as described previously.13 RNA from the BM cells, extracted by using Trizol reagent (Gibco BRL), was reverse transcribed with the Superscript preamplification system (180-890 11; Gibco BRL) according to the manufacturer's instructions. A region spanning the cloning site was amplified on the MSCVneo Egr-1 vector by using primers corresponding to base pairs 1096 to 2020 of the MSCVneo vector (5′TTCTGCTCTGCAGAATGGCCAACC3′) and base pairs 748 to 769 of the Egr-1 insert (5′AAGCAGCTGGAGAAGGCGCCG3′).
BM transplantation into irradiated mice
For BM transplantation studies, 4- to 5-week-old Balb/c mice were irradiated lethally with a total of 935 Gy or sublethally with a total of 700 Gy (delivered at the rate of 1.95 Gy/minute [195 rad/minute]) by using a cesium 137 source irradiator. Lethally irradiated mice were injected with 0.2 × 106 and sublethally irradiated animals with 2 × 106 infected or control BM cells in a volume of 300 μL through the lateral tail vein immediately after irradiation. The mice were maintained in microisolator cages in a barrier animal facility and fed sterilized food and acidified water. To prevent infection in lethally irradiated animals, their drinking water was supplemented with neomycin (1.1 g base/L; Sigma) and polymyxin B (106 U/L; Sigma). The number and type of colony-forming units–spleen were determined 8 or 13 days after transplantation, or both, essentially as described previously.28,29 The number of BM cells obtained from femurs of injected mice22 was determined at the indicated times and was used to assess the ability of injected cells to repopulate the BM of the irradiated mice.
Results
Egr-1 promotes macrophage differentiation and inhibits granulocyte differentiation of myeloid-enriched BM cells
Using a variety of differentiation-inducible myeloid cell lines, we previously showed that Egr-1 is a positive modulator of macrophage differentiation whose functions vary according to the state of lineage commitment for differentiation of the hematopoietic cell type. To assess how Egr-1 may modulate normal hematopoietic cell development, we here used high-efficiency retroviral transduction to infect BM cells. Because Egr-1 is a positive modulator of myeloid cell differentiation, initially myeloblast-enriched BM cultures were used as the source of normal cells. These were obtained from femurs of Balb/c mice injected intraperitoneally with SC, which initiates an inflammatory response that increases myelopoiesis.22 The BM cells isolated from mice injected with SC consisted primarily of cells of the myeloid lineage (95% ± 4%), with 33% ± 3% myeloid precursors at the myeloblast-to-promyelocyte stage, compared with 76% ± 4% of myeloid cells, with 18% ± 3% of myeloid precursors for normal BM cells obtained from untreated animals. Retroviral particles were generated by transfecting the pMSCV retroviral vectors into the high-efficiency Bosc23 packaging line. The resulting virus was used to infect the myeloblast-enriched BM cells. The infection efficiency of hematopoietic progenitor cells with pMSCVneo ranged from 25% to 50%, determined by the number of G418-resistant BM colonies generated in methylcellulose supplemented with IL-3, SCM, G-CSF, GM-CSF, or M-CSF.
Ectopic Egr-1 expression in infected BM cells
Evidence for the presence of transduced pMSCV Egr-1 in myeloid progenitor–enriched BM cells was obtained with RT-PCR using RNA obtained from BM cells infected with MSCVneo Egr-1 retrovirus (BMEgr-1), with the PCR primers corresponding to sequences on both the MSCVneo vector and the Egr-1 insert. This resulted in amplification of a 1-kb fragment spanning the cloning site (data not shown).
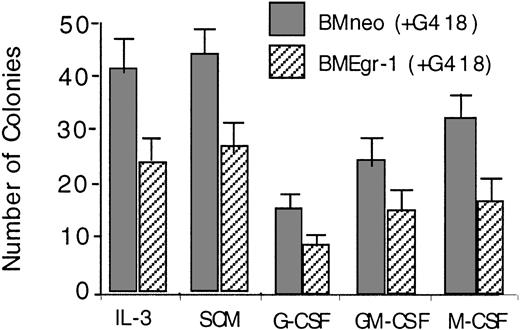
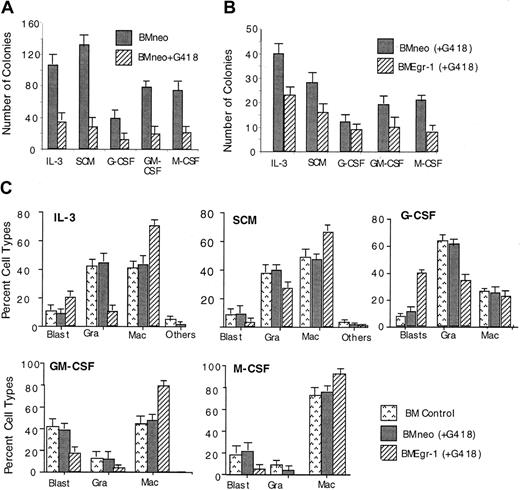
Transduction of myeloid progenitor–enriched BM cultures with pMSCV Egr-1 showed that ectopic expression of Egr-1 reduced the colony-forming ability of progenitor cells cultured in methylcellulose supplemented with either IL-3, SCM, G-CSF, GM-CSF, or M-CSF (Figure1). In all cases, Egr-1–infected colonies were also smaller in size (≤ 500 cells/colony) than neo-infected controls (800-1500 cells/colony; data not shown).
Effect of ectopic expression of Egr-1 on colony-forming ability of myeloid-enriched BM cell cultures.
After infection of BM cells with MSCVneo (BMneo) or MSCVneo Egr-1 (BMEgr-1), cells were assayed in methylcellulose supplemented with IL-3 (10% WEHI-3B–conditioned medium), SCM (10%), G-CSF (100 ng/mL), GM-CSF (100 ng/mL), or M-CSF (10% L-cell–conditioned medium as a source of M-CSF). Cells were seeded at concentrations of 0.5 × 105 cells/mL in 35-mm tissue-culture dishes in the presence of G418 (650 μg/mL), and colonies were scored after 8 days. Values are mean (± SD) results from 3 independent experiments.
Effect of ectopic expression of Egr-1 on colony-forming ability of myeloid-enriched BM cell cultures.
After infection of BM cells with MSCVneo (BMneo) or MSCVneo Egr-1 (BMEgr-1), cells were assayed in methylcellulose supplemented with IL-3 (10% WEHI-3B–conditioned medium), SCM (10%), G-CSF (100 ng/mL), GM-CSF (100 ng/mL), or M-CSF (10% L-cell–conditioned medium as a source of M-CSF). Cells were seeded at concentrations of 0.5 × 105 cells/mL in 35-mm tissue-culture dishes in the presence of G418 (650 μg/mL), and colonies were scored after 8 days. Values are mean (± SD) results from 3 independent experiments.
To test whether the smaller colony size of Egr-1–transduced BM cells was due to reduced proliferative capacity of the cells, secondary colony assays were done. Equal numbers of cells obtained from primary colonies were seeded in methylcellulose supplemented with IL-3. As shown in Figure 2, the ability of Egr-1–transduced progenitors to form secondary colonies was significantly impaired compared with that of neo-infected controls. All the colonies formed by the Egr-1–transduced BM cells in the secondary cultures were small (< 100 cells/colony) and had dispersed macrophage-colony morphologic characteristics. Cytologic analysis of May-Grünwald–stained cytospin smears of the secondary BMEgr-1 colonies revealed exclusively well-differentiated macrophage phenotypes. In contrast, secondary colonies formed by BMneo cells were larger (> 500 cells/colony) and contained different myeloid cell types, most of which had mature macrophage or granulocyte morphologic features (Figure 2). Similar results were obtained in secondary colony assays with other cytokines, such as SCM, G-CSF, GM-CSF, and M-CSF (data not shown).
Effect of Egr-1 on the proliferation of BM progenitor cells of myeloid- enriched BM in secondary clonogenic assays.
(A) Eight-day-old G418-resistant BMneo and BMEgr-1 colonies formed in the primary methylcellulose cultures supplemented with IL-3 (10% WEHI-3B–conditioned medium) were each pooled, 1000 cells/dish were seeded in methylcellulose supplemented with 10% IL-3 and G418 (650 μg/mL), and incubation was allowed to proceed for another 8 days before colony scoring. Values are mean (± SD) results from 3 independent experiments. Phenotypes of cells in methylcellulose secondary colonies were differentiated. In BMneo colonies, 52% of cells were macrophages, 39% granulocytes, and 9% other cell types. In BMEgr-1 colonies, 100% of cells were macrophages. Cell phenotype was determined by cytologic analysis of May-Grünwald–stained cytospin smears prepared with cells obtained from pooled methylcellulose colonies.
Effect of Egr-1 on the proliferation of BM progenitor cells of myeloid- enriched BM in secondary clonogenic assays.
(A) Eight-day-old G418-resistant BMneo and BMEgr-1 colonies formed in the primary methylcellulose cultures supplemented with IL-3 (10% WEHI-3B–conditioned medium) were each pooled, 1000 cells/dish were seeded in methylcellulose supplemented with 10% IL-3 and G418 (650 μg/mL), and incubation was allowed to proceed for another 8 days before colony scoring. Values are mean (± SD) results from 3 independent experiments. Phenotypes of cells in methylcellulose secondary colonies were differentiated. In BMneo colonies, 52% of cells were macrophages, 39% granulocytes, and 9% other cell types. In BMEgr-1 colonies, 100% of cells were macrophages. Cell phenotype was determined by cytologic analysis of May-Grünwald–stained cytospin smears prepared with cells obtained from pooled methylcellulose colonies.
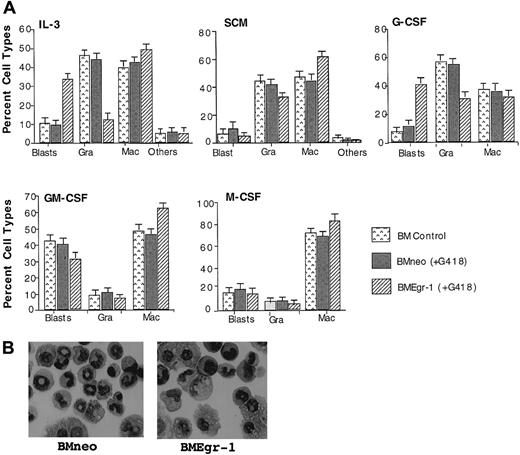
To determine the effect of ectopic expression of Egr-1 on myeloid progenitor development, cytologic examinations were done on May-Grünwald–stained cytospin smears of cells obtained from 8-day-old BM colonies formed in methylcellulose cultures (Figure3A). Representative pictures of neo-transduced and Egr-1–transduced cells obtained from colonies generated in the presence of IL-3 are shown in Figure 3B. Ectopic Egr-1 expression altered the profile of differentiated cellular phenotypes of BM progenitor cells. Regardless of the cytokines that were used, Egr-1 progenitors had an increased proportion of cells differentiated into macrophages (except when cultured with G-CSF) and reduced granulocytic differentiation. Thus, ectopic Egr-1 promoted monocytic differentiation and inhibited granulocytic differentiation of normal myeloid progenitors.
Effect of Egr-1 on macrophage/granulocyte differentiation of myeloid-enriched BM cells.
Uninfected BM cells (BM control), BM cells infected with MSCVneo (BMneo), and BM cells infected with MSCVneo Egr-1 (BMEgr-1) were seeded in methylcellulose supplemented with the indicated cytokines with or without G418, at the same concentrations indicated in the legend for Figure 1. To determine different cell types, colonies generated after 8 days were isolated, pooled, and used for cytospin smears. At least 300 May-Grünwald–stained cells were scored for each sample. (A) Percentage (± SD) of cell types in 3 independent determinations.P values (Student t test) for the difference in the percentage of granulocytes (Gra) or macrophages (Mac) of BMneo compared with BMEgr-1 cells were IL-3/Gra, P < .001; IL-3/Mac, P < .01; SCM/Gra, P < .05; SCM/Mac, P < .05; G-CSF/Gra, P < .001; G-CSF/Mac, P > .05 (not significant); GM-CSF/Gra,P > .05 (not significant); GM-CSF/Mac,P < .05; M-CSF/Gra, P > .05 (not significant); and M-CSF/Mac, P < .05. (B) Representative photomicrographs of May-Grünwald–stained cytospin smears of BM cells obtained from 8-day methylcellulose colonies plus IL-3 (with G418) that were generated by BM cells infected with either MSCVneo (BMneo) or MSCVneo Egr-1 (BMEgr-1).
Effect of Egr-1 on macrophage/granulocyte differentiation of myeloid-enriched BM cells.
Uninfected BM cells (BM control), BM cells infected with MSCVneo (BMneo), and BM cells infected with MSCVneo Egr-1 (BMEgr-1) were seeded in methylcellulose supplemented with the indicated cytokines with or without G418, at the same concentrations indicated in the legend for Figure 1. To determine different cell types, colonies generated after 8 days were isolated, pooled, and used for cytospin smears. At least 300 May-Grünwald–stained cells were scored for each sample. (A) Percentage (± SD) of cell types in 3 independent determinations.P values (Student t test) for the difference in the percentage of granulocytes (Gra) or macrophages (Mac) of BMneo compared with BMEgr-1 cells were IL-3/Gra, P < .001; IL-3/Mac, P < .01; SCM/Gra, P < .05; SCM/Mac, P < .05; G-CSF/Gra, P < .001; G-CSF/Mac, P > .05 (not significant); GM-CSF/Gra,P > .05 (not significant); GM-CSF/Mac,P < .05; M-CSF/Gra, P > .05 (not significant); and M-CSF/Mac, P < .05. (B) Representative photomicrographs of May-Grünwald–stained cytospin smears of BM cells obtained from 8-day methylcellulose colonies plus IL-3 (with G418) that were generated by BM cells infected with either MSCVneo (BMneo) or MSCVneo Egr-1 (BMEgr-1).
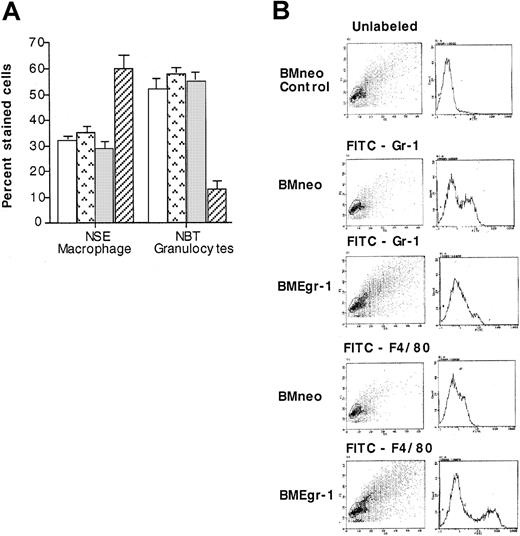
To corroborate the results of the cytologic examinations, neo-transduced and Egr-1–transduced cells obtained from 8-day-old colonies grown with IL-3 were analyzed for expression of macrophage-specific and granulocyte-specific differentiation markers. These markers included the cytochemical markers NSE and NBT and the cell-surface markers F4/80 and Gr-1 specific for macrophages and granulocytes, respectively. As shown in Figure4A, 65% of the BMEgr-1 cells stained for macrophage-specific NSE, whereas only 30% of the neo control cells did. Conversely, ectopic expression of Egr-1 resulted in a reduction in the percentage of cells expressing the granulocyte-specific marker NBT; only 15% of BMEgr-1 colony cells stained for NBT, whereas 60% of the control BMneo colony cells showed such staining (Figure 4A). Consistent with expression of macrophage and granulocyte cytochemical markers, flow cytometry analysis with FITC antibodies revealed that expression of the macrophage cell-surface marker F4/80 was greatly increased, whereas expression of the granulocyte cell-surface marker Gr-1 was greatly decreased in BMEgr-1 cells compared with BMneo controls (Figure4B).
Effect of Egr-1 on macrophage/granulocyte differentiation markers of myeloid-enriched BM cells.
Uninfected BM cells (BM control, □), mock-infected BM controls (BM mock,  ), and BM infected with either MSCVneo (BMneo, ░) or MSCVneo Egr-1 (BMEgr-1, ▨) were seeded in methylcellulose supplemented with IL-3 (10% WEHI-3B–conditioned medium) with or without G418 (650 μg/mL). After 8 days, colonies were isolated, pooled, and washed, and the cells were resuspended in PBS. (A) Cells were used to prepare cytospin smears to assay for NSE staining and NBT reduction. Values are mean (± SD) results from 3 independent experiments. (B) For FACS analysis, cells were stained with FITC granulocyte-specific Gr-1 antibodies (antimouse Ly-6G) or macrophage-specific F4/80 antibodies (rat antimouse macrophage). Cell scattering and the intensity of fluorescence staining are shown. Three independent experiments were performed; results were similar.
), and BM infected with either MSCVneo (BMneo, ░) or MSCVneo Egr-1 (BMEgr-1, ▨) were seeded in methylcellulose supplemented with IL-3 (10% WEHI-3B–conditioned medium) with or without G418 (650 μg/mL). After 8 days, colonies were isolated, pooled, and washed, and the cells were resuspended in PBS. (A) Cells were used to prepare cytospin smears to assay for NSE staining and NBT reduction. Values are mean (± SD) results from 3 independent experiments. (B) For FACS analysis, cells were stained with FITC granulocyte-specific Gr-1 antibodies (antimouse Ly-6G) or macrophage-specific F4/80 antibodies (rat antimouse macrophage). Cell scattering and the intensity of fluorescence staining are shown. Three independent experiments were performed; results were similar.
Effect of Egr-1 on macrophage/granulocyte differentiation markers of myeloid-enriched BM cells.
Uninfected BM cells (BM control, □), mock-infected BM controls (BM mock,  ), and BM infected with either MSCVneo (BMneo, ░) or MSCVneo Egr-1 (BMEgr-1, ▨) were seeded in methylcellulose supplemented with IL-3 (10% WEHI-3B–conditioned medium) with or without G418 (650 μg/mL). After 8 days, colonies were isolated, pooled, and washed, and the cells were resuspended in PBS. (A) Cells were used to prepare cytospin smears to assay for NSE staining and NBT reduction. Values are mean (± SD) results from 3 independent experiments. (B) For FACS analysis, cells were stained with FITC granulocyte-specific Gr-1 antibodies (antimouse Ly-6G) or macrophage-specific F4/80 antibodies (rat antimouse macrophage). Cell scattering and the intensity of fluorescence staining are shown. Three independent experiments were performed; results were similar.
), and BM infected with either MSCVneo (BMneo, ░) or MSCVneo Egr-1 (BMEgr-1, ▨) were seeded in methylcellulose supplemented with IL-3 (10% WEHI-3B–conditioned medium) with or without G418 (650 μg/mL). After 8 days, colonies were isolated, pooled, and washed, and the cells were resuspended in PBS. (A) Cells were used to prepare cytospin smears to assay for NSE staining and NBT reduction. Values are mean (± SD) results from 3 independent experiments. (B) For FACS analysis, cells were stained with FITC granulocyte-specific Gr-1 antibodies (antimouse Ly-6G) or macrophage-specific F4/80 antibodies (rat antimouse macrophage). Cell scattering and the intensity of fluorescence staining are shown. Three independent experiments were performed; results were similar.
Together, these data show that Egr-1 expression in hematopoietic progenitor cells promoted monocytic differentiation and inhibited granulocytic differentiation, regardless of the cytokines used.
Egr-1 enhances macrophage differentiation in stem cell–enriched myeloid BM cells
Using myeloid-enriched BM cultures, we confirmed our previous finding, obtained by employing differentiation-inducible cell lines, that Egr-1 is a positive modulator of differentiation of myeloid cells along the macrophage lineage and that it negatively regulates differentiation along the granulocytic lineage. To expand on these findings, Egr-1 was transduced into stem cell–enriched BM cell cultures obtained from mice given 5-FU.
As shown in Figure 5A, with stem cell–enriched BM cell cultures, MSCVneo transduction efficiencies of up to 40% were achieved. As with myeloid-enriched BM, Egr-1 also reduced by 30% to 50% the colony-forming ability of progenitors in stem cell–enriched BM cells that were seeded in methylcellulose supplemented with either IL-3, SCM, G-CSF, GM-CSF, or M-CSF by 30% to 50% (Figure 5B). In addition, the colonies were smaller than those with the neo controls and the ability of Egr-1–expressing progenitors to form secondary colonies was greatly reduced, a finding indicative of impaired proliferative capacity (data not shown).
Effect of Egr-1 on clonogenicity and macrophage/granulocyte differentiation of stem cell–enriched BM cells.
(A) Infection efficiency of 5-FU BM cells with pMSCVneo. The 5-FU BM cells were cocultivated for 4 days with Bosc23 cells transfected with MSCVneo or MSCVneo Egr-1 retroviral-expression constructs. After infection, control and infected BM cells were assayed in methylcellulose supplemented with IL-3 (10% WEHI-3B–conditioned medium), SCM (10%), G-CSF (100 ng/mL), GM-CSF (100 ng/mL), or M-CSF (10% L-cell–conditioned medium as a source of M-CSF). Cells were seeded at concentrations of 0.5 × 105 cells/mL in 35-mm tissue-culture dishes in the presence or absence of G418 (650 μg/mL), and colonies were scored after 8 days. Values are mean (± SD) results from 3 independent experiments. (B) Effect of Egr-1 on clonogenicity of stem cell–enriched BM cells. After infection of 5-FU BM cells with either MSCVneo (BMneo) or MSCVneo Egr-1 (BMEgr-1), the cells were assayed in methylcellulose supplemented with the indicated cytokines and G418 (650 μg/mL) as described above. Colonies were scored after 8 days. Values are mean (± SD) results from 3 independent experiments. (C) Effect of Egr-1 on differentiation of stem cell–enriched BM cells. Uninfected bone marrow cells (BM control) or cells infected with MSCVneo (BMneo) or MSCVneo Egr-1 (BMEgr-1) were seeded in a methylcellulose culture supplemented with the indicated cytokines with or without G418, as described above. Colonies generated after 8 days were isolated and pooled, and cytospin smears were prepared and stained with May-Grünwald stain. To determine cell types, at least 300 May-Grünwald–stained cells were scored. Values are percentages (± SD) of cell types from 3 independent determinations.
Effect of Egr-1 on clonogenicity and macrophage/granulocyte differentiation of stem cell–enriched BM cells.
(A) Infection efficiency of 5-FU BM cells with pMSCVneo. The 5-FU BM cells were cocultivated for 4 days with Bosc23 cells transfected with MSCVneo or MSCVneo Egr-1 retroviral-expression constructs. After infection, control and infected BM cells were assayed in methylcellulose supplemented with IL-3 (10% WEHI-3B–conditioned medium), SCM (10%), G-CSF (100 ng/mL), GM-CSF (100 ng/mL), or M-CSF (10% L-cell–conditioned medium as a source of M-CSF). Cells were seeded at concentrations of 0.5 × 105 cells/mL in 35-mm tissue-culture dishes in the presence or absence of G418 (650 μg/mL), and colonies were scored after 8 days. Values are mean (± SD) results from 3 independent experiments. (B) Effect of Egr-1 on clonogenicity of stem cell–enriched BM cells. After infection of 5-FU BM cells with either MSCVneo (BMneo) or MSCVneo Egr-1 (BMEgr-1), the cells were assayed in methylcellulose supplemented with the indicated cytokines and G418 (650 μg/mL) as described above. Colonies were scored after 8 days. Values are mean (± SD) results from 3 independent experiments. (C) Effect of Egr-1 on differentiation of stem cell–enriched BM cells. Uninfected bone marrow cells (BM control) or cells infected with MSCVneo (BMneo) or MSCVneo Egr-1 (BMEgr-1) were seeded in a methylcellulose culture supplemented with the indicated cytokines with or without G418, as described above. Colonies generated after 8 days were isolated and pooled, and cytospin smears were prepared and stained with May-Grünwald stain. To determine cell types, at least 300 May-Grünwald–stained cells were scored. Values are percentages (± SD) of cell types from 3 independent determinations.
Analysis of cell types in Egr-1–infected, stem cell–enriched BM confirmed what was observed with myeloid-enriched BM cell cultures. Namely, regardless of the cytokines used, Egr-1 altered the profile of progenitor development, favoring macrophage development at the expense of development along other lineages (Figure 5C). It is notable that this ability of Egr-1 was even more pronounced in stem cell–enriched BM compared with myeloid-enriched BM (Figure 5C [IL-3] and Figure 3A [IL-3]).
Egr-1 stimulates macrophage development at the expense of granulocyte or erythroid development
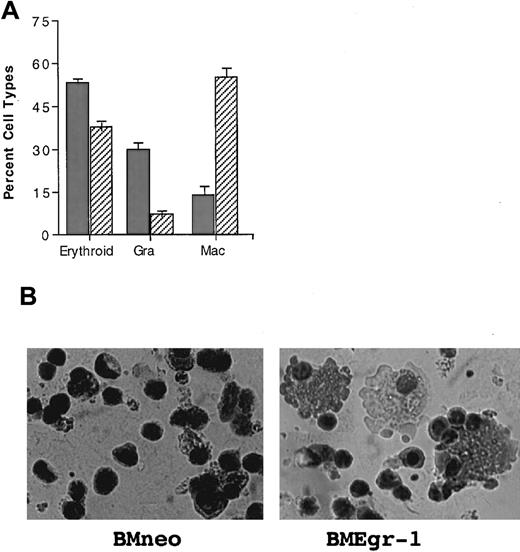
The pronounced ability of Egr-1 to enhance hematopoietic progenitor development along the macrophage lineage raised the possibility that Egr-1 expression, in addition to overriding granulocyte differentiation in favor of macrophage differentiation, might also stimulate macrophage differentiation in other blood cell lineages. To test this idea, we analyzed the effect of Egr-1 on the ability of IL-3 plus Epo to stimulate progenitor cells derived from mice treated with 5-FU to differentiate along both the myeloid and erythroid lineages.30 As shown in Figure6, Egr-1 expression resulted in a significant reduction in the percentage of erythroid cells and an increase in the percentage of macrophages, compared with cells obtained from colonies of neo controls.
Effect of Egr-1 on differentiation of stem cell–enriched BM cells stimulated for erythroid, macrophage, and granulocyte differentiation by IL-3 plus Epo.
(A) Effect of Egr-1 on differentiation of 5-FU BM cells stimulated with IL-3 plus Epo. After infection of 5-FU BM cells with MSCVneo (BMneo, ░) or MSCVneo Egr-1 (BMEgr-1, ▨), cells were assayed in methylcellulose supplemented with IL-3 (3% WEHI-3B–conditioned medium) and Epo (10 U). Cells were seeded at concentrations of 0.5 × 105 cells/mL in 35-mm tissue-culture dishes in the presence of G418 (650 μg/mL). Colonies generated after 8 days in the methylcellulose were isolated, pooled, washed, and resuspended in 1 × PBS, and cytospin smears were prepared. To determine different cell types, at least 300 May-Grünwald–stained cells were scored for each sample. The percentage of erythroid cells was also determined by benzidine staining. (B) Representative photomicrographs of May-Grünwald–stained cytospin smears of 5-FU BM cells obtained from 8-day colonies in methylcellulose with IL-3 plus Epo that were generated by uninfected BM cells (control BM without G418) and cells infected with MSCVneo (BMneo plus G418) or MSCVneo Egr-1 (BMEgr-1 plus G418).
Effect of Egr-1 on differentiation of stem cell–enriched BM cells stimulated for erythroid, macrophage, and granulocyte differentiation by IL-3 plus Epo.
(A) Effect of Egr-1 on differentiation of 5-FU BM cells stimulated with IL-3 plus Epo. After infection of 5-FU BM cells with MSCVneo (BMneo, ░) or MSCVneo Egr-1 (BMEgr-1, ▨), cells were assayed in methylcellulose supplemented with IL-3 (3% WEHI-3B–conditioned medium) and Epo (10 U). Cells were seeded at concentrations of 0.5 × 105 cells/mL in 35-mm tissue-culture dishes in the presence of G418 (650 μg/mL). Colonies generated after 8 days in the methylcellulose were isolated, pooled, washed, and resuspended in 1 × PBS, and cytospin smears were prepared. To determine different cell types, at least 300 May-Grünwald–stained cells were scored for each sample. The percentage of erythroid cells was also determined by benzidine staining. (B) Representative photomicrographs of May-Grünwald–stained cytospin smears of 5-FU BM cells obtained from 8-day colonies in methylcellulose with IL-3 plus Epo that were generated by uninfected BM cells (control BM without G418) and cells infected with MSCVneo (BMneo plus G418) or MSCVneo Egr-1 (BMEgr-1 plus G418).
Taken together, these observations indicate that ectopic expression of Egr-1 can stimulate the development of hematopoietic progenitors along the macrophage lineage at the expense of erythroid development.
Egr-1–transduced progenitors fail to prevent death in lethally irradiated mice
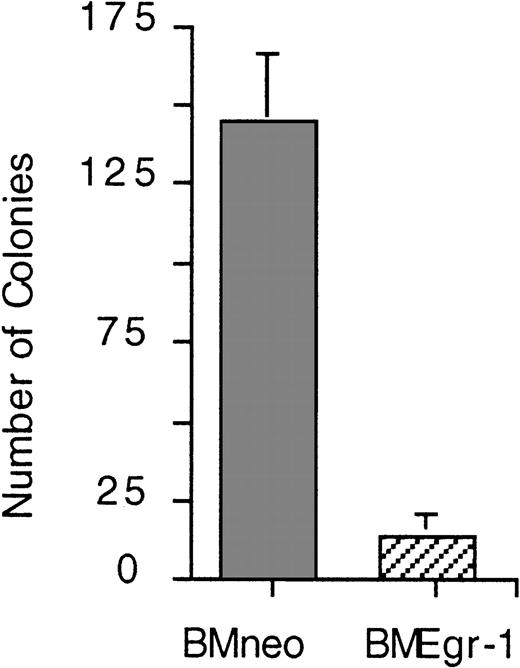
To examine how ectopic Egr-1 may modulate hematopoiesis in vivo, we tested the ability of Egr-1–transduced progenitor cells to repopulate the BM and thereby prevent death in lethally irradiated mice. Lethally irradiated mice given transplants of mock-infected BM cells formed spleen colonies and survived for the 4-week period of observation (Table 1). During this period, only 10% of the mice given neo-transduced cells died; the remaining 90% survived and formed macroscopic spleen colonies (Table1). In contrast, all the mice given transplants of Egr-1–transduced cells died by 10 days after transplantation. Autopsies of these animals revealed that they had shrunken spleens without spleen colonies (Table1). Furthermore, femurs of mice given transplants of Egr-1–transduced progenitors, unlike femurs of healthy control mice or control mice given neo transplants, contained a greatly reduced number of BM cells on days 7 and 9 after transplantation and the cells obtained failed to generate colonies in methylcellulose (Table 1). Similar results were obtained in mice given transplants of stem cell–enriched BM cell cultures (Figure 7 and data not shown). These observations indicate that the Egr-1–infected hematopoietic progenitors failed to repopulate the BM and thereby prevent death in lethally irradiated mice.
Results indicating failure of early growth response gene 1–infected, myeloid-enriched bone marrow cells to prevent death in lethally irradiated mice
| Cells transplanted* . | Mice surviving (%)† . | No. of spleen colonies‡ . | No. of femoral BM cells (×106)1-153 . | Results of colony assays according to cell type1-154 . | ||||
|---|---|---|---|---|---|---|---|---|
| No. of colonies1-155 . | Blast (%) . | Granulocytes (%) . | Macrophages (%) . | Others (%) . | ||||
| BM normal | 100 | 14 | 4.0 | 60 | 30 | 43 | 25 | 2 |
| BM mock | 90 | 10 | 2.9 | 65 | 60 | 25 | 11 | 3 |
| BMneo | 90 | 12 | 2.2 | 50 | 65 | 22 | 12 | 1 |
| BMEgr-1 | 0 | 0 | 0.001 | — | — | — | — | |
| Cells transplanted* . | Mice surviving (%)† . | No. of spleen colonies‡ . | No. of femoral BM cells (×106)1-153 . | Results of colony assays according to cell type1-154 . | ||||
|---|---|---|---|---|---|---|---|---|
| No. of colonies1-155 . | Blast (%) . | Granulocytes (%) . | Macrophages (%) . | Others (%) . | ||||
| BM normal | 100 | 14 | 4.0 | 60 | 30 | 43 | 25 | 2 |
| BM mock | 90 | 10 | 2.9 | 65 | 60 | 25 | 11 | 3 |
| BMneo | 90 | 12 | 2.2 | 50 | 65 | 22 | 12 | 1 |
| BMEgr-1 | 0 | 0 | 0.001 | — | — | — | — | |
BM indicates bone marrow; Egr-1, early growth response gene 1.
Each BM type was transplanted into 13 lethally irradiated mice (0.2 × 106 cells/mouse).
For each BM type, 10 mice were monitored for survival for 4 weeks. All mice injected with BMEgr-1 died within 10 days after injection.
Spleen colonies were counted on day 8. Values are mean (with SDs up to ±20%) results for spleens obtained from 3 mice.
BM cells were collected from femurs 7 days after transplantation. Values are mean numbers of BM cells (with SDs up to ±15%) obtained from femurs of 3 mice.
Seven days after transplantation, BM cells were recovered from femurs and 0.05 × 106 cells (except in the BMEgr-1 group, in which 0.001 × 106 cells were used) were plated in 1 mL methylcellulose supplemented with interleukin 3 (10% WEHI-3B–conditioned medium). Colony number was determined after 8 days. Values are mean (SD up to ±15%) results in triplicate plates.
Colonies generated after 8 days in methylcellulose were pooled and used to prepare cytospin smears. At least 300 May-Grünwald–stained cells were scored. Percentage values are mean (SD up to ±15%; ie, 30% ± 4.5%) results from 3 smears prepared from triplicate methylcellulose cultures.
Survival in lethally irradiated mice given transplants of stem cell–enriched BM control cells, MSCVneo–infected cells, or MSCVneo Egr-1–infected cells.
After irradiation of Balb/c mice, infection of BM cells, and selection in G418, 10 mice in each group were given injections in the tail vein with the indicated infected BM cells (0.2 × 106 cells per 300 μL PBS per mouse). Cell-free control animals were given injections of 300 μL PBS. Mice were housed and fed in a sterile environment and monitored for survival for up to 28 days.
Survival in lethally irradiated mice given transplants of stem cell–enriched BM control cells, MSCVneo–infected cells, or MSCVneo Egr-1–infected cells.
After irradiation of Balb/c mice, infection of BM cells, and selection in G418, 10 mice in each group were given injections in the tail vein with the indicated infected BM cells (0.2 × 106 cells per 300 μL PBS per mouse). Cell-free control animals were given injections of 300 μL PBS. Mice were housed and fed in a sterile environment and monitored for survival for up to 28 days.
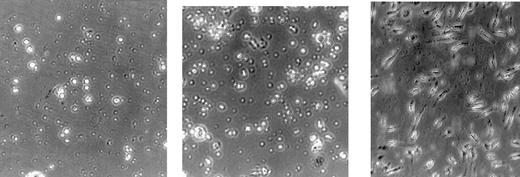
To determine whether the failure of Egr-1–infected hematopoietic progenitors to repopulate the BM may have been due to a homing defect or alteration of the terminal-differentiation program, Egr-1–transduced progenitors were transplanted into sublethally irradiated mice. Sublethally irradiated mice given transplants of either PBS (no BM cells), mock-infected, neo-infected, or Egr-1–infected BM cells survived for the 6-week observation period. Groups of these mice were killed 7 days after transplantation and autopsies done. These revealed that mice given transplants of Egr-1–infected BM cells formed a reduced number of spleen colonies compared with control mice given mock-infected or neo BM cells. Femurs of mice given transplants of Egr-1–transduced progenitors, unlike femurs of healthy control mice or control mice given neo transplants, also contained a reduced number of BM cells on day 7 after transplantation (Table 2). In addition, BM cells of mice given transplants of Egr-1 progenitors had a significant reduction in the percentage of blast and granulocytic cells and an increase in the percentage of macrophages adhering to the surface of the tissue-culture plates (in liquid culture supplemented with IL-3; Figure 8), compared with BM cells obtained from controls given mock-infected or neo-infected cells.
Results indicating that early growth response gene 1 enhances macrophage differentiation in bone marrow cells of sublethally irradiated mice
| Cells transplanted* . | Mice surviving (%)† . | No. of spleen colonies‡ . | No. of femoral BM cells (×106)2-153 . | Adherent cells (%)2-155 . | Cell type (%)2-154 . | |||
|---|---|---|---|---|---|---|---|---|
| Blast . | Granulocytes . | Macrophages . | Others . | |||||
| BM normal | 100 | 13 | 7.0 | 24 | 31 | 46 | 22 | 1 |
| BM mock | 100 | 12 | 5.5 | 25 | 33 | 44 | 24 | 2 |
| BMneo | 100 | 13 | 5.0 | 24 | 39 | 37 | 22 | 2 |
| BMEgr-1 | 100 | 6 | 3.0 | 50 | 23 | 25 | 51 | 0 |
| Cells transplanted* . | Mice surviving (%)† . | No. of spleen colonies‡ . | No. of femoral BM cells (×106)2-153 . | Adherent cells (%)2-155 . | Cell type (%)2-154 . | |||
|---|---|---|---|---|---|---|---|---|
| Blast . | Granulocytes . | Macrophages . | Others . | |||||
| BM normal | 100 | 13 | 7.0 | 24 | 31 | 46 | 22 | 1 |
| BM mock | 100 | 12 | 5.5 | 25 | 33 | 44 | 24 | 2 |
| BMneo | 100 | 13 | 5.0 | 24 | 39 | 37 | 22 | 2 |
| BMEgr-1 | 100 | 6 | 3.0 | 50 | 23 | 25 | 51 | 0 |
For abbreviations, see Table 1.
Each BM type was transplanted into 13 sublethally irradiated mice (.2 × 106 cells/mouse).
For each BM type, 10 mice were monitored for survival for 6 weeks.
Spleen colonies were counted on day 8. Values are mean (with SDs up to ±20%) results for spleens obtained from 3 mice.
BM cells were collected from femurs 8 days after transplantation. Values are mean numbers of BM cells (with SDs up to ±15%) obtained from femurs of 3 mice.
See Figure 8.
Eight days after transplantation, BM cells were recovered from femurs and 0.3 × 106 cells/mL were seeded in liquid culture medium supplemented with interleukin 3 (10% WEHI-3B–conditioned medium). After 3 days, cytospin smears were prepared. At least 300 May-Grünwald–stained cells were scored. Values are mean (SD up to ±15%) results from 3 smears prepared from triplicate cultures.
Egr-1 enhances macrophage differentiation in BM cells of sublethally irradiated mice.
Eight days after transplantation, BM cells were recovered from femurs and 0.3 × 106 cells/mL were seeded in liquid culture medium supplemented with IL-3 (10% WEHI-3B–conditioned medium). After 3 days, the percentage of adherent cells was determined (see Table 2, “Adherent cells”). Values are the average of triplicate plates (SD up to ± 15%).
Egr-1 enhances macrophage differentiation in BM cells of sublethally irradiated mice.
Eight days after transplantation, BM cells were recovered from femurs and 0.3 × 106 cells/mL were seeded in liquid culture medium supplemented with IL-3 (10% WEHI-3B–conditioned medium). After 3 days, the percentage of adherent cells was determined (see Table 2, “Adherent cells”). Values are the average of triplicate plates (SD up to ± 15%).
Taken together, these observations are consistent with the idea that Egr-1–transduced progenitors have a greatly enhanced probability of undergoing macrophage differentiation after transplantation into irradiated mice, and as a result, they fail to repopulate the BM and spleen and prevent death in the irradiated animals.
Discussion
In previous studies, a variety of myeloid-differentiation–inducible cell lines were used to show that Egr-1 is a positive modulator of macrophage differentiation. In this study, to increase our understanding of the role Egr-1 plays in hematopoiesis, we analyzed how Egr-1 modulates hematopoietic development of normal myeloid progenitor cells in vitro and in vivo.
High-efficiency retroviral transduction was used to ectopically express Egr-1 in myeloid-enriched and stem cell–enriched BM cell cultures. Regardless of the differentiating stimulus, Egr-1 significantly increased the proportion of progenitor cells that developed along the macrophage lineage, often at the expense of developing along either the granulocyte or the erythroid lineage. Thus, ectopic Egr-1 appears to have a remarkable ability to stimulate hematopoietic cell development to the macrophage lineage at the expense of development along other lineages. Consistent with this idea, Egr-1 progenitors failed to repopulate the BM and spleen and therefore prevent death in lethally irradiated mice. In addition, an increase in the proportion of macrophage progenitors was observed in BM cells obtained from sublethally irradiated mice. These findings support our previous findings11-13 that Egr-1 is a positive modulator of macrophage differentiation that overrides development of normal myeloid progenitor cells along other hematopoietic lineages.
Using 2-dimensional gel analysis, we previously identified rapid changes in protein expression, including induction and suppression of specific proteins, that were rapidly induced in HL-60 cells on cell attachment associated with TPA-induced macrophage differentiation.31 These protein changes did not occur on induction of HL-60 granulocytic differentiation by dimethyl sulfoxide (DMSO). The same set of protein changes was also observed in normal human peripheral blood monocytes after attachment to the surface of tissue-culture plates and was not observed in human peripheral blood granulocytes,31 indicating the relevance of the observed changes in protein expression to normal myelopoiesis. Furthermore, using this set of protein changes as a diagnostic tool, we showed that HL-60 cells treated with both TPA and DMSO attached and underwent even more rapid macrophage differentiation than cells treated with TPA alone. Moreover, after induction of the granulocyte program by DMSO, stimulation with TPA resulted in rapid cell attachment and a switch from granulocyte to macrophage development. These observations led to the conclusion that cytokine-regulated cell adhesion plays a major role in determining the developmental program of myeloid progenitor cells.31 In accordance with this idea, it is possible that Egr-1 target genes that encode for or regulate expression of cell-surface adhesion proteins play a crucial role in the remarkable ability of Egr-1 to divert the development of progenitors from other myeloid lineages toward macrophage differentiation.
Previous studies identified several genes encoding for adhesion molecules involved in both cellular attachment to the extracellular matrix and cell-cell interactions as potential targets for Egr-1. These include intracellular adhesion molecule 1, which was shown to be subject to Egr-1 regulation in B cells32; CD44, which was found to be a target for Egr-1 in B cells33 and the endothelial cell line ECV-30434; and CD31, whose promoter was shown to contain Sp-1 and Egr-1 elements that conferred phorbol myristate acetate inducibility of a reporter gene in myeloid cells.35 The possibility that these and other genes that encode for cell-adhesion molecules are direct targets for Egr-1, which dictates monocytic development of hematopoietic cells, is being investigated.
Our results here indicate that Egr-1 not only dictated development of myeloid progenitors along the macrophage lineage but also accelerated this process by suppressing the proliferative phase of the macrophage growth-to-differentiation developmental program. This was evident from the smaller colony size of Egr-1–transduced progenitors compared with controls as well as their greatly reduced ability to yield secondary colonies. This interesting unexpected finding highlights the increasing evidence of a role for Egr-1 in growth suppression and suppression of transformation of many cell types of both hematopoietic and nonhematopoietic origins.7 This finding is also consistent with previous studies demonstrating that ectopic expression of Egr-1 impairs the leukemogenicity of M1 myeloblastic leukemia cells in vivo.12
It has been reported that macrophage differentiation was not affected in mice lacking the Egr-1 gene.36 There is increasing evidence, however, that the other Egr family members—Egr-2, Egr-3, and Egr-4—share a high degree of structural and functional homology with Egr-1.7 Recently, we observed that depending on the cytokines used and the hematopoietic cell type, either 1, 2, or all 4 members of the Egr family are induced during myeloid differentiation (unpublished data). These observations strongly suggest that other members of the Egr family of transcription factors are capable of compensating for the function of Egr-1 in macrophage development. Determining the role of these Egr transcription factors in hematopoiesis should be instrumental in addressing this issue.
Supported by National Institutes of Health grants 1R01CA59774 (D.A.L.) and 1R01CA51162 (B.H.), the Core Program on Carcinogenesis (5P3CA12227), and Amgen.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
D. A. Liebermann, Fels Institute for Cancer Research and Molecular Biology and the Department of Biochemistry, Temple University School of Medicine, 3307 N Broad St, Philadelphia, PA 19140; e-mail: lieberma@unix.temple.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal