BAD is a proapoptotic member of the BCL-2 family of proteins, which play a major role in regulating apoptosis in cytokine-dependent hematopoietic cells. The function of BAD is regulated by reversible phosphorylation. Deprivation of survival factors induces BAD dephosphorylation, resulting in apoptosis. Serine–threonine phosphatase activity dephosphorylated BAD in interleukin-3–dependent FL5.12 lymphoid cells. Inhibition of PP2A activity by treatment of cells with PP2A-selective inhibitors, okadaic acid and fostriecin, prevented BAD dephosphorylation in these cells. Conversely, BAD dephosphorylation was not inhibited by the PP1-selective inhibitor tautomycin. In cell-free extracts, BAD phosphatase activity was also inhibited by the PP2A-selective inhibitors okadaic acid and fostriecin, but not by the PP1-specific protein inhibitor I-2. Dissociation of 14-3-3 from BAD was a prerequisite for BAD dephosphorylation in vitro, suggesting a mechanism by which 14-3-3 can regulate the activation of the proapoptotic function of BAD in vivo. Significantly, the inhibition of BAD phosphatase activity rescued cell death induced by survival factor withdrawal in FL5.12 cells expressing wild-type BAD but not phosphorylation-defective mutant BAD. These data indicate that PP2A, or a PP2A-like enzyme, dephosphorylates BAD and, in conjunction with 14-3-3, modulates cytokine-mediated survival.
Introduction
Apoptosis is a major mechanism regulating hematopoietic development and homeostasis. The prototypical apoptosis regulator BCL-2 was discovered as the gene involved in a lymphoma translocation, and its oncogenic role was proven in animal models of lymphomagenesis.1-3 The BCL-2 family of proteins, consisting of prosurvival members (including BCL-2, BCL-XL, MCL-1, A1) and proapoptotic members (including BAX, BAD, BCL-XS, BAK, BID) play key roles in integrating signals from survival factors and death stimuli.4-6 The mechanisms of apoptosis have been extensively studied in cytokine-dependent hematopoietic systems, such as interleukin-3 (IL-3)–dependent progenitor cells.
Reversible phosphorylation is a key regulatory mechanism for cell survival and cell death in response to extracellular survival factors, such as IL-3,7 insulin-like growth factor-1 (IGF-1),8 and erythropoietin,9,10 and is highly relevant to hematopoietic cell biology. IL-3–induced phosphorylation of BCL-2 at Ser70 correlates with its antiapoptotic function,11,12 and mitochondrial serine–threonine phosphatase PP2A has been implicated in the dephosphorylation of BCL-2.13 FKHRL1, a forkhead transcription factor that can regulate apoptosis in erythroid9,10 and other cell types,14 is phosphorylated by Akt and sequestered in the cytosol by 14-3-3 in the presence of the survival factors.14 On activation of apoptosis, FKHRL1 is dephosphorylated and translocates to the nucleus, where it transactivates cell death genes such as the Fas ligand gene.
BAD, a proapoptotic member of the BCL-2 family, is highly regulated by phosphorylation. In IL-3–dependent FL5.12 lymphoid progenitor cells, dephosphorylated BAD counters the survival function of BCL-2 and BCL-XL by binding to BCL-2 or BCL-XL. BAX is displaced from BCL-2 or BCL-XL, and BAX homo-dimerization increases, leading to cell death.15 In the presence of survival factor IL-3, BAD is phosphorylated at Ser112 and Ser136. Phosphorylated BAD does not bind mitochondrial BCL-2 or BCL-XL and is sequestered in the cytosol by 14-3-3.7 BAD is rapidly dephosphorylated on IL-3 withdrawal, and its proapoptotic function is activated.
Several signaling pathways impact BAD phosphorylation. Both mitochondria-anchored cyclic adenosine monophosphate-dependent protein kinase A (PKA)16 and p21-activated protein kinase 1 (PAK1)17 can phosphorylate BAD at Ser112 and inactivate its proapoptotic activity in IL-3–dependent FL5.12 cells. Recently, PKA was also reported to phosphorylate Ser155 in the BH3 domain, disrupting the binding of BAD to BCL-XL and BCL-2 and promoting cell survival.18-21 In other cell types, Akt phosphorylates BAD at Ser136 in response to survival factors such as IGF-1.8 IGF-1 also protects murine hematopoietic 32D cells from IL-3 withdrawal-induced death by at least 3 alternative signaling pathways,22 including the PI3-kinase–Akt pathway, the mitogen-activated protein kinase pathway, and a third pathway resulting in the mitochondrial translocation of Raf. These pathways all lead to BAD phosphorylation and inactivation of its proapoptotic function. Thus, the reversible phosphorylation of BAD represents a point of convergence for diverse survival–apoptosis signaling pathways.
The integrated actions of kinases and phosphatases are likely to regulate the phosphorylation of BAD in response to survival and apoptosis signaling. Serine–threonine phosphatases have been shown to be involved in the regulation of BAD phosphorylation. PP2B (calcineurin) acts as a BAD phosphatase in neuronal cells.23 In a murine T-cell line, BAD was shown to be a substrate for PP1α.24
We addressed the role of serine–threonine phosphatases in regulating BAD dephosphorylation in IL-3–dependent FL5.12 lymphoid cells—the cells in which the proapoptotic function of BAD was originally reported. We used a panel of phosphatase inhibitors with differential selectivity against PP1 and PP2A, including okadaic acid (OA), fostriecin, calyculin A, and tautomycin, and inhibitors against PP2B, cyclosporin A, and FK506. BAD dephosphorylation was investigated in cells and in cell-free extracts, and the role of BAD phosphatase in cell death was examined. Our findings revealed that the BAD phosphatase activity in FL5.12 cells was PP2A or a PP2A-like enzyme. We found that the dissociation of 14-3-3 from BAD was required for BAD dephosphorylation in vitro, suggesting that 14-3-3 actively protects BAD against dephosphorylation in vivo. Moreover, the inhibition of BAD phosphatase activity rescued FL5.12 cells from IL-3 deprivation-induced cell death. These data indicate that PP2A, or a PP2A-like phosphatase, catalyzes BAD dephosphorylation and regulates its proapoptotic activity in this model of lymphoid progenitor cells, and they implicate 14-3-3 binding in the regulation of BAD dephosphorylation.
Materials and methods
Materials
Mouse anti-BAD monoclonal antibodies B31420 and B36420 were purchased from Transduction Laboratory (Lexington, KY). Rabbit anti-BAD antibody (Ab) (C20) and anti-14-3-3β (K-19) Ab were purchased from Santa Cruz Biotechnology (La Jolla, CA). Antiphospho Ser112 Ab was from Upstate Biotechnology (Lake Placid, NY). Serine–threonine phosphatase inhibitors, OA, calyculin A, and tautomycin were purchased from Alexis Biochemicals (San Diego, CA). Rabbit muscle phosphorylase a, phosphorylase kinase (rabbit muscle), protein kinase C, histone H1 (type III-S calf thymus), and microcystin-LR are from Sigma (St Louis, MO). A23187 was purchased from Calbiochem (San Diego, CA). Cyclosporin A and FK506 were obtained from the pharmacy of Vanderbilt University Medical Center, Nashville, TN. Fostriecin was a kind gift from Parke Davis Pharmaceuticals (Ann Arbor, MI). Recombinant human PP1 inhibitor I-2 was purified as described.25
Cells and cultures
FL5.12 cells were cultured in Iscove modified Dulbecco medium supplemented with 10% fetal bovine serum and IL-3. FL5.12 BCL-XL/BAD and FL5.12BCL-XL/BAD112A136A cell lines were constructed by infecting FL5.12 BCL-XL cells with retroviruses containing pBabeBAD or pBabeBAD112A136A. Two days later cells were selected with 4 μg/mL puromycin, and single-cell clones were isolated after 4 days. In IL-3 deprivation experiments, FL5.12 BCL-XL/BAD cells were washed with phosphate-buffered saline 3 times and recultured in media lacking IL-3. For treatment of cells in culture, each phosphatase inhibitor or the same volume of vehicle was added to the media, as stated in the figures.
Plasmid and retrovirus preparation
The cDNAs of wild-type or phosphorylation-defective mutant BAD112A136A with serine-to-alanine substitutions at Ser112 and Ser136 were cloned in the retroviral expression vector pBabepuro26 and transfected into BOSC or φNX cells by the calcium phosphate method, as described.27 Viral supernatants were collected 2 days later and filtered through 0.45-μm filters.
Western blotting and immunoprecipitation
Cell lysates for Western blotting were prepared in radioimmunoprecipitation assay (RIPA) buffer (100 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholic acid, 0.1% sodium dodecyl sulfate (SDS), 50 mM Tris-Cl, pH 8.0, and 1 mM EDTA). For immunoprecipitation, 107 cells were lysed in isotonic immunoprecipitation (IP) buffer (142.5 mM KCl, 5 mM MgCl2, 10 mM HEPES, and 0.25% Nonidet P-40) with or without 0.1 μM phosphatase inhibitor microcystin-LR, incubated with polyclonal anti-BAD Ab C20, precipitated with protein A–Sepharose, fractionated by 12.5% SDS-PAGE, and transferred to polyvinylidene difluoride membrane. To immunoprecipitate BAD without 14-3-3, 1% empigen BB or 25 μM R18 peptide (PHCVPRDLSWLDLEANMCLP)28 29 was added to the IP buffer. For sequential experiments, R18 was added to the BAD immunoprecipitate, followed by 3 washes with isotonic IP buffer. Western blots were developed using enhanced chemiluminescence.
In vitro BAD phosphatase assay
For analyses of BAD dephosphorylation in whole cell homogenates, cell extracts were prepared in phosphatase assay buffer A (50 mM Tris-Cl, 150 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.25% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 10 μg/mL aprotinin, and 10 μg/mL leupeptin),13 and incubated on ice with or without empigen BB for 30 minutes. Cell lysates were centrifuged at 14 000 rpm for 30 minutes at 4°C, and the supernatant was further incubated at 30°C for 30 minutes with or without the addition of R18 peptide. For dephosphorylation of BAD by PP2A, BAD was immunoprecipitated from 107 FL5.12BCL-XL/BAD cells and incubated in phosphatase assay buffer B (20 mM imidazole, 150 mM NaCl, 1% empigen BB, 14.4 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, 10 μg/mL aprotinin, and 10 μg/mL leupeptin)30 at 4°C for 30 minutes. Purified PP2A catalytic subunit (0.01 mg/mL) from bovine testes was added to the reaction mixture and incubated at 30°C for 30 minutes. Phosphatase reactions were terminated by adding 4 × SDS sample buffer and boiling for 5 minutes. For endogenous BAD phosphatase activity, 40 μg FL5.12 cell lysates prepared by sonication in phosphatase assay buffer B without empigen were preincubated with or without PP1 inhibitor 2 (I-2) at 1.6 μM final concentration, then added to immunoprecipitated BAD. Phosphatase reactions were performed as described above.
PP1 and PP2A phosphatase activity assays
Rabbit muscle phosphorylase a labeled by phosphorylase kinase and protein kinase C-phosphorylated histone H1 (type III-S calf thymus) were prepared as described.31,32 The phosphatase activity against labeled phosphorylase a or histone H1 was measured by quantitating the release of 32P by scintillation counting. The assay was limited to a maximum of 20% release of total 32P to ensure linearity of the phosphatase reaction. FL5.12 BCL-XL/BAD cells were treated with 500 nM OA, 5 μM fostriecin, or vehicle alone for 3 hours and were sonicated in phosphatase buffer B without empigen; 2.5 μg or 0.25 μg cell lysate was used for the dephosphorylation of labeled phosphorylase a or histone H1, respectively. Lysates were incubated with an additional 5 nM or 1 μM OA for 15 minutes on ice before the labeled substrates were added. PP2A activity is defined as the activity inhibited by 5 nM OA, and PP1 activity is defined as the activity insensitive to 5 nM OA but inhibited by 1 μM OA.33
Cell survival assay
Calyculin A (5 nM) was added to the media of FL5.12 BCL-XL/BAD cells or FL5.12 BCL-XL/BAD112A136A cells for 4 hours in the presence of IL-3, followed by IL-3 withdrawal. Cell viability was measured by trypan blue exclusion.
Results
Okadaic acid inhibits BAD phosphatase activity
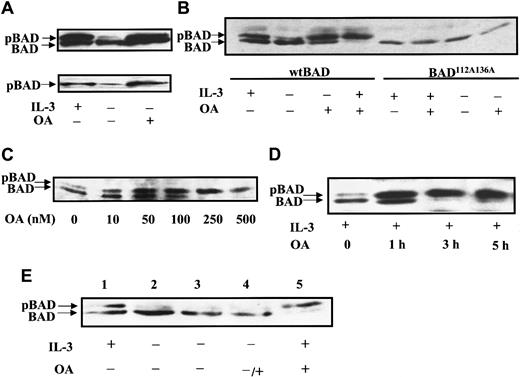
In the presence of survival factors, both phosphorylated and dephosphorylated BAD are present, forming a doublet on SDS–polyacrylamide gel electrophoresis (PAGE). The slower migrating band represents BAD phosphorylated at Ser112 and Ser136 (hyper-phosphorylated BAD), and the faster migrating band contains mainly the dephosphorylated form and a minor amount of BAD phosphorylated at a single site.7 In the absence of survival factor IL-3, FL5.12 cells undergo apoptosis, and BAD is dephosphorylated. Because high levels of BAD cause cell death, cells overexpressing both BCL-XL and BAD were used in our studies. To explore which serine–threonine phosphatases are involved in BAD dephosphorylation, we treated FL5.12 cells expressing BCL-XL and BAD (FL5.12BCL-XL/BAD) with the potent PP1 and PP2A phosphatase inhibitor okadaic acid. During IL-3 withdrawal, BAD was seen on SDS-PAGE as mostly the faster migrating, dephosphorylated band (Figure 1A, upper panel). When OA was added to the culture media, the slower migrating hyper-phosphorylated species of BAD persisted even during IL-3 starvation. An antibody specific to phosphorylated Ser112 of BAD confirmed that the slower migrating BAD was phosphorylated and that the faster migrating BAD was not phosphorylated at Ser112 (Figure 1A, lower panel). In the presence of both IL-3 and OA, only hyper-phosphorylated BAD was observed (Figure 1B). The inhibition of BAD dephosphorylation suggested the presence of an OA-sensitive BAD phosphatase activity. A mutant BAD with alanine substitutions at residues Ser112 and Ser136 cannot be phosphorylated at these sites and is more potent than wild-type BAD in promoting cell death. OA treatment of a stable FL5.12 cell line expressing BCL-XL and this phosphorylation-defective mutant BAD did not result in mobility shifts indicative of phosphorylation (Figure 1B). This confirmed that the enhanced BAD phosphorylation caused by OA occurred at Ser112 and Ser136, sites involved in the regulation of apoptosis.
Okadaic acid inhibits BAD dephosphorylation.
(A) FL5.12BCL-XL/BAD cells were treated with 0.5 μM OA or dimethyl sulfoxide (DMSO) for 4 hours in the presence (+) or absence (−) of IL-3. Cell lysates were analyzed by Western blotting. The blot was first probed with monoclonal anti-BAD Ab 31420 (upper panel), then reprobed with antiphospho serine 112 BAD Ab (lower panel). pBAD, phosphorylated BAD. (B) FL5.12BCL-XL/BAD (wt BAD) and cells expressing the phosphorylation-defective BAD mutant (BAD112A136A) were treated with 0.5 μM OA with (+) or without (−) IL-3 for 3 hours, and analyzed by Western blotting using polyclonal anti-BAD Ab C20. (C) FL5.12BCL-xL/BAD cells were treated with indicated doses of OA in the presence of IL-3 for 3 hours and analyzed by Western blotting using anti-BAD Ab 31420. (D) Cells were treated with 0.5 μM OA for the times indicated and analyzed by Western blotting as above. (E) BAD phosphorylation induced by OA is the result of inhibition of BAD phosphatase activity, not indirect activation of kinases. FL5.12BCL-XL/BAD cells were grown in the presence (lanes 1, 5) or absence of IL-3 (lanes 2-4) for 3 hours. Cells in lanes 3 to 5 were then treated with DMSO (lane 3) or 0.5 μM OA (lanes 4, 5) for another 3 hours. Lysates were analyzed by Western blotting as above.
Okadaic acid inhibits BAD dephosphorylation.
(A) FL5.12BCL-XL/BAD cells were treated with 0.5 μM OA or dimethyl sulfoxide (DMSO) for 4 hours in the presence (+) or absence (−) of IL-3. Cell lysates were analyzed by Western blotting. The blot was first probed with monoclonal anti-BAD Ab 31420 (upper panel), then reprobed with antiphospho serine 112 BAD Ab (lower panel). pBAD, phosphorylated BAD. (B) FL5.12BCL-XL/BAD (wt BAD) and cells expressing the phosphorylation-defective BAD mutant (BAD112A136A) were treated with 0.5 μM OA with (+) or without (−) IL-3 for 3 hours, and analyzed by Western blotting using polyclonal anti-BAD Ab C20. (C) FL5.12BCL-xL/BAD cells were treated with indicated doses of OA in the presence of IL-3 for 3 hours and analyzed by Western blotting using anti-BAD Ab 31420. (D) Cells were treated with 0.5 μM OA for the times indicated and analyzed by Western blotting as above. (E) BAD phosphorylation induced by OA is the result of inhibition of BAD phosphatase activity, not indirect activation of kinases. FL5.12BCL-XL/BAD cells were grown in the presence (lanes 1, 5) or absence of IL-3 (lanes 2-4) for 3 hours. Cells in lanes 3 to 5 were then treated with DMSO (lane 3) or 0.5 μM OA (lanes 4, 5) for another 3 hours. Lysates were analyzed by Western blotting as above.
To test whether OA inhibited BAD dephosphorylation in a dose- and time-dependent manner, FL5.12BCL-XL/BAD cultures were treated with increasing concentrations of OA and assayed for BAD phosphorylation by Western blot analysis (Figure 1C). A predominant dephosphorylated band and a minor hyper-phosphorylated band were seen in untreated cultures. In the presence of IL-3 and 10 nM OA, a significant increase in hyper-phosphorylated BAD was observed. Maximal BAD hyper-phosphorylation occurred in cells treated with 250 nM OA. Okadaic acid also inhibited BAD dephosphorylation in a time-dependent manner (Figure 1D). The intensity of the hyper-phosphorylated band relative to the dephosphorylated band increased significantly after 1 hour of incubation with 500 nM OA. After 3 hours, only hyper-phosphorylated BAD could be detected.
Phosphatases are known to regulate kinases, and the inhibition of phosphatases by OA can affect multiple cellular signaling pathways.34-37 To test the hypothesis that BAD hyper-phosphorylation induced by OA is due to the inhibition of BAD phosphatase activity and not to the indirect activation of kinases, we asked whether OA promoted the phosphorylation of BAD that had previously been dephosphorylated. FL5.12BCL-XL/BAD cells were deprived of IL-3 for 3 hours until BAD was completely dephosphorylated and then were treated with OA for another 3 hours (Figure 1E). No increase in hyper-phosphorylated BAD was detected. In contrast, OA treatment of cells grown in the presence of IL-3 shifted BAD to its fully phosphorylated form. Thus, OA could only promote the conversion of BAD to its hyper-phosphorylated form when BAD kinases were activated by survival factors such as IL-3. This suggests that OA did not activate BAD kinases but that it inhibited a phosphatase that dephosphorylates BAD.
PP2A-selective inhibitors enhanced BAD phosphorylation in vivo
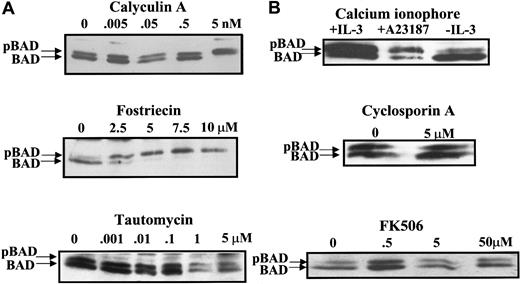
Three major serine–threonine phosphatases—PP1, PP2A, and PP2B—are found in eukaryotic cells. Although OA inhibits both PP1 and PP2A, it inhibits PP2A at concentrations 10 to 100 times lower than those required to inhibit PP1.35 To characterize the BAD phosphatase activity in FL5.12 cells, an array of phosphatase inhibitors was investigated as modulators of BAD phosphorylation. Calyculin A is an inhibitor that shows a slight preference for PP1 over PP2A in vitro. At a concentration as low as 5 nM in the media, calyculin A promoted the complete phosphorylation of BAD (Figure2A, upper panel). Fostriecin, an antitumor antibiotic,38-40 is a highly selective inhibitor of PP2A-like enzymes and inhibits PP2A at 10 000 to 40 000 times lower concentration than that required for PP1 inhibition in vitro.41,42 Fostriecin increased BAD phosphorylation at 2.5 μM. At 5 μM, fostriecin caused all detectable BAD to be phosphorylated (Figure 2A, middle panel). In contrast, tautomycin, a selective inhibitor against PP1 in vitro43 and in vivo,33 44 showed no effect on BAD phosphorylation at concentrations up to 5 μM (Figure 2A, bottom panel). Results from these studies suggested that the BAD phosphatase in FL5.12 cells is most likely PP2A or a PP2A-like enzyme.
PP2A-selective inhibitors block BAD dephosphorylation in vivo.
(A) FL5.12BCL-XL/BAD cells were treated with PP1 and PP2A inhibitors calyculin A, fostriecin, and tautomycin in the presence of IL-3 for 3 hours. BAD was analyzed by Western blotting as in Figure1. (B) Cells were treated with an activator (2 μM A23187) or an inhibitor (cyclosporin A or FK506) of PP2B (calcineurin) and analyzed for BAD by Western blotting.
PP2A-selective inhibitors block BAD dephosphorylation in vivo.
(A) FL5.12BCL-XL/BAD cells were treated with PP1 and PP2A inhibitors calyculin A, fostriecin, and tautomycin in the presence of IL-3 for 3 hours. BAD was analyzed by Western blotting as in Figure1. (B) Cells were treated with an activator (2 μM A23187) or an inhibitor (cyclosporin A or FK506) of PP2B (calcineurin) and analyzed for BAD by Western blotting.
Calcium-induced apoptosis in hippocampal neurons was reported to occur through calcineurin (PP2B)-mediated dephosphorylation of BAD.23 Thus, we addressed the possible role of calcineurin in BAD dephosphorylation in FL5.12 cells (Figure 2B). The calcium ionophore A23187, which activates calcineurin, did not promote BAD dephosphorylation. Conversely, treatment of FL5.12 cells with the calcineurin inhibitors cyclosporin A and FK506 did not increase BAD phosphorylation. Similarly, no change in BAD phosphorylation was observed in mouse embryo fibroblasts treated with high extracellular calcium, the calcium ionophore A23187, or calcineurin inhibitors, whereas OA caused a complete shift of BAD to the hyper-phosphorylated form (data not shown). These findings indicated that calcineurin does not play a significant role in BAD dephosphorylation in FL5.12 cells and mouse embryo fibroblasts.
Inhibition of BAD phosphatase activity in FL5.12 cells is coincident with PP2A inhibition
When OA and other serine–threonine phosphatase inhibitors are used to treat cells44 or are injected into mice,45 the inhibitors have been shown to form stable complexes with their target enzymes in cell-free extracts or tissue homogenates. The tight association between phosphatase inhibitors and their target enzymes has facilitated in vitro measurement of the PP1 and PP2A activities in lysates of cells treated with different inhibitors.33,44 To confirm that treatment of FL5.12 cultures with PP2A inhibitors resulted in selective inhibition of the expected phosphatase activity in cells, PP1 and PP2A activities were measured in lysates of FL5.12BCL-XL/BAD cells treated with inhibitors using either 32P-labeled phosphorylase a or32P-labeled histone H1 as substrates. PP2A and PP2A-like activities are defined as the phosphatase activity inhibited by 5 nM OA, and PP1 activity is defined as the phosphatase activity resistant to 5 nM OA but sensitive to 1 μM OA.33
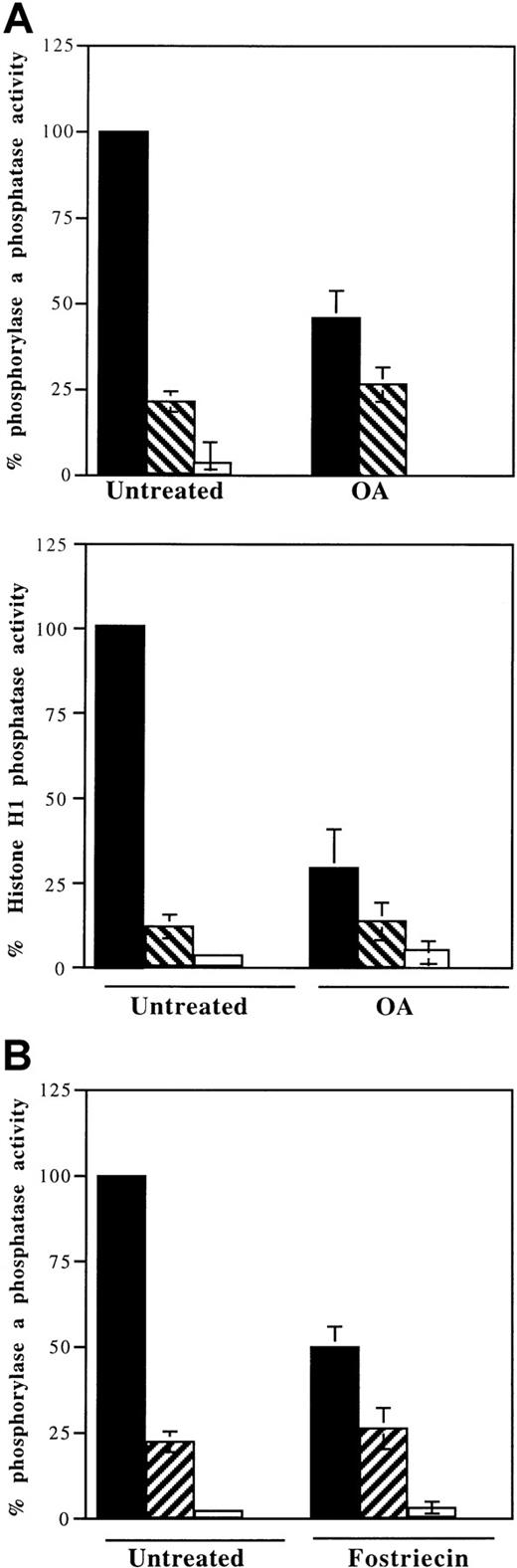
Using phosphorylase a, a substrate for both PP1 and PP2A, we found that most of the serine–threonine phosphatase activity in untreated FL5.12BCL-XL/BAD cells was inhibited by adding 5 nM OA to the lysate. This represented the activity of PP2A and PP2A-like enzymes (Figure 3A, upper panel, left). The remainder of the total phosphatase activity in untreated cells was inhibited by 1 μM OA and represented PP1 activity. When FL5.12BCL-XL/BAD cultures were treated with 0.5 μM OA for 3 hours, which effectively inhibited BAD dephosphorylation, the phosphorylase phosphatase activity was reduced by 54% (Figure 3A, upper panel, right). Addition of 5 nM OA to the lysates decreased the activity by another 20%, indicating that most of the PP2A activity was inhibited by OA treatment. The remaining phosphatase activity was similar to the PP1 activity in lysates from untreated cells and was completely inhibited by the further addition of 1 μM OA, indicating that OA treatment did not significantly affect PP1 activity. Therefore, treatment of FL5.12 cells with 0.5 μM OA, the concentration which abrogated BAD dephosphorylation, inhibited most PP2A activity while not significantly affecting PP1.
PP2A activity is significantly inhibited by in vivo OA and fostriecin treatment.
(A) Phosphatase activity in lysates of cells treated with 0.5 μM OA (OA) using either 32P-labeled phosphorylase a (upper panel) or 32P-labeled histone H1 (lower panel) as substrate. (B) Phosphatase activity in lysates of cells treated with 5 μM fostriecin (fostriecin) using 32P-labeled phosphorylase a as substrate. Cells exposed to vehicle alone (untreated) were used as controls. Assays were performed with further addition of vehicle alone (▪), 5 nM OA (▨), or 1 μM OA (■) to the lysates. Activities were expressed as a percentage of 32P release in untreated lysates. Data shown were mean ± SD of triplicate assays from at least 2 separate experiments.
PP2A activity is significantly inhibited by in vivo OA and fostriecin treatment.
(A) Phosphatase activity in lysates of cells treated with 0.5 μM OA (OA) using either 32P-labeled phosphorylase a (upper panel) or 32P-labeled histone H1 (lower panel) as substrate. (B) Phosphatase activity in lysates of cells treated with 5 μM fostriecin (fostriecin) using 32P-labeled phosphorylase a as substrate. Cells exposed to vehicle alone (untreated) were used as controls. Assays were performed with further addition of vehicle alone (▪), 5 nM OA (▨), or 1 μM OA (■) to the lysates. Activities were expressed as a percentage of 32P release in untreated lysates. Data shown were mean ± SD of triplicate assays from at least 2 separate experiments.
Phosphatase activity was also measured using PKC-phosphorylated histone H1 as a PP2A-selective substrate31 (Figure 3A, bottom panel). In lysates of cells treated with 0.5 μM OA, 70% of total histone H1 phosphatase activity was inhibited (Figure 3A, bottom panel, right). Only approximately 15% more was inhibited by the further addition of 5 nM OA to the lysates (Figure 3A, bottom panel), again indicating that most PP2A activity was inhibited when BAD dephosphorylation was maximally inhibited in FL5.12 cells.
Next, phosphorylase phosphatase activity was measured in lysates from fostriecin-treated cells (Figure 3B). More than 50% of total cellular phosphatase activity was inhibited after fostriecin treatment, and 25% more activity was inhibited by the addition of 5 nM OA to the lysates (Figure 3B, right). Further addition of 1 μM OA reduced the phosphatase activity to background (Figure 3B, right). This indicated that treatment with fostriecin, at the dose that blocked BAD dephosphorylation, also inhibited most PP2A and PP2A-like activities but left PP1 activity largely unaffected.
In summary, the concentrations of PP2A inhibitors used to inhibit BAD dephosphorylation in vivo effectively were shown to selectively inhibit most intracellular PP2A, but not PP1, activity in FL5.12 cells. These results established the BAD phosphatase as PP2A, or a member of the PP2A subfamily, in FL5.12 cells.
14-3-3 inhibits BAD dephosphorylation in vitro
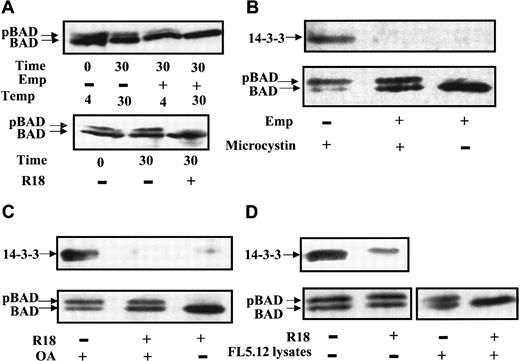
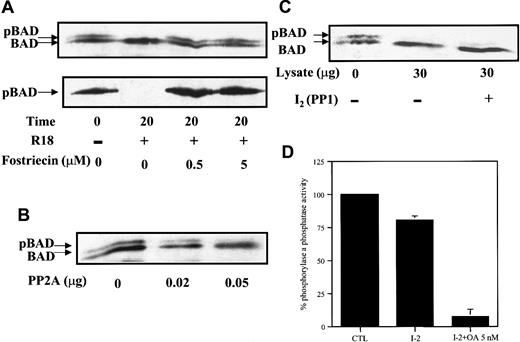
In examining BAD dephosphorylation in vitro, cell lysates were prepared in buffer containing a mild detergent but no phosphatase inhibitors, followed by incubation at 30°C, SDS-PAGE, and Western blot analysis. Surprisingly, no significant BAD dephosphorylation was observed under these conditions (data not shown). Because 14-3-3 binds to the phosphorylated sites of BAD, we asked whether 14-3-3 played a role in preventing BAD dephosphorylation. Our BAD phosphatase assay was thus modified by adding either empigen BB (Emp), a zwitterionic detergent that has been used to dissociate 14-3-3 from phosphorylated keratins,46 or R18, a peptide (PHCVPRDLSWLDLEANMCLP) shown to displace 14-3-3 targets by interacting with basic residues in the amphipathic groove of 14-3-3.28 29 Both empigen (Figure4A, upper panel) and R18 (Figure 4A, bottom panel) enhanced BAD dephosphorylation, suggesting that the removal of 14-3-3 from BAD facilitated phosphatase activity on BAD. Immunoprecipitations of BAD from FL5.12 lysates confirmed that 14-3-3 dissociated from BAD under these conditions (Figure 4B,C). In the absence of empigen, abundant 14-3-3 coprecipitated with BAD (Figure 4B, upper panel). In the presence of 1% empigen and the potent serine–threonine phosphatase inhibitor microcystin, no 14-3-3 was detected in the BAD immunocomplexes, and phosphorylation of BAD was maintained (Figure 4B, upper panel). In contrast, in the presence of empigen but in the absence of a phosphatase inhibitor, significant BAD dephosphorylation occurred (Figure 4B, lower panel). Similarly, the specific 14-3-3 displacement peptide R18 facilitated BAD dephosphorylation in the absence of a phosphatase inhibitor (Figure 4C).
14-3-3 prevents BAD dephosphorylation in vitro.
(A) FL5.12BCL-XL/BAD lysates were prepared in phosphatase buffer A with or without 1% empigen BB (Emp) (upper panel) or 25 μM R18 peptide (lower panel). BAD dephosphorylation was carried out for 30 minutes at 4°C or 30°C, followed by Western blotting for BAD. (B) BAD was immunoprecipitated by polyclonal C20 Ab from FL5.12BCL-XL/BAD cells in isotonic IP buffer with or without 1% empigen BB (Emp), in the presence or absence of 0.1 μM microcystin, and the immunocomplexes were analyzed by Western blotting. BAD was detected by monoclonal Ab 31420. The same blot was probed for 14-3-3 using polyclonal Ab K-19. (C) BAD was immunoprecipitated in isotonic buffer with or without 25 μM R18 peptide, in the presence or absence of 1 μM okadaic acid (OA), and analyzed by Western blotting as in panel B. (D) BAD was immunoprecipitated in isotonic buffer, R18 peptide was or was not added to the BAD immunocomplex, which was washed, and 40 μg FL5.12 lysate was added and incubated at 30°C in phosphatase buffer B. Reaction products were analyzed by Western blotting for BAD and 14-3-3 as in panel B.
14-3-3 prevents BAD dephosphorylation in vitro.
(A) FL5.12BCL-XL/BAD lysates were prepared in phosphatase buffer A with or without 1% empigen BB (Emp) (upper panel) or 25 μM R18 peptide (lower panel). BAD dephosphorylation was carried out for 30 minutes at 4°C or 30°C, followed by Western blotting for BAD. (B) BAD was immunoprecipitated by polyclonal C20 Ab from FL5.12BCL-XL/BAD cells in isotonic IP buffer with or without 1% empigen BB (Emp), in the presence or absence of 0.1 μM microcystin, and the immunocomplexes were analyzed by Western blotting. BAD was detected by monoclonal Ab 31420. The same blot was probed for 14-3-3 using polyclonal Ab K-19. (C) BAD was immunoprecipitated in isotonic buffer with or without 25 μM R18 peptide, in the presence or absence of 1 μM okadaic acid (OA), and analyzed by Western blotting as in panel B. (D) BAD was immunoprecipitated in isotonic buffer, R18 peptide was or was not added to the BAD immunocomplex, which was washed, and 40 μg FL5.12 lysate was added and incubated at 30°C in phosphatase buffer B. Reaction products were analyzed by Western blotting for BAD and 14-3-3 as in panel B.
To establish that the dissociation of 14-3-3 was required before BAD dephosphorylation, the BAD immunocomplex was first incubated without or with R18 to remove 14-3-3, then subjected to dephosphorylation using FL5.12 lysates as the source of cellular phosphatases (Figure 4D). When 14-3-3 was present in the BAD immunocomplex, the addition of FL5.12 lysate did not stimulate dephosphorylation of BAD. However, the dissociation of most 14-3-3 from the BAD immunocomplex by R18, followed by incubation with FL5.12 extracts, resulted in BAD dephosphorylation. These experiments demonstrated that dissociation of 14-3-3 from BAD is essential for efficient BAD dephosphorylation.
PP2A-selective inhibitor, but not PP1-specific inhibitor, inhibits BAD dephosphorylation in vitro
Using either empigen or R18 to dissociate 14-3-3, we were able to assess the sensitivity of the BAD phosphatase to selective inhibitors in whole cell homogenates. In the presence of R18, BAD was completely dephosphorylated within 20 minutes (Figure5A, upper panel). Addition of 0.5 μM fostriecin to lysates containing R18 completely inhibited BAD dephosphorylation (Figure 5A, upper panel), as confirmed by the antiphospho–Ser112-specific antibody (Figure 5A, bottom panel). The minimal effective concentration of fostriecin required to inhibit BAD phosphatase activity under these conditions was approximately 100 times higher than the IC50 for the purified PP2A catalytic subunit but was at least approximately 200 times lower than the IC50 for PP1.42 Similarly, 100 nM OA completely inhibited BAD phosphatase in vitro (data not shown). The response of the BAD phosphatase activity in these studies is consistent with that of PP2A, or a PP2A-like enzyme. The ability of PP2A to dephosphorylate BAD in vitro was confirmed using purified PP2A catalytic subunit on immunoprecipitated BAD (Figure 5B).
PP2A-selective inhibitor, but not PP1-specific inhibitor, inhibits BAD dephosphorylation in vitro.
(A) FL5.12BCL-XL/BAD lysates were prepared in phosphatase buffer A with or without fostriecin, in the presence or absence of 25 μM R18 peptide, incubated at 30°C for 20 minutes, and analyzed by Western blotting—first for BAD with monoclonal Ab 31420 (upper panel) and then reprobed with polyclonal antiphosphoserine 112 Ab (lower panel). (B) PP2A dephosphorylated BAD in vitro. Immunoprecipitated BAD was incubated with purified PP2A and analyzed by Western blotting. (C) FL5.12 lysates with or without the addition of 1.6 μM of the PP1 inhibitor I-2 were used as the source of phosphatase to react with immunoprecipitated BAD. (D) FL5.12 lysates preincubated with 1.6 μM inhibitor I-2 for 15 minutes without (I-2) or with the addition of 5 nM OA (I-2+OA 5 nM) were assayed for phosphatase activity using 32P-labeled phosphorylase a as substrate. Results were expressed as a percentage of 32P release in lysates with no additions (CTL) and represent mean ± SD of triplicate assays.
PP2A-selective inhibitor, but not PP1-specific inhibitor, inhibits BAD dephosphorylation in vitro.
(A) FL5.12BCL-XL/BAD lysates were prepared in phosphatase buffer A with or without fostriecin, in the presence or absence of 25 μM R18 peptide, incubated at 30°C for 20 minutes, and analyzed by Western blotting—first for BAD with monoclonal Ab 31420 (upper panel) and then reprobed with polyclonal antiphosphoserine 112 Ab (lower panel). (B) PP2A dephosphorylated BAD in vitro. Immunoprecipitated BAD was incubated with purified PP2A and analyzed by Western blotting. (C) FL5.12 lysates with or without the addition of 1.6 μM of the PP1 inhibitor I-2 were used as the source of phosphatase to react with immunoprecipitated BAD. (D) FL5.12 lysates preincubated with 1.6 μM inhibitor I-2 for 15 minutes without (I-2) or with the addition of 5 nM OA (I-2+OA 5 nM) were assayed for phosphatase activity using 32P-labeled phosphorylase a as substrate. Results were expressed as a percentage of 32P release in lysates with no additions (CTL) and represent mean ± SD of triplicate assays.
To exclude the possibility that PP1 was involved in BAD dephosphorylation in whole cell extracts, we added the PP1-specific protein inhibitor I-2. I-2, an endogenous PP1 regulator found in most mammalian cells, inhibits PP1 activity with a Ki≈ 3.1 nM and has little or no effect on PP2A activity.47 Using immunoprecipitated BAD as a substrate and FL5.12 lysates as the source of phosphatases, we assessed the dephosphorylation of BAD by Western blot analysis. Addition of the PP1 inhibitor I-2 to FL5.12 lysates had no effect on BAD dephosphorylation (Figure 5C), indicating that the major BAD phosphatase was an I-2–insensitive phosphatase. We used32P-labeled phosphorylase as substrate to confirm that PP1 was effectively inhibited in FL5.12 lysates treated with I-2 (Figure5D). Approximately 20% of the total phosphorylase phosphatase activity was inhibited by I-2 (Figure 5D, I-2), consistent with the amount of PP1 activity shown in earlier experiments to be present in FL5.12 cells (see Figure 3). Further addition of 5 nM OA reduced the phosphatase activity to background level (Figure 5D, I-2+OA 5 nM), indicating that the remaining activity was owing to PP2A and PP2A-like enzymes. Thus, in vivo and in vitro data suggested that PP1 was not a significant BAD phosphatase in FL5.12 cells.
Inhibition of serine–threonine phosphatases enhanced the survival of FL5.12 cells expressing wild-type BAD, but not phosphorylation-defective mutant BAD, after IL-3 withdrawal
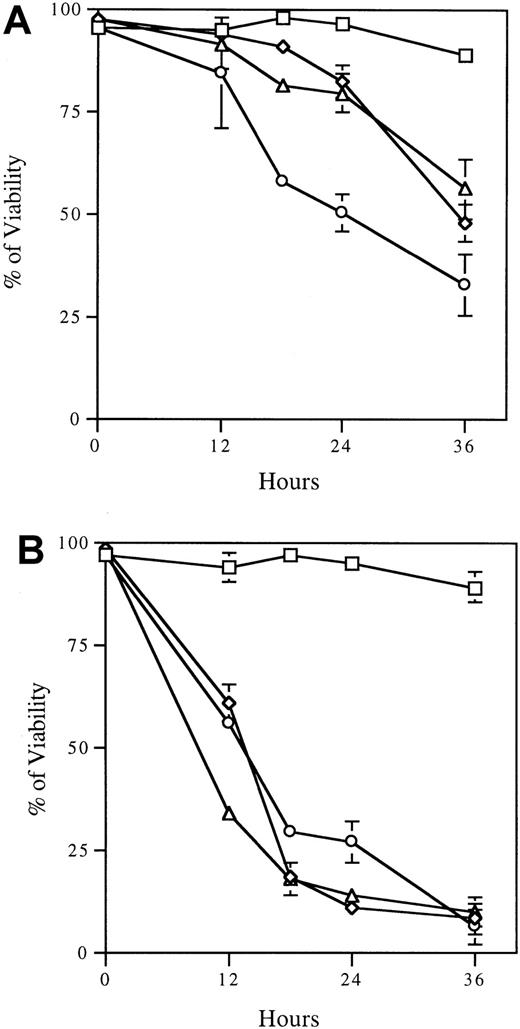
Phosphorylation inactivates the proapoptotic function of BAD by causing it to dissociate from mitochondrial BCL-2 or BCL-XLand to associate with 14-3-3 in the cytosol.7 To assess the physiologic relevance of the BAD phosphatase in apoptosis, we asked whether the inhibition of BAD dephosphorylation enhances cell survival in the absence of survival factors. We initially observed rescue of cells by OA (data not shown); however, interpretation of those results was complicated by the cytotoxicity of OA during prolonged incubation. Therefore, we used calyculin A, which causes less death in these cells (Figure 6A). FL5.12 BCL-XL/BAD cells treated with 5 nM calyculin A and deprived of IL-3 exhibited a survival curve similar to that of cells treated with calyculin A in the presence of IL-3 (Figure 6A). Thus, a phosphatase inhibitor, at a dose shown to inhibit BAD phosphatase activity and to promote BAD hyper-phosphorylation (Figure 2A), rescued cells from IL-3 deprivation-induced apoptosis. In contrast, the phosphorylation-defective mutant FL5.12 BCL-XL/BAD112A136A was not rescued by calyculin A treatment during IL-3 withdrawal (Figure6B), confirming that rescue of survival was specific to the inhibition of BAD dephosphorylation. Importantly, rescue of IL-3 withdrawal-induced cell death by calyculin A suggested that BAD phosphatase plays a critical role in the induction of apoptosis in FL5.12 cells.
Serine/threonine phosphatase inhibitor calyculin A rescues FL5.12 cells expressing BAD from apoptosis after IL-3 withdrawal.
Cells expressing wild-type BAD (A) or the phosphorylation-defective mutant BAD (B) were treated with 5 nM calyculin A (⋄, ▵) or vehicle alone (■, ○) for 4 hours. Cells were then cultured in the presence (⋄, ■) or absence (▵, ○) of IL-3. Viability was assayed by trypan blue exclusion at the indicated times after IL-3 withdrawal. Data are mean ± SD of triplicate assays and are representative of 3 experiments.
Serine/threonine phosphatase inhibitor calyculin A rescues FL5.12 cells expressing BAD from apoptosis after IL-3 withdrawal.
Cells expressing wild-type BAD (A) or the phosphorylation-defective mutant BAD (B) were treated with 5 nM calyculin A (⋄, ▵) or vehicle alone (■, ○) for 4 hours. Cells were then cultured in the presence (⋄, ■) or absence (▵, ○) of IL-3. Viability was assayed by trypan blue exclusion at the indicated times after IL-3 withdrawal. Data are mean ± SD of triplicate assays and are representative of 3 experiments.
Discussion
The dephosphorylation of BAD activates its proapoptotic function. In IL-3–dependent FL5.12 cells, PP2A or a PP2A-like enzyme is found to regulate cell death by dephosphorylating BAD. Mammalian serine–threonine phosphatases have been classified into several subfamilies, including PP1, PP2A, Ca++-dependent PP2B, and Mg2+-dependent PP2C, based on primary structure, substrate specificity, cation requirements, and sensitivity to inhibitors.34-36 These phosphatases have been shown to regulate diverse cellular functions such as metabolism, cell cycle progression, T-lymphocyte activation,34-36 long-term potentiation of hippocampal neurons,48 and apoptosis.23,24,49-51 PP2A, in particular, represents a complicated group of enzymes that share biochemical properties but have different regulatory subunits that influence substrate specificity and intracellular targeting.52-54 The PP2A subfamily includes PP4, PP6, and other less well-characterized enzymes. In this study, we have characterized a BAD phosphatase activity based on its sensitivities to several different serine–threonine phosphatase inhibitors. Treatment of FL5.12 BCL-XL/BAD cells with OA, at a dose that inhibited PP2A but not PP1, promoted the complete phosphorylation of BAD in the presence of a survival factor and sustained BAD phosphorylation during IL-3 withdrawal (Figure 1). Fostriecin, a more selective inhibitor than OA against PP2A, also caused complete BAD phosphorylation in vivo and inhibited BAD phosphatase activity in vitro at concentrations at least approximately 200 times lower than the IC50 for PP1 (Figures 2, 5). Calyculin A, a highly permeable phosphatase inhibitor, blocked BAD dephosphorylation in vivo at 5 nM (Figure 2). In contrast, tautomycin, which preferentially inhibits PP1 both in vitro and in vivo,33 44 did not affect BAD dephosphorylation in FL5.12 cells (Figure 2). Finally, inhibitor-2 (I-2), the physiologic regulator of PP1 activity, failed to inhibit BAD phosphatase activity in FL5.12 cell lysates (Figure 5). Taken together, these results suggest that the BAD phosphatase is either PP2A or a member of the PP2A subfamily.
PP2B (calcineurin) was shown to catalyze BAD dephosphorylation in hippocampal neurons in response to Ca++ influx and to promote apoptosis.23 In our studies, known calcineurin modulators had no significant effect on BAD phosphorylation or dephosphorylation in FL5.12 cells or mouse embryo fibroblasts. Moreover, BAD dephosphorylation was not impaired in vitro in buffers containing 0.2 mM EGTA or lacking calcium or calmodulin, suggesting that, in FL5.12 cells, PP2B is not the principal BAD phosphatase. More recently, BAD was also shown to be associated with, and dephosphorylated by, PP1α in the IL-2–dependent TS1αβ T-cell line.24 Our experiments using tautomycin in vivo and I-2 of PP1 in vitro indicated that PP1 also was not a major regulator of BAD activity in FL5.12 cells. In IL-3–dependent FL5.12 lymphoid cells, PP2A or a PP2A-like phosphatase is most likely the major enzyme responsible for BAD dephosphorylation. Identification of PP2A as a BAD phosphatase was unlikely to have been influenced by the abundance of PP2A in these cells because the treatment of NIH 3T3 cells, in which PP1 is the major phosphatase, with OA also caused BAD hyper-phosphorylation (data not shown). Moreover, the phosphorylation of CREB, a known PP1 target, was significantly increased in FL5.12 cells treated with the PP1-selective inhibitor tautomycin but not with OA (data not shown). These results support the specificity of PP2A or a PP2A-like enzyme as a BAD phosphatase in FL5.12 cells. The existence of distinct BAD phosphatases in different cells may be consistent with the ability of each cell type to respond to different survival factors or apoptotic stimuli.
In preliminary coimmunoprecipitations, PP2A was detected in the BAD immunocomplex; however, it was not consistently present when more stringent immunoprecipitation conditions were applied. In addition, BAD was not detected in microcystin pull-down assays. Because phosphorylated BAD is bound to 14-3-3 and becomes more susceptible to phosphatase activity only early in the apoptotic process, the BAD–phosphatase interaction is likely transient and may elude detection. It is also possible that the BAD phosphatase we characterized is a novel phosphatase belonging to the PP2A subfamily.
In characterizing the BAD phosphatase activity in vitro, we found that displacement of 14-3-3 from BAD was essential for its dephosphorylation by cellular phosphatases. This result provides data for a previously hypothesized role for 14-3-3 in protecting BAD from phosphatases.7,14,55 The 14-3-3 proteins may also influence cell survival by inhibiting dephosphorylation of apoptotic activators in addition to BAD, such as FKHRL1, which is also regulated by survival factor stimulation through phosphorylation and 14-3-3 binding.14 A similar role has been shown for 14-3-3 in protecting Raf-1 from inactivation by phosphatases in vitro.29,56 Dissociation of 14-3-3 from Raf-1, resulting from subsequent phosphorylation of 14-3-3 by PKC,57attenuates Raf-1 activation. Our data suggest that competitive binding between 14-3-3 and the cellular phosphatases for Ser112 may represent a coordinate mechanism for regulating BAD function. Our finding that dissociation of 14-3-3 was necessary before BAD dephosphorylation is also consistent with a model in which these events occur sequentially in vivo on apoptosis stimulation.
Inhibiting BAD dephosphorylation by blocking cellular phosphatase activity rescued FL5.12 cells from IL-3 withdrawal-induced cell death, suggesting that phosphatases play an active role in regulating cell death. Reversible phosphorylation of BAD is regulated by kinases such as Akt and PKA on survival factor stimulation and by phosphatases in cell death. In FL5.12 cells, IL-3 withdrawal causes the rapid inactivation of BAD kinases, but the rate at which phosphorylated BAD disappears in the absence of IL-3 exceeds its half-life in the presence of IL-3 (data not shown), arguing that BAD is actively dephosphorylated in the absence of IL-3.
In the presence of IL-3, both phosphorylated and dephosphorylated BAD are present, suggesting a balance between kinase and phosphatase activities. When BAD cannot be phosphorylated at Ser112 and Ser136, moderate stable expression can only be maintained by overexpression of a heterodimerizing partner, BCL-XL or BCL-2. The recent finding that Ser155 phosphorylation results in dissociation of BAD from BCL-XL and BCL-218-21 offers the possibility that the 112A136A mutant is phosphorylated at Ser155 in the presence of IL-3 and allows cell survival. Survival of cells expressing the phosphorylation defective 112A136A mutant in the presence of IL-3 indicates that BAD dephosphorylation is insufficient to cause cell death and suggests that other pathways are also involved. However, the ability of phosphatase inhibitors to rescue cells expressing wild-type BAD, but not phosphorylation-defective BAD, from apoptosis suggests that the BAD pathway is a major, but not an exclusive, one in IL-3 deprivation-induced cell death in these cells. Similarly, BAD is unlikely to be the only substrate of PP2A in cell death. Other relevant targets include BCL-2, which is dephosphorylated by mitochondrial PP2A.13 PP2A has also been implicated in the negative regulation of the survival kinase Akt,58 which likely also plays a role in the IL-3 signal transduction pathway.59
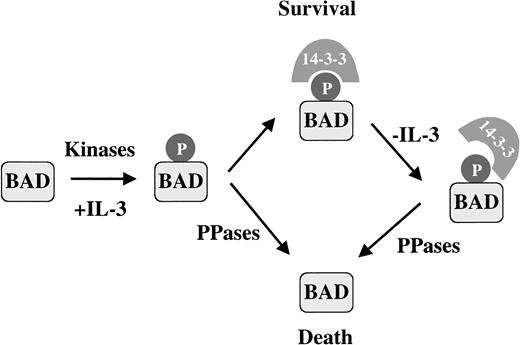
The BAD phosphatase activity we characterized is present constitutively, but the functional consequences of this activity may depend on the complex interactions between 14-3-3, BAD, kinases, and phosphatases in survival and apoptotic conditions. In vivo, 14-3-3 protects phosphorylated BAD from phosphatase activity when IL-3 is present. When apoptosis is stimulated by IL-3 withdrawal, changes in the interaction between 14-3-3 and BAD may favor the dephosphorylation of BAD. Although the mechanism of 14-3-3 dissociation is unknown, death signals could disrupt the 14-3-3–BAD complex by the modification of 14-3-3, perhaps through phosphorylation57 or cleavage of 14-3-3 by caspases.60 Additionally, increases in other 14-3-3 binding proteins could compete for BAD interaction,61 thus accelerating BAD dephosphorylation. Regulation of BAD phosphorylation involves a number of kinases and phosphatases that integrate various signal transduction pathways in different cell types to promote survival or to activate apoptosis. We propose that in FL5.12 cells, a death signal increases the susceptibility of BAD to the phosphatase by destabilizing the 14-3-3–BAD complex, leading to rapid dephosphorylation of BAD and in turn causing cell death (Figure 7).
Activation of the proapoptotic function of BAD by phosphatase in IL-3–dependent FL5.12 cells is regulated by 14-3-3 binding.
In the presence of survival factor IL-3, BAD is phosphorylated and binds 14-3-3. Dephosphorylated BAD is also present, presumably because of constitutively active phosphatases. We propose that an apoptotic stimulus such as IL-3 withdrawal destabilizes the 14-3-3–BAD interaction, increasing the susceptibility of BAD to PP2A or a PP2A-like phosphatase, promoting BAD dephosphorylation and cell death.
Activation of the proapoptotic function of BAD by phosphatase in IL-3–dependent FL5.12 cells is regulated by 14-3-3 binding.
In the presence of survival factor IL-3, BAD is phosphorylated and binds 14-3-3. Dephosphorylated BAD is also present, presumably because of constitutively active phosphatases. We propose that an apoptotic stimulus such as IL-3 withdrawal destabilizes the 14-3-3–BAD interaction, increasing the susceptibility of BAD to PP2A or a PP2A-like phosphatase, promoting BAD dephosphorylation and cell death.
We thank Dr Roger Colbran for advice on phosphatase inhibitor studies and Dr Haian Fu for helpful discussions on 14-3-3 and for kindly providing the R18 peptide. We thank Drs Jeffrey Rottman, Bart Lutterbach, and Lilin Wang for critical reading of the manuscript. We thank Ms Anuja Chattopadhyay for excellent technical assistance. We thank Dr Jeffrey Rottman for expert computer assistance.
Supported by a Grant-in-Aid from the American Heart Association, and supported in part by National Institutes of Health grant RO1CA78443 and grants from the V Foundation and Pfizer, Inc. Supported also by National Institutes of Health grant GM53165 (S.C.M., R.S.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Elizabeth Yang, Department of Cell Biology, Vanderbilt-Ingram Cancer Center, 525 MRB II, Vanderbilt Medical Center, Nashville, TN 37232; e-mail: elizabeth.yang@mcmail.vanderbilt.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal