Abstract
Human porphobilinogen deaminase (PBGD) is, reportedly, encoded by 2 distinct messenger RNAs (mRNAs) transcribing from a single gene. The ubiquitous form of the PBGD gene product is often used as an endogenous reference in gene expression studies because it is pseudogene free and has minimal transcriptional variability among tissues. A distinct erythroid-specific gene product has also been described because of the alternate splicing of the gene. Here is reported the existence of an additional erythroid-specific isoform of PBGD mRNA in primary cells.
Introduction
The third enzyme of the heme synthesis pathway, porphobilinogen deaminase (PBGD), is expressed as 2 isoforms: the so-called “housekeeping” form present in all cell types, and a second isoform expressed only in erythroid cells.1-4 The autosomal dominant disorder acute intermittent porphyria (AIP) is caused by mutations in the PBGD gene.5Clinically, AIP gene variants may or may not affect enzymatic levels within erythroid cells.3,6,7 One study reported low erythrocyte PBGD activity in blood donors, whereas lymphocyte PBGD activity was within the normal range.8 A possible mutation within the erythroid-specific part of thePBGD gene was proposed to explain this finding, but was not further explored. On the basis of the extensive genetic studies of AIP, genomic analyses have also shown that PBGD is a pseudogene-free, single-copy gene, thus making it a useful control for RNA analyses.9-14 We sought to use PBGD expression as both an erythroid-specific and housekeeping control for our studies of erythroid-cell gene expression patterns. Unexpectedly, our efforts revealed the existence of 2 distinct erythroid-specific PBGD messenger RNA (mRNA) transcripts.
Study design
Reverse transcriptase–polymerase chain reaction
Total RNA was purified from CD34+ cells collected on specified days after growing in erythropoietin-supplemented media using TRIzol reagent (Life Technologies, Rockville, MD) according to the manufacturer's directions. Reverse transcription (RT) was performed using oligo- (dT) primer and Superscript Reverse Transcriptase (Life Technologies) as suggested by the manufacturer at 42°C for 50 minutes. One microliter of RT reaction was used in 50 μL polymerase-chain reaction (PCR) amplification with appropriate primers as follows: initial denaturation at 95°C for 1 minute, denaturing at 95°C for 15 seconds, annealing at 68°C for 15 seconds, and extension at 72°C for 20 seconds for 28 cycles. PCR products were separated in 1.2% agarose gel and stained with ethidium bromide. For PBGD amplification, the following primers were used: 5′-ACACAGCCTACTTTCCAAGCGGAGCCAT-3′ (primer 1), 5′-TGGATCCCTGAGGAGGGCAGAAG-3′ (primer 2), 5′-TCTTGTCCCCTGTGGTGGACATAGCAAT-3′ (primer 3). Primers 1 and 3 specifically amplify the housekeeping PBGD. Primers 2 and 3 specifically amplify an erythroid PBGD. The PCR products were completely sequenced using a dRhodamine DNA sequencing kit (PE Biosystems, Foster City, CA).
Northern blotting
Human Immune System Multiple Tissue Northern (MTN) Blot II was processed as recommended by the manufacturer (Clontech, Palo Alto, CA). Briefly, the membrane was prehybridized in ULTRAhyb hybridization buffer (Ambion, Austin, TX) at 42°C for 30 minutes. The buffer was replaced with the fresh ULTRAhyb containing 32P-labeled probe and hybridized at 42°C for 16 hours. The membrane was washed in low- and high-stringency buffers (Ambion) according to the manufacturer's protocol and subjected to autoradiography. Probes specific for the housekeeping (probe H) and an erythroid alternative (probe EA) part of PBGD used in Northern blotting were generated by PCR, purified by QIAquick PCR Purification Kit (Qiagen, Valencia, CA) and labeled using the DNA-labeling beads (−dCTP) (Amersham Pharmacia Biotech, Piscataway, NJ). Primers 5′-GGTACTGAGGAAGGTTAAAGGGA -3′ and 5′-AGGATTACCCCTCGAACACCTGCAGAAAAAGTCCCCAGGT-3′ were used to amplify the 4138-4315 portion (corresponding to intron between exons 2 and 3) of the PBGD gene (GenBank accession number M95623). Primers 5′-GGCTCTGCGGAGACCAGGAGTCAG-3′ and 5′-TTACCAGACATGGCTCCGCTTGGAA-3′ were used to amplify the 959-1101 region (in exon 1) of the gene. Genomic DNA from K562 cells was used as a template for PCR amplification.
Results and discussion
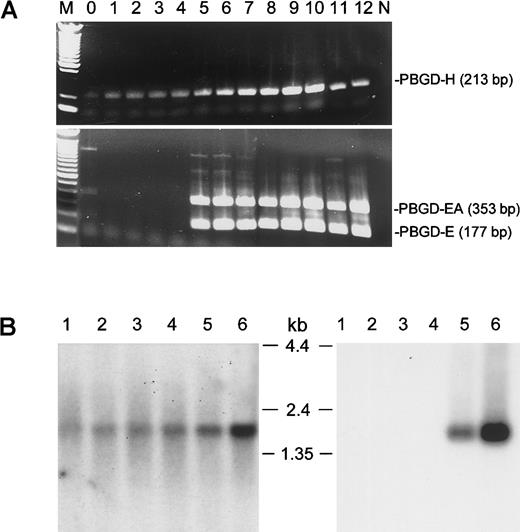
The PBGD gene comprises 15 exons encoded within a 10-kilobase (kb) region of chromosome 11q23.3.4 Figure1 depicts the structure of the human PBGD mRNA. Different isoforms are produced through alternative splicing of exon 1 and exon 2. Splicing exon 1 to exon 3 results in expression of the housekeeping form of PBGD (PBGD-H). The combination of exon 2 and exon 3 produces an erythroid form of PBGD (PBGD-E). Figure2A shows the results of PT-PCR amplification of PBGD gene products in CD34+cells grown in the presence of erythropoietin. Cells were collected over 12 days, total RNA was purified, reverse transcribed with oligo- (dT) primer, and amplified using housekeeping or erythroid-specific primers for PBGD. Primers 1 and 3 were used to amplify housekeeping PBGD (expected product size is 213 base pairs [bp]), and primers 2 and 3 were used to amplify erythroid PBGD (expected product size is 177 bp). Although the PBGD-H product was present in all samples, the PBGD-E9 PCR product (sequence confirmed) was amplified only after several days in culture consistent with erythroid cell commitment and differentiation. Additionally, a 353-bp product was amplified with primers 2 and 3 (PBGD-EA). Complete sequencing of PBGD-EA revealed an alternatively spliced form of erythroid PBGD containing the intron between exons 2 and 3, thus extending the 5′ untranslated region of the erythroid transcript by 176 bp. Two downstream introns between exons 3, 4, and 5 were correctly spliced in both 177-bp and 353-bp PR-PCR products.
Structure of human PBGD transcripts.
Probe H: Probe specific for housekeeping PBGD. Probe EA: Probe specific for erythroid alternative PBGD. Relevant exons are shown. AUG and UAA depict start and stop codons, respectively.
Structure of human PBGD transcripts.
Probe H: Probe specific for housekeeping PBGD. Probe EA: Probe specific for erythroid alternative PBGD. Relevant exons are shown. AUG and UAA depict start and stop codons, respectively.
Confirmation of existence of the PBGD erythroid alternative transcript.
(A) RT-PCR assay of PBGD transcipts. Total RNA from CD34+cells collected on indicated (0-12) days after addition of erythropoietin was used for RT-PCR as described in study design. M: 100-bp DNA ladder (Life Technologies). N: No DNA added control. (B) Northern blot analysis of PBGD-H (left panel) and PBGD-EA (right panel) expression in human hematologic tissues. Lanes 1 to 6 contain, in order, 2 μg polyA+ RNA from human spleen, lymph node, thymus, peripheral blood leukocytes, bone marrow, and fetal liver.32P-labeled probe H (panel A) or probe EA (panel B) was used for hybridization. RNA size marker bands are indicated in between the blots.
Confirmation of existence of the PBGD erythroid alternative transcript.
(A) RT-PCR assay of PBGD transcipts. Total RNA from CD34+cells collected on indicated (0-12) days after addition of erythropoietin was used for RT-PCR as described in study design. M: 100-bp DNA ladder (Life Technologies). N: No DNA added control. (B) Northern blot analysis of PBGD-H (left panel) and PBGD-EA (right panel) expression in human hematologic tissues. Lanes 1 to 6 contain, in order, 2 μg polyA+ RNA from human spleen, lymph node, thymus, peripheral blood leukocytes, bone marrow, and fetal liver.32P-labeled probe H (panel A) or probe EA (panel B) was used for hybridization. RNA size marker bands are indicated in between the blots.
Because PBGD-EA has not been reported elsewhere, we attempted to confirm the existence of this transcript in primary tissues (Figure2B). Human Immune System MTN Blot II (Clontech) was hybridized with probes specific either for the housekeeping (probe H) or the alternate erythroid (probe EA) forms of PBGD. The probe EA (Figure 1) corresponds to intron between exons 2 and 3. Hybridization with the probe H produced a band of expected size (about 1.5 kb) in all 6 tested tissues. In contrast, a distinct 1.5-kb band was observed after hybridization with the probe EA only in bone marrow and fetal spleen. This confirms the erythroid-specific expression of PBGD-EA and provides evidence for its presence in primary tissues. A computer-assisted BLAST query of the expressed sequence tag (EST) database with the 176-bp intronic sequence identified 3 additional human ESTs (GenBank accession nos. T84131, R06321, and T57423) that retain this intronic fragment in processed transcripts. All 3 ESTs were derived from fetal liver or spleen libraries supporting our hypothesis that PBGD-EA represents an erythroid-specific isoform. BLAST queries failed to identify any EST currently deposited in GenBank that match the human PBGD-E isoform.
Erythroid-specific transcription of the PBGD gene has been examined previously in the context of promoter transactivation.15,16 We report here that human PBGD is expressed as 3 different transcripts, 2 being erythroid specific. The additional 176-bp fragment of PBGD-EA isoform contains at least one known polymorphism in humans.5 The existence of this second erythroid PBGD isoform has not been reported elsewhere, despite extensive analyses of this gene over the last 15 years. Interestingly, a previous study of erythroid-specific PBGD expression failed to reveal an alternate erythroid transcript in HEL cells.9 Despite our clear demonstration of PBGD-EA in primary erythroid cell cultures, bone marrow, and fetal liver, we were unable to show expression of the alternate transcript in HEL cells (not shown). Our discovery highlights the prospects of identifying unexpected, primary tissue-specific gene expression patterns even among extensively studied genes. Erythroid-specific isoforms involving the first 3 exons of the second (5-aminolevulinate dehydratase)17 and the fourth (uroporphyringen III synthase)18 heme biosynthesis enzymes have also been described. Transcripts from an alternate promoter within the first intron of the NF-E2 are also associated with erythroid specificity.19,20 Similarly, alternate transcriptional start sites and splicing result in the erythroid band 4.1 gene products.21 In all cases, the erythroid-specific mRNAs contain an alternate 5′ region. These data suggest erythroid-specific cell development is accomplished at the genomic level via multiple mechanisms, including shared patterns of gene organization, transactivation, and RNA maturation.
Acknowledgment
We thank the National Institutes of Health Department of Transfusion Medicine for providing the primary cells used in this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
J. L. Miller, Laboratory of Chemical Biology, Bldg 10, Rm 9B17, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892; email: jm7f@nih.gov.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal