Abstract
The genes at the INK4A/ARF locus at 9p21 are frequently involved in human cancer. Virtually allp16INK4A exon 2 (henceforth calledp16) inactivation in pediatric acute lymphoblastic leukemia (ALL) occurs by gene deletion. The results of this study illustrate that real-time quantitative polymerase chain reaction is capable of detecting gene deletion in primary patient specimens with a precision not previously achieved by conventional methods. Importantly, this assay includes the detection of hemizygous deletions. The study revealed, strikingly, that the risk ratio for relapse for hemizygous deletion compared with no deletion was 6.558 (P = .00687) and for homozygous deletion was 11.558 (P = .000539). These results confirm and extend the authors' previous findings that homozygous deletion of p16 in pediatric ALL patients is an independent prognostic indicator of outcome from therapy.
Introduction
The assessment of deletion of certain genes requires the detection of hemizygosity in primary patient specimens contaminated with normal cells. Meeting this challenge in cancer screening is becoming increasingly critical with the recent identification of several tumor suppressor genes that are haplo-insufficient rather than classically recessive.1 The genes at the INK4A/ARF locus2 act as tumor suppressors via 2 proteins, p16INK4A and p14ARF(p19ARF in the mouse), while the role of p15 in leukemogenesis remains unresolved. The p16INK4A is a cyclin-dependent kinase inhibitor that acts upstream of the retinoblastoma (RB) protein to control cell cycle arrest.3 The p19ARF is translated in an alternative reading frame from p16INK4A and activates p53 by interfering with its negative regulator, MDM2.4 Hence, mutations at theINK4A/ARF locus can disrupt both the RB1 and thep53 tumor suppressor pathways.3,4Essentially all p16INK4A inactivation in pediatric acute lymphoblastic leukemia (ALL) occurs by gene deletion (reviewed in Kees et al5 and Drexler6). The evidence for an independent prognostic role ofp16INK4A, exon 2 (henceforth calledp16) deletion in pediatric ALL, is inconclusive (reviewed in Drexler6 and Tsihlias et al7). All of these studies were based on detecting p16 deletion by either Southern blotting or conventional polymerase chain reaction (PCR) analysis. Bone marrow specimens from leukemia patients at diagnosis invariably contain some normal cells that cause problems with accurate quantitation of deletion of any gene. This prompted us to examine whether gene deletions can be detected by real-time quantitative PCR and whether p16 zygosity presents a simple and reliable test for leukemia prognosis.
Study design
Patients
Diagnosis bone marrow specimens were studied from 45 ALL patients, on the basis of the availability of cryopreserved specimens (Table 1). The Princess Margaret Hospital, Perth, Australia, provided 30 patients, and the Children's National Medical Center, Washington, DC, provided 15. There was no selection on the basis of either p16 genotype or time-to-relapse. Informed consent was obtained from all patients or their guardians to use specimens for research. Specimens were collected between 1981 and 1997. The leukemia immunophenotype (B-lineage or T-cell ALL) was determined with the use of a panel of monoclonal antibodies.5 Cytogenetic results revealed that none of the patients were known to have either t(4;11) or t(9;22). For all but 2 patients studied, therapy was administered according to risk-adjusted protocols of the Children's Cancer Group,8 most of which were based upon modifications of the Berlin-Frankfurt-Munster trials; the exception consisted of 2 patients treated on a previously reported intensive therapy protocol for high-risk patients.8
Clinical characteristics and p16 status of study group
| Parameter . | Number (percentage) . |
|---|---|
| Total no. | 45 |
| Sex | |
| Female | 14 (31) |
| Male | 31 (69) |
| Age | |
| <1 y | 1 (2) |
| 1 y-10 y | 30 (67) |
| >10 y | 14 (31) |
| Immunophenotype | |
| T-ALL | 13 (29) |
| B-lineage ALL | 32 (71) |
| WCC at diagnosis | |
| <50 000 × 106/L | 26 (58) |
| ≥50 000 × 106/L | 19 (42) |
| P16genotype | |
| G/G | 28 (62) |
| D/D | 11 (25) |
| G/D | 6 (13) |
| Parameter . | Number (percentage) . |
|---|---|
| Total no. | 45 |
| Sex | |
| Female | 14 (31) |
| Male | 31 (69) |
| Age | |
| <1 y | 1 (2) |
| 1 y-10 y | 30 (67) |
| >10 y | 14 (31) |
| Immunophenotype | |
| T-ALL | 13 (29) |
| B-lineage ALL | 32 (71) |
| WCC at diagnosis | |
| <50 000 × 106/L | 26 (58) |
| ≥50 000 × 106/L | 19 (42) |
| P16genotype | |
| G/G | 28 (62) |
| D/D | 11 (25) |
| G/D | 6 (13) |
T-ALL indicates T-lineage acute lymphoblastic leukemia: WCC, white cell count; G/G, germline p16; D/D p16deletion; G/D, hemizygous p16.
Real-time PCR analysis in multiplex format
Genomic DNA was isolated from cryopreserved specimens and control cell lines by standard methodology.5 All primers and probes were designed by means of Perkin-Elmer Primer Express software (Perkin-Elmer, Foster City, CA), and primers were supplied by Geneworks (Adelaide, Australia). The sequences were as follows: p16 exon 2 forward (F): ggctctacacaagcttcctttcc; p16 exon 2 reverse (R): tcatgacctgccagagagaaca; β-actin F: agcgcggctacagcttca; andβ-actin R: cgtagcacagcttctccttaatgtc. The probe forp16 had the sequence cccccaccctggctctgacca and was labeled with FAM, whereas the probe for β-actin, atttcccgctcggccgtggt, was labeled with VIC (both probes manufactured by Perkin-Elmer). The reactions were optimized first individually and then for multiplexing. The reaction was performed in a final volume of 50 μL. The final concentrations of primers and probes were as follows: p16 F 50 ng/μL,p16 R 50 ng/μL, p16 probe 200 nM,β-actin F 50 ng/μL, β-actin R 200 ng/μL, and β-actin probe 200 nM. Each reaction contained 50 ng DNA as template, and the Taqman Universal Master Mix (Perkin-Elmer) was used. The standard thermal cycling conditions of the ABI PRISM 7700 Sequence Detection instrument were applied. A standard calibration curve was included with each experiment with the use of a range of concentrations of DNA extracted from Raji B cells (range, 1.56-100 ng DNA).
P16 gene deletion analysis in specimens containing normal cells
We simulated normal cell contamination by using mixtures of DNA from Raji B cells, which are wild type for p16 (G/G), and K562 cells, which show homozygous deletion of p16(D/D). The experimentally determined ratio forp16/β-actin was expressed as a function of the input ratio of Raji B/K562 cells. The test yielded a linear graph with a correlation coefficient of 0.9687, indicating that normal cell contamination (here simulated by Raji B cells) in ap16 D/D sample can be accurately measured by this technique. On the basis of this result, the bone marrow specimens were interpreted as follows. Ratio for p16/β-actin less than 0.4:p16 deletion (D/D); ratio 0.4 to 0.8: hemizygousp16 (G/D); ratio exceeding 0.8: germline p16(G/G). The method was compared with Southern blot analysis, and the 2 methods agreed in all 11 cases tested, including G/D specimens. All but one of the 45 specimens contained fewer than 25% normal cells, according to an independent review by a hematologist, and the experimentally determined ratio for p16/β-actin was used to determine the genotype directly. The remaining specimen contained more than 50% normal cells, a factor that was taken into account.
Statistical analysis
The main analysis was based on methods appropriate for censored failure times. The primary time scale was calendar time from diagnosis; the primary response was relapse. Univariate analysis was based upon Kaplan-Meier survival functions9 and the Mantel-Cox (log-rank) test statistic.9 Multivariate analysis was based on the Cox proportional hazards regression model9 and the likelihood-ratio test.10Covariates known to modulate the risk of relapse were included in the primary model whether they were statistically significant or not. Secondary modeling demonstrated that removal of the nonsignificant covariates did not modify substantive conclusions. Final models were subjected to (and passed) standard tests of goodness of fit.10 Analysis was undertaken in SAS version 6.12 (Cary, NC) for Unix.
Results and discussion
Deletion analysis of p16 was performed on 45 pediatric ALL patients at diagnosis, and the results are summarized in Table 1. Of the 45 patients, 11 (25%) demonstrated a homozygous deletion; 6 (13%) were hemizygous; and 28 (62%) were wild type for thep16 gene. In a previous study using Southern blot analysis performed in this laboratory, the incidence of homozygousp16 deletions at diagnosis was 18.3% (9/48).5These findings for homozygous deletions are in agreement with the frequency of homozygous deletion reported for pediatric ALL patients: 23% for B-lineage and 64% for T-lineage ALL (T-ALL) (reviewed by Drexler6). The combined frequency for hemizygous and homozygous deletions determined here is 38%. When the distribution of T-ALL versus B-lineage ALL cases in our study is taken into account, the observed frequency is higher than expected, most likely owing to the higher sensitivity of the PCR technique.
Hemizygous and homozygous p16 deletions are independent prognostic indicators for poor outcome
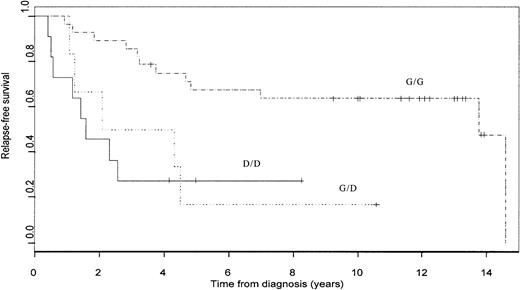
Figure 1 illustrates the Kaplan-Meier curves for relapse-free survival stratified byp16 status. Among those patients who were “censored” (ie, had not relapsed at the end of follow-up), the minimum follow-up time was 3 years, and all but 3 such patients were followed for at least 8 years. With the use of the log-rank test for any differences between the 3 groups, the P value was .0021. Multivariate analysis was performed by means of Cox proportional hazards regression, including the known risk factors for pediatric ALL patients: gender, immunophenotype, age, and white cell count at diagnosis. When we adjusted simultaneously for the influence of these risk factors, both hemizygous and homozygous deletions remained highly statistically significant predictors of poor outcome; compared with G/G patients, the risk ratio was 11.558 (P = .000539) for patients with homozygous deletions and 6.558 (P = .00687) for hemizygous deletions (Table 2). This comprehensive adjustment for potential confounding variables showed that any p16 deletion (D/D or G/D) is a major independent risk factor for relapse. These results confirm and extend previous findings from our laboratory and others5,11 regarding the prognostic significance of p16 deletion in pediatric ALL patients. Of the patients in this analysis, 11 were also included in our earlier study.5 If these are excluded, the estimated adjusted-risk ratios for G/D versus G/G and D/D versus G/G are 5.325 (P = .0462) and 9.856 (P = .0027), respectively. This represents a completely independent test of the hypothesis that the p16 deletion is associated with prognosis in childhood ALL. Nevertheless, a prospective study on a larger number of uniformly treated patients should be conducted to confirm the prognostic significance of the p16 deletion, and such a study should include a test for loss of p14.
Kaplan-Meier survivor function.
Kaplan-Meier survivor function for G/G (_ . _ . _ . _ .), G/D (· · · · ·) and D/D (––––) patients.
Kaplan-Meier survivor function.
Kaplan-Meier survivor function for G/G (_ . _ . _ . _ .), G/D (· · · · ·) and D/D (––––) patients.
Hemizygous and homozygous p16 deletions are independent prognostic indicators for poor outcome
| Genotype . | Risk ratio* . | Confidence interval . | P value . |
|---|---|---|---|
| G/D versus G/G | 6.5 | 1.858 -23.880 | .00687 |
| D/D versus G/G | 11.5 | 2.825 -47.284 | .000539 |
| Genotype . | Risk ratio* . | Confidence interval . | P value . |
|---|---|---|---|
| G/D versus G/G | 6.5 | 1.858 -23.880 | .00687 |
| D/D versus G/G | 11.5 | 2.825 -47.284 | .000539 |
See Table 1 footnote for explanation of abbreviations.
Multivariate analysis was performed to determine risk ratios.
Hemizygous status as determined in this study could be due to a mixture of leukemia cells (D/D, G/D, and G/G) or true hemizygosity in all leukemia cells. Owing to lack of material, it was not possible to study the G/D specimens by means of the fluorescence in situ hybridization technique. However, the clonality could be assessed by analyzing the rearrangement of the T-cell receptor and immunoglobulin heavy chain genes.12 13 Examination of the 5 G/D specimens from patients who relapsed revealed that in 3 cases there was clear evidence for clonal disease as only one rearranged band was detected whereas the 2 remaining cases showed 2 bands suggesting biclonal disease (data not shown). This test does not exclude the possibility that leukemia cells in these specimens were also hemizygous forp16. Clearly, 3 patients were diagnosed with a monoclonal disease showing hemizygous deletion of p16, excluding the potential for false hemizygous readout due to a mixture of G/G with D/D leukemic cells in the specimens.
In the current study, we identify hemizygous and homozygous loss of p16 as a major independent negative prognostic indicator in pediatric ALL. These results are consistent with the reported general sensitivity of pediatric ALL to current chemotherapy. Unlike the majority of good-prognosis pediatric ALL patients, who appear to have an intact apoptosis pathway, the subpopulation that is refractory to treatment with apoptosis-inducing drugs may have bypassed normal regulation by mutation of key regulators such asp16.14 Independent evidence thatINK4A/ARF mutations promote resistance to chemotherapeutic drugs has recently been reported in a transgenic lymphoma model.15 The findings from this animal model provide direct evidence that mutations at the INK4A/ARF locus have a negative impact on the outcome of cancer therapy. The quantitative PCR method used in our study is suitable for high-throughput screening of patient specimens and has many clinical applications as it can be adapted to the deletion analysis of other tumor suppressor genes and other cancers.
Supported by the Children's Leukaemia and Cancer Research Foundation, Western Australia; the National Childhood Cancer Foundation; and the Children's Cancer Group, Arcadia, CA.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Ursula R. Kees, Division of Children's Leukaemia and Cancer Research, TVWT Institute for Child Health Research, PO Box 855, West Perth WA 6872, Australia; e-mail: ursula@ichr.uwa.edu.au.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal