To investigate the potential for functional interactions between heterologous receptors, the cytoplasmic domains of 2 different receptors (c-Kit and Flt-3) were coexpressed in the interleukin-3–dependent cell line Ba/F3. The receptor signaling domains were presented in the context of fusion proteins, with c-Kit linked to the FK506 binding protein (FKBP12) and Flt-3 linked to the FRB domain of the FKBP12–rapamycin-associated protein. The fusions were brought into apposition with the use of chemical inducers of dimerization (CIDs). Two classes of CID were employed. FK1012 and its synthetic analogue AP1510 bring together 2 copies of the FKBP12 domain, thereby inducing homodimerization of the c-KitFKBP12fusion. A second type of CID, rapamycin, brings together one FKBP12 domain and one FRB domain, resulting in heterodimerization of the c-KitFKBP12 and Flt-3FRB fusions. Ba/F3 cell growth was promoted not only by FK1012- or AP1510-induced homodimerization of the c-KitFKBP12 fusion (as reported previously), but also by rapamycin-induced c-KitFKBP12–Flt-3FRB heterodimerization. These findings demonstrate the potential for a direct functional interaction between c-Kit and Flt-3.
Introduction
A variety of receptors coexist on the surface membrane of hemopoietic cells. To test for potential functional interactions between different receptors, we have employed a system that allows the signaling domains of 2 different receptors to be forcibly juxtaposed. The system relies on coexpressing 2 fusion proteins in the interleukin-3 (IL-3)–dependent cell line Ba/F3.1 The first fusion protein contains the signaling domain of one receptor linked to the FK506 binding protein (FKBP12), whereas the second fusion protein contains the signaling domain of another receptor linked to the FKBP12–rapamycin-binding (FRB) domain of the FKBP12–rapamycin-associated protein (FRAP). Dimerization is achieved by the synthetic ligands FK10122,3 (or its synthetic analogue AP15104,5) and rapamycin.6FK1012 and AP1510 induce homodimerization of the FKBP12 fusions, whereas rapamycin directs heterodimerization between the FKBP12 and FRB fusions. Signaling is reflected in Ba/F3 cell growth in the absence of IL-3.
Study design
Plasmid construction
The c-Kit3FKBP12 and c-Kit1FKBP12constructs have been described previously.5 FRB domain–containing constructs were generously provided by Steffan Ho and were generated by inserting a XhoI/SalI linkered fragment of FRAP (residues 2026-2114) downstream of a myristylation-targeting sequence from c-Src, as described previously.2 Two FRB-containing constructs were made, one with 3 tandem copies of FRB and one with a single FRB domain, and were named pMFRB3E and pMFRB1E, respectively. A 1296-bp fragment encoding the cytoplasmic domain of murine c-Kit was freed from a pBS-based vector5 by XhoI digestion. The fragment was gel purified and subcloned into the SalI site of the pMFRB1E and pMFRB3E constructs to generate c-Kit1FRB and c-Kit3FRB. A 1313-bp fragment encoding the cytoplasmic portion of Flt-3 was amplified by polymerase chain reaction (PCR) using Pfu polymerase and a plasmid cDNA template (a gift from Ihor Lemishcka, Princeton University, NJ). The following oligonucleotide sequences were used for the amplification reaction: 5′-CCC CTC GAG TGC CAC AAA TAC AAA AAG C-3′ and 5′-CCC CTC GAG CTA ACT TCT TTC TCT GTG A-3′. Each primer contains added sequence to incorporate a XhoI restriction site at each end of the PCR product. The amplicon was digested with XhoI, gel purified, and subcloned into XhoI-digested pBluescript. After sequence confirmation using the PRISM system (Applied Biosystems, Foster City, CA), the fragment was released from pBluescript byXhoI digestion, gel purified, and subcloned into theSalI site of the pMFRB1E and pMFRB3E constructs. The plasmids generated were named Flt-31FRB and Flt-33FRB.
Electroporation, Western blot analysis, and cell proliferation assays
Electroporation, Western blot analysis, and cell proliferation assays were performed as described previously.3
Results and discussion
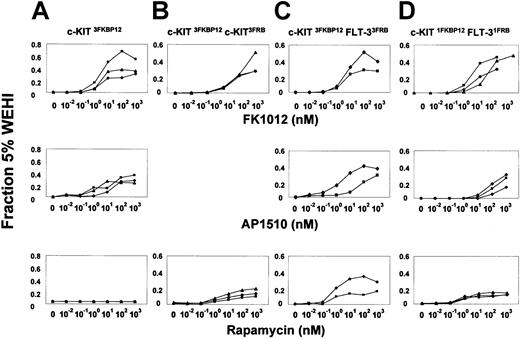
Studies were directed at determining whether forced apposition of c-Kit and Flt-3 could induce Ba/F3 cell growth. Constructs were generated in which sequences encoding the intracellular portions of c-Kit and Flt-3 were linked to one or more copies of either FKBP12 or FRB. We have previously reported that FK1012 and AP1510 induce proliferation in Ba/F3 cells that express a fusion protein containing 3 tandem copies of FKBP12 linked to the intracellular portion of c-Kit (c-Kit3FKBP12) (Figure 1A, upper and middle panels). In contrast to the proliferative effects of FK1012/AP1510, these cells failed to proliferate in response to rapamycin (Figure 1A, lower panel), confirming that rapamycin is unable to activate the c-Kit3FKBP12 fusion and lacks nonspecific proliferative effects in Ba/F3 cells. In fact, as expected,7 increasing concentrations of rapamycin were inhibitory to IL-3–dependent Ba/F3 cell growth (data not shown).
Forced engagement of c-Kit and Flt-3 induces Ba/F3 cell proliferation.
The effects of different CIDs on representative Ba/F3 cell clones expressing various combinations of the dimerizable fusion proteins were tested in transient cell-proliferation assays using FK1012 (upper panels), AP1510 (middle panels), or rapamycin (lower panels). The degree of proliferation is indicated as a fraction of that obtained using 5% of IL-3–containing WEHI-3–conditioned medium, which induces more than 50% of maximal IL-3–dependent cell growth.3 Each line depicts a different cell clone. (A) Ba/F3 cells expressing the c-Kit3FKBP12 fusion proliferate in response to FK1012 and AP1510, but fail to proliferate in the presence of rapamycin. (B) Clones coexpressing c-Kit3FKBP12and c-Kit3FRB proliferate in response to FK1012, but also proliferate in response to rapamycin. (C) Ba/F3 cells expressing both the c-Kit3FKBP12 and the Flt-33FRB fusions exhibit proliferative responses to FK1012, AP1510, and rapamycin. (D) Likewise, clones coexpressing c-Kit1FKBP12 and Flt-31FRB were capable of responding to all 3 drugs, with the effects of rapamycin indicating that heterodimerization of c-Kit and Flt-3 can induce cell growth.
Forced engagement of c-Kit and Flt-3 induces Ba/F3 cell proliferation.
The effects of different CIDs on representative Ba/F3 cell clones expressing various combinations of the dimerizable fusion proteins were tested in transient cell-proliferation assays using FK1012 (upper panels), AP1510 (middle panels), or rapamycin (lower panels). The degree of proliferation is indicated as a fraction of that obtained using 5% of IL-3–containing WEHI-3–conditioned medium, which induces more than 50% of maximal IL-3–dependent cell growth.3 Each line depicts a different cell clone. (A) Ba/F3 cells expressing the c-Kit3FKBP12 fusion proliferate in response to FK1012 and AP1510, but fail to proliferate in the presence of rapamycin. (B) Clones coexpressing c-Kit3FKBP12and c-Kit3FRB proliferate in response to FK1012, but also proliferate in response to rapamycin. (C) Ba/F3 cells expressing both the c-Kit3FKBP12 and the Flt-33FRB fusions exhibit proliferative responses to FK1012, AP1510, and rapamycin. (D) Likewise, clones coexpressing c-Kit1FKBP12 and Flt-31FRB were capable of responding to all 3 drugs, with the effects of rapamycin indicating that heterodimerization of c-Kit and Flt-3 can induce cell growth.
To evaluate the feasibility of achieving rapamycin-inducible cell growth, we tested Ba/F3 cells expressing both c-Kit3FKBP12and a second fusion containing the same c-Kit fragment linked to 3 copies of the FRB domain of FRAP (c-Kit3FRB).8Fusions containing 3 copies of the drug-binding domain were expected to allow for the generation of both dimers as well as higher-order oligomers by chemical inducers of dimerization (CID). C-kit3FKBP12 homo-oligomers, generated by the addition of FK1012, induced cell growth as expected (Figure 1B, upper panel). Rapamycin-mediated juxtaposition of the c-Kit3FKBP12 and c-Kit3FRB fusions also induced Ba/F3 cell growth (Figure1B, lower panel), although to a lesser extent than observed in response to FK1012. The relatively less intense cell growth observed in response to rapamycin may be due to the well-characterized antiproliferative effects of rapamycin.7 Alternatively, signaling by c-Kit3FKBP12–c-Kit3FRBhetero-oligomers may be less efficient, possibly as a consequence of suboptimal alignment between the 2 c-Kit signaling domains.
To test whether c-Kit can functionally interact with Flt-3, we generated Ba/F3 cell clones expressing both c-Kit3FKBP12and Flt-33FRB fusions. Of note, fusions containing Flt-3 linked to 3 copies of FKBP12 produced factor-independent growth in Ba/F3 cells and were therefore not characterized further (data not shown). As illustrated in Figure 1C (lower panel), 2 clones coexpressing the c-Kit3FKBP12 and Flt-33FRBfusions demonstrated proliferative responses to rapamycin.
As noted above, rapamycin-induced aggregation of the c-Kit3FKBP12 and Flt-33FRB fusions would be expected to produce hetero-oligomeric complexes. To determine whether c-Kit–Flt-3 heterodimers were sufficient to induce Ba/F3 cell growth, we made 2 additional constructs in which c-Kit was linked to a single FKBP12 domain (c-Kit1FKBP12) and Flt-3 was linked to a single FRB domain (Flt-31FRB). As shown in Figure 1D (lower panel), Ba/F3 cell clones coexpressing the c-Kit1FKBP12 and Flt-31FRB fusions exhibited dose-dependent proliferative responses to rapamycin.
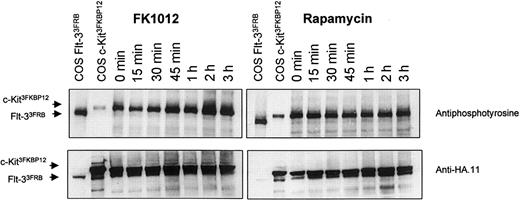
Studies directed at examining tyrosine phosphorylation following stimulation with either FK1012 or rapamycin were performed. Ba/F3 cells coexpressing the c-Kit3FKBP12 fusion and the Flt-33FRB fusion exhibited constitutive tyrosine phosphorylation, including phosphorylation of the c-Kit3FKBP12 fusion, as we have reported previously.5 FK1012 induced a modest increase in tyrosine phosphorylation, whereas rapamycin had no discernible effect (Figure2). We interpret the disparity between our cell-proliferation assays and signaling studies to indicate that although rapamycin-triggered signals are sufficient to induce cell proliferation, they fall below the threshold needed for inducing appreciable changes in tyrosine phosphorylation.
Changes in tyrosine phosphorylation are discernible following stimulation with FK1012 but not rapamycin.
Western blot analysis of whole-cell lysates from Ba/F3 cells coexpressing c-Kit3FKBP12 and Flt-33FRB and stimulated with either FK1012 (left) or rapamycin (right). Lysates from COS cells expressing either fusion (COS c-Kit3FKBP12 and COS Flt-33FRB) have been included for size comparisons. Top panels show antiphosphotyrosine antibody labeling (4G10; Upstate Biotechnology, Lake Placid, NY) at various times in response to either FK1012 (100 nM) or rapamycin (100 nM). Bottom panels show the same blots after being stripped and reprobed with the anti-HA.11 antibody (Berkeley Antibody Company, Berkeley, CA) to show equivalent loading.
Changes in tyrosine phosphorylation are discernible following stimulation with FK1012 but not rapamycin.
Western blot analysis of whole-cell lysates from Ba/F3 cells coexpressing c-Kit3FKBP12 and Flt-33FRB and stimulated with either FK1012 (left) or rapamycin (right). Lysates from COS cells expressing either fusion (COS c-Kit3FKBP12 and COS Flt-33FRB) have been included for size comparisons. Top panels show antiphosphotyrosine antibody labeling (4G10; Upstate Biotechnology, Lake Placid, NY) at various times in response to either FK1012 (100 nM) or rapamycin (100 nM). Bottom panels show the same blots after being stripped and reprobed with the anti-HA.11 antibody (Berkeley Antibody Company, Berkeley, CA) to show equivalent loading.
Receptor tyrosine kinases are segregated into approximately 20 structurally related subfamilies, and interactions between different members within a subfamily are well described, as exemplified by the 4 members of the epidermal growth factor receptor (EGFR) subfamily. The formation of heterodimers between different members of the EGFR family is a key mechanism for achieving signal diversity.9 Using the same system as described here, Muthuswamy and colleagues10 showed that c-Cbl phosphorylation is induced by ErbB1 homodimers, but not by ErbB1–ErbB2 heterodimers. In the hemopoietic system, functional interactions have been documented between c-Kit and the erythropoietin receptor.11Additionally, the erythropoietin receptor has been found to interact directly with the common β chain of the granulocyte-macrophage colony-stimulating factor and IL-3 receptors.12
This method provides a general approach for identifying productive interactions between different growth factor receptor signaling domains. In the present study, we show that functional interactions between c-Kit and Flt-3 are possible. Whether similar interactions occur in nature is unknown. Once the potential for functional interactions between different receptors is revealed, these interactions can be further explored for their physiologic relevance.
We thank Ihor Lemischka for providing the Flt-3 cDNA, Steffan Ho for the FRB cDNA, Mike Gilman and Tim Clackson for providing FK1012 and AP1510, Joachim Deeg for providing rapamycin, and Barbara Richard for providing expert technical support.
Supported by grants 5R01DK52997, 1R01DK57525, 2P01HL53750, and 2P01DK47754 from the National Institutes of Health; an American Society of Hematology Junior Faculty Scholar Award; and an award from the Fanconi Anemia Research Fund.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
C. Anthony Blau, Mailstop 357710, Health Sciences Building, University of Washington, Seattle, WA 98195; e-mail: tblau@u.washington.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal