Abstract
Hodgkin lymphoma (HL) is characterized by the abnormal expression of multiple cytokines, accounting for its unique clinicopathologic features. We have previously shown that interleukin-13 (IL-13) is secreted by HL cell lines and may serve as an autocrine growth factor. To determine the frequency of IL-13 expression in lymphoma patients, tissue sections from 36 patients with classical HL, 5 patients with nodular lymphocyte predominance HL (NLPHL), and 23 patients with non-Hodgkin lymphoma (NHL) were subjected to in situ hybridization. In 31 of 36 cases (86%) of classical HL patients of all histologic subtypes, between 25% to almost 100% of Hodgkin and Reed Sternberg (HRS) cells were positive for IL-13 expression. In contrast, in no case of NLPHL and in only 4 of 23 NHL cases (1 of 5 T-cell–rich B-cell lymphomas, 2 of 5 anaplastic large cell lymphomas, and 1 of 5 peripheral T-cell lymphomas) did the neoplastic cells express IL-13. The expression of the IL-13 receptor chain α1 (IL-13Rα1) was also analyzed by in situ hybridization. In 24 of 27 (89%) cases of classical HL, between 25% to 75% of HRS cells, as well as a high frequency of lymphocytes and histiocytes, were positive for IL-13Rα1 expression. These results were confirmed by the construction of complementary DNA libraries from single HRS cells, followed by polymerase chain reaction analysis, in which IL-13Rα1 transcripts were found to be present in all 6 cases of HL. These data indicate that expression of IL-13 and IL-13Rα1 is a common feature of HRS cells in HL, consistent with the hypothesis that IL-13 may play a role in autocrine growth in classical HL.
Introduction
Hodgkin lymphoma (HL) is unusual among malignancies in that the neoplastic cells, the Hodgkin and Reed-Sternberg (HRS) cells, make up only a small proportion of the clinically detectable mass. The bulk of the tumor is composed of a reactive infiltrate of lymphocytes, eosinophils, plasma cells, histiocytes, and fibroblasts. Although HL has distinct clinical features and responses to treatment that warrant its distinction from the non-Hodgkin lymphomas (NHL), within HL there are variations in the composition of the inflammatory infiltrate, the morphology of the HRS cells, and the presence of the Epstein-Barr virus (EBV).1 The clinicopathologic features of HL are consistent with an abnormal pattern of cytokine expression of the HRS cells. These cells can express a variety of cytokines, including interleukin-2 (IL-2), IL-5, IL-6, IL-7, IL-9, IL-10, granulocyte-macrophage colony-stimulating factor, lymphotoxin-α, and transforming growth factor-β (TGF-β).2 Some cytokines show variable expression that correlates with specific clinicopathologic features of HL. For example, IL-5 expression is associated with tissue eosinophilia,3 IL-10 is associated with EBV-positive HL,4 and TGF-β with sclerosis.5 However, abnormal expression of no one cytokine has been consistently demonstrated in all forms of HL.
IL-13 is a pleiotropic cytokine secreted by activated T cells that shares immunoregulatory characteristics with IL-4.6,7IL-13 exerts its effects primarily on B cells and monocytes, stimulating growth and immunoglobulin (Ig) class switching in B cells, and up-regulation of the low-affinity IgE receptor and MHC class II on B cells and monocytes. We have recently shown that IL-13 is expressed by HL cell lines and in HRS cells from 4 cases of nodular sclerosis HL (NSHL).8 In that study, antibody-mediated neutralization of IL-13 inhibited proliferation of an HL cell line, suggesting that IL-13 might function as an autocrine growth factor for HL. As well as IL-13, the HL cell lines examined expressed NF-IL3. Expression of the NF-IL3 gene is up-regulated by IL-4,9 a cytokine that shares signaling pathways with IL-13.10 The expression of NF-IL3 by HL cells thus supports the existence of autocrine signaling by IL-13 in HL.
IL-13 signals are transduced via the IL-13 receptor (IL-13R) complex, which varies in its composition among cell types.11,12 Two distinct IL-13R chains have been identified, IL-13Rα1 and IL-13Rα2.11 IL-13Rα1 associates with IL-4Rα to form a high-affinity IL13R complex, engagement of which triggers proliferation through activation of JAK1 and subsequent phosphorylation of STAT6.13 This sharing of the IL-4α chain accounts for the overlapping biologic effects of IL-13 and IL-4.10,13Little is known about the IL-13Rα2 chain, which was not expressed in several hematopoietic cell lines12 and which appears to be a decoy receptor.14 In the mouse, IL-13 can elicit a mitogenic response through IL-13Rα1 but not through IL-13Rα2.15
To extend our investigations of the role of IL-13 in the pathogenesis of HL, we analyzed the expression patterns of IL-13 and IL-13Rα1 in a series of lymphomas, including those from patients with HL and NHL. We report here that IL-13 was expressed in 86% of patients with HL examined, a substantially higher frequency than was observed in patients with NHL. Furthermore, IL-13Rα1 was expressed in 89% of patients with HL, reinforcing the potential for IL-13 to act as an autocrine growth factor in HL.
Materials and methods
Tissues and cell lines
Formalin-fixed paraffin-embedded biopsy specimens representing the following diseases were investigated: 36 cases of classical HL (23 cases of nodular sclerosis HL [NSHL], including 6 cases of syncytial NSHL,16 10 cases of mixed cellularity HL [MCHL], and 3 cases of lymphocyte-depletion HL [LDHL]), 5 cases of nodular lymphocyte predominance HL (NLPHL), 5 cases of T-cell–rich B-cell lymphoma (TCRBCL), 8 cases of diffuse large B-cell lymphoma (DLBCL), including 4 cases with anaplastic morphology (3 of which were CD30+), 5 cases of anaplastic large-cell lymphoma (ALCL) with a T/null cell phenotype, and 5 cases of peripheral T-cell lymphoma, unspecified (PTCL). All cases were diagnosed according to the Revised European-American Lymphoma Classification.17 The cases of ALCL and TCRBCL were selected from previous studies.18 19
HL cell lines L540Cy, L428, HD-LM2, and KM-H2 were provided by Dr H. G. Drexler (Braunschweig, Germany). HL cell line L1236 and Burkitt lymphoma (BL) cell line BL-31 were provided by Prof V. Diehl (Cologne, Germany). A previous study12 demonstrated the presence of IL-13Rα1 messenger RNA (mRNA) in a BL cell line; BL-31 was therefore used as a positive control. Complementary DNA (cDNA) was prepared from cell lines using an mRNA purification kit (Pharmacia, Freiburg, Germany) and subjected to polyA amplification as described.20
Probes
The IL-13 probe was a 388-base pair (bp) fragment that has been previously described.8 The IL-13Rα1 probe was an 810-bpEcoRI-ScaI fragment cloned from the IMAGE consortium clone number 755037.21 The cDNA was subcloned into pBluescript SK (Stratagene, La Jolla, CA), and sense and antisense probes were synthesized from the linearized vector with T3 or T7 RNA polymerase, labeled with [33P]-UTP and processed as previously described.22
In situ hybridization
The in situ hybridization procedure was performed as previously described.22 Sections were deparaffinized in xylene, rehydrated in series of decreasing ethanol concentrations, washed in saline for 5 minutes, and washed 2 × 5 minutes in phosphate-buffered saline (PBS). After fixation in 4% paraformaldehyde for 20 minutes and another 5-minute wash in PBS, sections were treated with 20 μg/mL proteinase K for 7.5 minutes at room temperature. After 5 minutes in 4% paraformaldehyde and another 2 × 5-minute washes in PBS, sections were acetylated for 10 minutes in a 0.1 M triethanolamine pH 8.0/0.25% acetic anhydride solution, washed for 5 minutes in PBS, and dehydrated. Approximately 150 μL hybridization solution (50% formamide, 0.3 M NaCl, 20 mM Tris-HCl, pH 8.0, 5 mM EDTA, 10 mM NaPO4, pH 8.0, 10% dextran sulfate, 1 × Denhardt's, 0.5 mg/mL yeast transfer RNA [tRNA], 10 mM DTT, and approximately 75 000 cpm/μL of labeled sense or antisense probe) was applied to each slide. Slides were overlain with coverslips and incubated overnight in a humid chamber at 55°C. The coverslips were removed by dipping slides into 5 × SSC containing 0.1% β-mercaptoethanol. The slides were then immersed for 30 minutes in washing buffer (50% formamide, 2 × SSC, 0.1% β-mercaptoethanol) at 62°C, rinsed 2 × 15 minutes in NTE (0.5 M NaCl, 10 mM Tris-HCl, pH 7.5, 5 mM EDTA) at 37°C, treated for 30 minutes at 37°C with 20 μg/mL RNaseA, and washed for 15 minutes in NTE. The slides were then incubated for 25 minutes in washing buffer at 62°C, 15 minutes in 2 × SSC at 37°C, 15 minutes in 0.1 × SSC at 37°C, and dehydrated. The slides were coated with Kodak NTB 2 emulsion (Eastman Kodak, Rochester, NY) and stored at 4°C. After exposure for 10 to 14 days, the slides were developed in Kodak D19 (Eastman Kodak) and counterstained with toluidine blue. The specificity of the hybridization signal was verified by the corresponding lack of signal when sense RNA probes were used. Cells were scored as positive if the number of silver grains was at least 4-fold over background. Control β-actin mRNA was detected using a commercially available kit (Dako, Carpinteria, CA), following the manufacturer's instructions.
Single-cell reverse transcriptase–polymerase chain reaction
Single-cell suspensions were prepared from pathologic lymph nodes freshly isolated from 6 patients with HL who were not included in the in situ hybridization analysis. Individual HRS cells were identified by their CD30 expression and distinctive morphologic features (Figure 1) as previously described.23 In this study,23 the correct identity of the HRS cells in an EBV+ HL case was supported by demonstrating an EBV gene expression pattern that is seen in HL, BL, and nasopharyngeal carcinoma.24 The cDNA of individual HRS cells was synthesized using the global polyA-polymerase chain reaction (PCR) method as previously described.20 Only cases with a positive β-actin signal were used for subsequent IL-13Rα1 PCR. IL-13Rα1 cDNA was amplified using the following primers: 5′ CAT GAA GAG GAT GCT GTG AAA TTC CCA ACA AAC 3′ and 5′ GTT AAA CAG AAA CAA TCC CTG GTT GAA GAC TAC C 3′. PCR analysis was performed in a 50 μL volume containing 200 nM of each primer, 1.5 mM MgCl2, 1.5 units AmpliTaq Gold (Perkin Elmer, Boston, MA), with 4% DMSO. PCR conditions were as follows: denaturing at 94°C for 1 minute, annealing at 58°C for 1 minute, and extension at 72°C for 2 minutes, for 40 cycles. Products were analyzed by agarose gel electrophoresis. PCR products were confirmed by hybridization with a [32P]–end-labeled internal oligonucleotide (5′ ATG GGA AAT CCA CTG ATA CAG ACA CCT CCA AGA GC 3′), as previously described.20 Appropriate positive (cDNA from BL-31 or KM-H2 cells) and negative (cells with no reverse transcriptase; H20) controls were included.
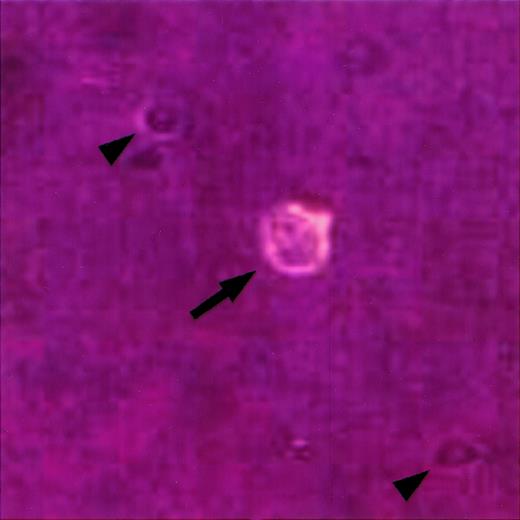
Identification of single HRS cells in cell suspension.
A single cell suspension was prepared from a lymph node involved by HL, stained with an anti-CD30 antibody coupled to phycoerythrin, and viewed under an inverted fluorescence microscope as previously described.23 A single HRS cell is identified by its large size, bilobed nucleus with prominent nucleoli, and CD30 expression (arrow). Note smaller CD30-negative cells in the background (arrowheads).
Identification of single HRS cells in cell suspension.
A single cell suspension was prepared from a lymph node involved by HL, stained with an anti-CD30 antibody coupled to phycoerythrin, and viewed under an inverted fluorescence microscope as previously described.23 A single HRS cell is identified by its large size, bilobed nucleus with prominent nucleoli, and CD30 expression (arrow). Note smaller CD30-negative cells in the background (arrowheads).
Results
IL-13 is expressed by HRS cells in a large majority of patients with HL
IL-13 expression was analyzed by in situ hybridization in tissue samples from 36 patients with classical HL, 5 patients with NLPHL, and 23 patients with NHL. IL-13 transcripts were detected in HRS cells in 31 of 36 (86%) patients with classical HL (Table1, Figure2A). In contrast, the lymphocytic and histiocytic (L&H) cells in all 5 patients with NLPHL were negative for IL-13 (Table 1; Figure 2B). Similarly, only 1 of 5 patients with TCRBCL demonstrated IL-13 expression in less than 25% of the malignant cell population, and only 2 of 5 patients with ALCL were IL-13 positive (25% to 50% of cells). Only 1 of 5 patients with PTCL had scattered malignant cells (less than 10%) that expressed IL-13, and all 8 patients with DLBCL were negative for IL-13.
Expression of IL-13 and IL-13Rα1 in Hodgkin lymphomas and non-Hodgkin lymphomas
| Diagnosis . | IL-13+ cases*/ total cases (%) . | IL-13Rα1+ cases*/total cases (%) . |
|---|---|---|
| Classical Hodgkin lymphoma | 31/36 (86%) | 24/27 (89%) |
| Nodular sclerosis | 21/23 (91%) | 16/17 (94%) |
| Mixed cellularity | 8/10 (80%) | 5/7 (71%) |
| Lymphocyte depletion | 2/3 (67%) | 3/3 (100%) |
| Nodular lymphocyte predominance Hodgkin lymphoma | 0/5 | ND |
| Diffuse large B-cell lymphoma | 0/8 | ND |
| T-cell-rich B-cell lymphoma | 1/5 (20%) | ND |
| Anaplastic large-cell lymphoma | 2/5 (40%) | ND |
| Peripheral T-cell lymphoma, unspecified | 1/5 (20%) | ND |
| Diagnosis . | IL-13+ cases*/ total cases (%) . | IL-13Rα1+ cases*/total cases (%) . |
|---|---|---|
| Classical Hodgkin lymphoma | 31/36 (86%) | 24/27 (89%) |
| Nodular sclerosis | 21/23 (91%) | 16/17 (94%) |
| Mixed cellularity | 8/10 (80%) | 5/7 (71%) |
| Lymphocyte depletion | 2/3 (67%) | 3/3 (100%) |
| Nodular lymphocyte predominance Hodgkin lymphoma | 0/5 | ND |
| Diffuse large B-cell lymphoma | 0/8 | ND |
| T-cell-rich B-cell lymphoma | 1/5 (20%) | ND |
| Anaplastic large-cell lymphoma | 2/5 (40%) | ND |
| Peripheral T-cell lymphoma, unspecified | 1/5 (20%) | ND |
Cases with IL-13 or IL-13Rα1 positive malignant cell population (HRS cells in classical HL, L&H cells in NLPHL, and large B-cells in TCRBCL); ND = not done.
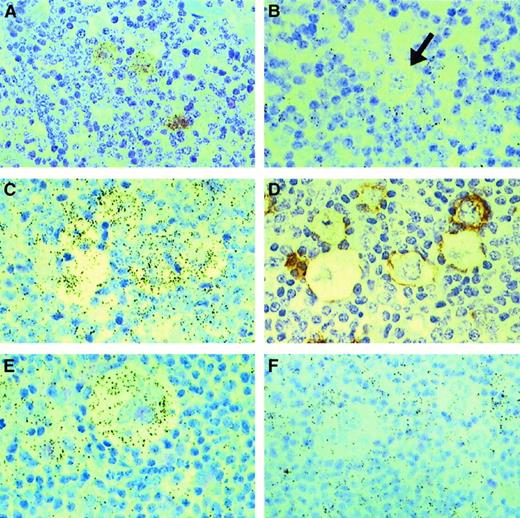
IL-13 and IL-13Rα1 expression in Hodgkin lymphoma.
In situ hybridization for IL-13 (A, B) and IL-13Rα1 (C, E, F); black-colored silver grains denote mRNA expression. HRS cells from a case of mixed cellularity HL (A) express IL-13, whereas an L&H cell (arrow) from a case of NLPHL (B) is negative for IL-13. HRS cells from 2 cases of nodular sclerosis HL (C, E) express IL-13Rα1, whereas HRS cells from a case of mixed cellularity HL (F) are negative for IL-13Rα1. Serial sections from one of the cases of nodular sclerosis (C, D) were hybridized with an IL-13Rα1 antisense probe (C) or stained for CD30 (D), illustrating IL-13Rα1 expression in CD30+ HRS cells. Note IL-13Rα1 expression in surrounding lymphocytes and histiocytes (C, F).
IL-13 and IL-13Rα1 expression in Hodgkin lymphoma.
In situ hybridization for IL-13 (A, B) and IL-13Rα1 (C, E, F); black-colored silver grains denote mRNA expression. HRS cells from a case of mixed cellularity HL (A) express IL-13, whereas an L&H cell (arrow) from a case of NLPHL (B) is negative for IL-13. HRS cells from 2 cases of nodular sclerosis HL (C, E) express IL-13Rα1, whereas HRS cells from a case of mixed cellularity HL (F) are negative for IL-13Rα1. Serial sections from one of the cases of nodular sclerosis (C, D) were hybridized with an IL-13Rα1 antisense probe (C) or stained for CD30 (D), illustrating IL-13Rα1 expression in CD30+ HRS cells. Note IL-13Rα1 expression in surrounding lymphocytes and histiocytes (C, F).
Among the classical HL cases, no significant differences in IL-13 expression were detected among subtypes of classical HL: 21 of 23 (91%) of nodular sclerosis cases, 8 of 10 (80%) of mixed cellularity cases, and 2 of 3 (67%) of lymphocyte depletion cases contained positive HRS cells (P = not significant) (Table2). Nearly all cells expressing IL-13 demonstrated the morphology of HRS cells, with only rare small lymphocytes in 19 of 36 (53%) cases also staining positively. Staining of serial sections with either anti-CD30 or anti-CD15 to identify HRS cells showed that the percentage of IL-13–expressing HRS cells varied from case to case, ranging from 25% to almost 100%. A study of the slides in the same hybridization batch and exposed for the same length of time revealed that there was no significant difference among the different subtypes of classical HL in the percentage of HRS cells staining positively for IL-13. Similarly, although the intensity of the IL-13 signal varied from case to case, there was no significant difference in the distribution of signal intensity among subtypes of classical HL.
IL-13 and IL-13Rα1 expression in patients with Hodgkin lymphoma
| Case no. . | Diagnosis . | Age/sex . | Stage . | IL-13 + HRS cells . | IL-13Rα1 + HRS cells . |
|---|---|---|---|---|---|
| 1 | NSHL | 16/M | IV B | + | + |
| 2 | NSHL | 24/F | II A | + | + |
| 3 | NSHL | 15/F | II A | + | + |
| 4 | NSHL | 43/M | IV B | + | + |
| 5 | NSHL | 33/M | III B | + | + |
| 6 | NSHL | 30/M | II A | + | + |
| 7 | NSHL | 32/F | II B | + | + |
| 8 | NSHL | 25/F | III B | + | ND |
| 9 | NSHL | 27/F | II A | + | + |
| 10 | NSHL | 22/M | III B | + | U |
| 11 | NSHL | 34/M | III A | + | + |
| 12 | NSHL | 19/M | III A | + | + |
| 13 | NSHL | 81/F | II A | + | ND |
| 14 | NSHL | 36/M | III B | + | + |
| 15 | NSHL | 31/F | IV A | + | ND |
| 16 | NSHL | NA | NA | + | ND |
| 17 | NSHL | NA | NA | + | + |
| 18 | NSHL, syn | 64/M | III B | − | − |
| 19 | NSHL, syn | 20/F | II A | + | + |
| 20 | NSHL, syn | 35/M | I B | − | + |
| 21 | NSHL, syn | 45/M | III A | + | + |
| 22 | NSHL, syn | 41/F | II A | + | U |
| 23 | NSHL, syn | 25/M | III B | + | + |
| 24 | MCHL | NA | NA | + | + |
| 25 | MCHL | NA | NA | − | − |
| 26 | MCHL | 28/M | II A | + | + |
| 27 | MCHL | 28/M | II B | + | ND |
| 28 | MCHL | 42/M | I A | + | + |
| 29 | MCHL | NA/M | II A | + | ND |
| 30 | MCHL | 42/F | III A | + | + |
| 31 | MCHL | NA/F | II A | + | + |
| 32 | MCHL | 45/M | I A | + | ND |
| 33 | MCHL | 88/M | I A | − | − |
| 34 | LDHL | 54/F | IV B | + | + |
| 35 | LDHL | 65/M | NA | − | + |
| 36 | LDHL | 72/F | IV B | + | + |
| Case no. . | Diagnosis . | Age/sex . | Stage . | IL-13 + HRS cells . | IL-13Rα1 + HRS cells . |
|---|---|---|---|---|---|
| 1 | NSHL | 16/M | IV B | + | + |
| 2 | NSHL | 24/F | II A | + | + |
| 3 | NSHL | 15/F | II A | + | + |
| 4 | NSHL | 43/M | IV B | + | + |
| 5 | NSHL | 33/M | III B | + | + |
| 6 | NSHL | 30/M | II A | + | + |
| 7 | NSHL | 32/F | II B | + | + |
| 8 | NSHL | 25/F | III B | + | ND |
| 9 | NSHL | 27/F | II A | + | + |
| 10 | NSHL | 22/M | III B | + | U |
| 11 | NSHL | 34/M | III A | + | + |
| 12 | NSHL | 19/M | III A | + | + |
| 13 | NSHL | 81/F | II A | + | ND |
| 14 | NSHL | 36/M | III B | + | + |
| 15 | NSHL | 31/F | IV A | + | ND |
| 16 | NSHL | NA | NA | + | ND |
| 17 | NSHL | NA | NA | + | + |
| 18 | NSHL, syn | 64/M | III B | − | − |
| 19 | NSHL, syn | 20/F | II A | + | + |
| 20 | NSHL, syn | 35/M | I B | − | + |
| 21 | NSHL, syn | 45/M | III A | + | + |
| 22 | NSHL, syn | 41/F | II A | + | U |
| 23 | NSHL, syn | 25/M | III B | + | + |
| 24 | MCHL | NA | NA | + | + |
| 25 | MCHL | NA | NA | − | − |
| 26 | MCHL | 28/M | II A | + | + |
| 27 | MCHL | 28/M | II B | + | ND |
| 28 | MCHL | 42/M | I A | + | + |
| 29 | MCHL | NA/M | II A | + | ND |
| 30 | MCHL | 42/F | III A | + | + |
| 31 | MCHL | NA/F | II A | + | + |
| 32 | MCHL | 45/M | I A | + | ND |
| 33 | MCHL | 88/M | I A | − | − |
| 34 | LDHL | 54/F | IV B | + | + |
| 35 | LDHL | 65/M | NA | − | + |
| 36 | LDHL | 72/F | IV B | + | + |
NSHL = nodular sclerosis Hodgkin lymphoma; syn = syncytial variant; MCHL = mixed cellularity Hodgkin lymphoma; LDHL = lymphocyte-depletion Hodgkin lymphoma; U = uninterpretable; ND = not done; NA = not available.
All 5 IL-13 negative cases showed a positive signal for IL-13Rα1 (see below), excluding the possibility of false-negative results due to degradation of RNA before fixation or during tissue processing. Selected NHL cases that were negative for IL-13 expression were analyzed for β-actin mRNA by in situ hybridization. All of these were positive for β-actin expression (data not shown), again suggesting that the RNA was intact.
IL-13Rα1 is expressed by HRS cells expressing IL-13
To determine whether IL-13–expressing HRS cells also expressed IL-13Rα1, and thus could become subject to autocrine stimulation, IL-13Rα1 expression was examined by in situ hybridization in 29 cases of classical HL and was interpretable in 27 cases (see below). Twenty-four cases (89%) contained HRS cells positive for IL-13Rα1 expression (Figure 2C-F). No significant differences in IL-13Rα1 expression were detected among subtypes of classical HL: 16 of 17 (94%) of nodular sclerosis cases, 5 of 7 (71%) of mixed cellularity cases, and 3 of 3 (100%) of lymphocyte-depletion cases contained positive HRS cells (P = NS). The percentage of IL-13Rα1 positive HRS cells varied from 25% to 75% in different cases. In all cases, IL13Rα1 expression was not limited to HRS cells, but was also present in a large proportion of cells in reactive infiltrates that could be morphologically defined as lymphocytes and histiocytes (Figure2C,F). IL-13Rα1 transcripts could not be detected in the fibroblasts within the sclerotic bands in cases of NSHL. As a positive control, a benign reactive tonsil was hybridized with the IL-13Rα1 probe. Positive signals were obtained for cells morphologically resembling histiocytes (including tingible-body macrophages in reactive germinal centers) and numerous lymphocytes outside the germinal center (data not shown). Two cases of HL that contained reactive germinal centers with tingible-body macrophages did not show the IL-13Rα1 signal in any cell population, and were therefore considered noninterpretable.
Of the 27 cases of classical HL analyzed for both IL-13 and IL-13Rα1, 22 cases were IL-13+ / IL-13Rα1+; 3 cases were IL-13−/ IL-13Rα1−; and 2 cases were IL-13−/ IL-13Rα1+ (Table 2). The relationship between IL-13 and IL-13Rα1 expression was statistically significant (P = .003).
Expression of IL-13Rα1 as determined by PCR of individual HRS cell cDNA libraries
To confirm the presence of IL-13Rα1 mRNA in individual HRS cells, cDNA libraries of mRNA from HRS cells were constructed and analyzed by PCR with IL-13Rα1–specific primers. First, the presence of IL-13Rα1 sequences in 5 HRS cell lines (KM-H2, HD-LM2, L1236, L540Cy, and L428) was examined. IL-13Rα1 transcripts were detected in all 5 cell lines (Figure 3). These studies were extended to construct cDNA libraries from single HRS cells from 6 patients with HL, including 4 patients with NSHL and 2 patients with MCHL. In all 6 patients, IL-13Rα1 sequences were detected in 30% to 85% of HRS cells (Table 3; Figure 4) and were confirmed by hybridization with an internal oligonucleotide (data not shown).
IL-13Rα1 expression by HL cell lines.
The cDNA was prepared from HL cell lines (KM-H2, HD-LM2, L1236, L540Cy, and L428) and amplified by PCR using IL-13Rα1–specific oligonucleotides, followed by agarose gel electrophoresis. The presence of the 430-bp PCR product indicates IL-13Rα1 gene expression in all 5 HL cell lines examined. Negative control (−): H20; positive control (+): cDNA from BL-31 cells. MW = 100-bp ladder.
IL-13Rα1 expression by HL cell lines.
The cDNA was prepared from HL cell lines (KM-H2, HD-LM2, L1236, L540Cy, and L428) and amplified by PCR using IL-13Rα1–specific oligonucleotides, followed by agarose gel electrophoresis. The presence of the 430-bp PCR product indicates IL-13Rα1 gene expression in all 5 HL cell lines examined. Negative control (−): H20; positive control (+): cDNA from BL-31 cells. MW = 100-bp ladder.
IL-13Rα1 polymerase chain reaction from single Hodgkin and Reed-Sternberg cell complementary DNA libraries
| Case no. . | Diagnosis . | Age/sex . | No. HRS cells examined . | No. IL-13Rα1 + HRS cells/no. HRS cells examined (%) . |
|---|---|---|---|---|
| 37 | NSHL | 25/F | 20 | 15/20 (75%) |
| 38 | NSHL | 28/F | 20 | 6/20 (30%) |
| 39 | NSHL | 33/M | 8 | 3/8 (37.5%) |
| 40 | NSHL | 26/M | 20 | 7/20 (37%) |
| 41 | MCHL | 29/M | 20 | 17/20 (85%) |
| 42 | MCHL | 30/M | 19 | 12/19 (63%) |
| Case no. . | Diagnosis . | Age/sex . | No. HRS cells examined . | No. IL-13Rα1 + HRS cells/no. HRS cells examined (%) . |
|---|---|---|---|---|
| 37 | NSHL | 25/F | 20 | 15/20 (75%) |
| 38 | NSHL | 28/F | 20 | 6/20 (30%) |
| 39 | NSHL | 33/M | 8 | 3/8 (37.5%) |
| 40 | NSHL | 26/M | 20 | 7/20 (37%) |
| 41 | MCHL | 29/M | 20 | 17/20 (85%) |
| 42 | MCHL | 30/M | 19 | 12/19 (63%) |
NSHL = nodular sclerosis Hodgkin lymphoma; MCHL = mixed cellularity Hodgkin lymphoma.
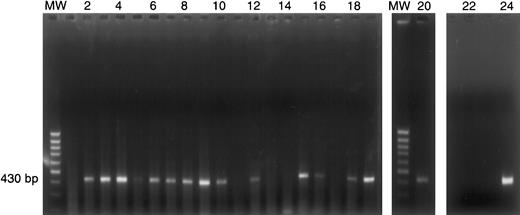
IL-13Rα1 expression by individual HRS cells.
Single HRS cells from case 37 were micromanipulated and subjected to polyA PCR as described in “Materials and methods.” PolyA cDNA products from 20 individual HRS cells (lanes 1-20) were amplified by PCR with IL-13Rα1–specific oligonucleotides. The 430-bp PCR product indicates IL-13Rα1 gene expression in 15 of 20 single HRS cells. The presence of IL-13Rα1 cDNA was confirmed by hybridization with an internal oligonucleotide. MW = 100-bp ladder; lanes 1-20: single HRS cells; lane 21: negative control (single HRS cell, no reverse transcriptase); lane 22: blank; lane 23: negative control (H20); lane 24: positive control (cDNA from KM-H2 cells).
IL-13Rα1 expression by individual HRS cells.
Single HRS cells from case 37 were micromanipulated and subjected to polyA PCR as described in “Materials and methods.” PolyA cDNA products from 20 individual HRS cells (lanes 1-20) were amplified by PCR with IL-13Rα1–specific oligonucleotides. The 430-bp PCR product indicates IL-13Rα1 gene expression in 15 of 20 single HRS cells. The presence of IL-13Rα1 cDNA was confirmed by hybridization with an internal oligonucleotide. MW = 100-bp ladder; lanes 1-20: single HRS cells; lane 21: negative control (single HRS cell, no reverse transcriptase); lane 22: blank; lane 23: negative control (H20); lane 24: positive control (cDNA from KM-H2 cells).
Discussion
We have shown that IL-13 is expressed in a variable percentage of HRS cells in 86% of the classical HL cases examined. Transcripts for IL-13 are present in HRS cells of NSHL, MCHL, and LDHL, with no significant differences in the intensity of signal or percentage of positive cells between HL subtypes. The HRS cells appear to be the dominant source of IL-13, with rare lymphocytes showing a positive signal in approximately half of the cases. There were no morphologic differences between IL-13 positive and IL-13 negative cases, either in the presence of fibrosis or composition of reactive infiltrate. It is possible that IL-13 is still present in these “negative” cases but at levels undetectable by in situ hybridization. Alternatively, some cases of HL may be truly negative for IL-13, in which case other cytokines presumably play a role in their pathogenesis. Our data suggest that IL-13 expression is a basic property of HRS cells in the majority of cases of HL, regardless of histologic subtypes.
IL-13 mRNA was not detected in the L&H cells of NLPHL, which shows clinical, morphologic, and immunophenotypic features distinct from those of classical HL.25,26 HRS cells in classical HL are typically CD15+CD30+ and are negative for B-cell markers in the majority of cases.1 Classical HL is believed in many cases to be a B-lineage lymphoproliferative disorder in which the HRS cells demonstrate clonal rearrangement and somatic mutations of the Ig heavy chain genes but do not express Ig.27 28 The L&H cells of NLPHL, on the other hand, demonstrate a B-cell phenotype, produce functional Ig, and are typically CD15−CD30−. The absence of IL-13 in NLPHL further emphasizes the difference between these 2 distinct disease entities.
IL-13 is produced predominantly by activated T cells and acts on several cell types.6,7 In B cells, IL-13 promotes survival and proliferation, as well as Ig class switching to IgG4 and IgE.6,7,29 IL-13 plays an important role in Th2 cell differentiation in the mouse,30 but its exact role in T-cell development in humans is unclear. Although some investigators have reported that human T cells lack surface IL-13 receptors,31 others have shown that T cells can respond to IL-13 in vitro.32 Human fibroblasts express the IL-13 receptor,10,14 and IL-13 has been shown to play a role in development of fibrosis in asthma33 and parasite-associated hepatic damage.34 In view of these effects, many features of HL, including the reactive infiltrate of T and B cells, histiocytes, as well as the fibrosis in NSHL, may be attributable to IL-13 expression. Indeed, it has been shown that in transgenic mice that overexpress IL-13 in the lung, elevated levels of IL-13 lead to the presence of a mononuclear infiltrate, eosinophilia, and fibrosis.33
Although the effects of IL-13 on target cells have been reasonably well studied, the receptor complex through which it acts is much less well understood. In the best characterized receptor complex, IL-13Rα1 associates with IL-4α, but the IL-13 receptor structure varies among different cell types.10-13 The precise function of the IL-13Rα2 chain has yet to be defined.14,15 In this study, in situ hybridization of tissue sections showed that HRS cells in 89% of classical HL cases expressed IL-13Rα1. IL-13Rα1 expression was also demonstrated using PCR of cDNA libraries made from single HRS cells picked from cell suspensions of lymph nodes with HL involvement. The presence of IL-13Rα1 on HRS cells that also express IL-13 suggests the potential existence of autocrine regulatory loops influencing HRS cell proliferation. These results support the findings of our previous study in which it was shown that HL cell lines both produce IL-13 and express the IL-13 receptor, and that neutralization of IL-13 can lead to inhibition of proliferation.8However, whereas cells in the majority of HL cases in this study expressed both IL-13 and IL-13Rα1, HRS cells in a few cases expressed neither the cytokine nor the receptor, implying that IL-13 does not act as an autocrine growth factor in all cases. The clinical significance of the absence of IL-13 and IL-13Rα1 in these cases remains to be determined.
In addition to HRS cells, many histiocytes and lymphocytes expressed IL-13Rα1. The presence of the receptor on those cells surrounding the IL-13 positive HRS cells would allow a paracrine mechanism to facilitate the accumulation of the mixed inflammatory infiltrate characteristic of HL. However, although IL-13Rα1 is expressed in human fibroblast cell lines,10 14 IL-13Rα1 expression could not be detected in fibroblasts in the sclerotic bands of NSHL cases examined in this study. The particular role that IL-13 plays in the development of fibrosis in NSHL remains unclear because there was no difference in IL-13 expression between cases of NSHL and MCHL, which by definition lack sclerotic collagenous bands.
IL-13 was also detected in 3 NHL cases that share some morphologic or immunophenotypic features with HL, including 1 of 5 cases of TCRBCL and 2 of 5 cases of ALCL. TCRBCL is an aggressive NHL that morphologically resembles HL because of its characteristic prominent reactive component of benign T cells and histiocytes,19,35 suggesting that it is also a malignancy with abnormal cytokine expression. Indeed, the neoplastic cells of both HL and TCRBCL have been shown to produce the CC chemokine TARC (thymus and activation-regulated chemokine), likely accounting for the prominent T-cell infiltration of both tumors.36 Although IL-13 transcripts were detected in the malignant cells of only one case of TCRBCL in this study, expression of IL-4, whose biologic effects overlap those of IL-13, was found by others in malignant cells of additional TCRBCL cases.37 HL shows an opposite expression pattern of these 2 related cytokines, namely, positive expression of IL-13 but negative expression of IL-4.4 These results suggest that IL-4 and IL-13 may contribute to a malignant phenotype to different degrees in different types of lymphomas.
Because T cells are the predominant source of IL-13 secretion, we analyzed 9 cases of T-cell lymphomas for the presence of IL-13, including cases of ALCL and PTCL. ALCL shares some features with HL, including pleomorphic morphology of the neoplastic cells and CD30 expression.18 In situ hybridization studies showed that 2 cases of ALCL and one case of PTCL contained IL-13 positive cells. Unlike HL, these cases contained a homogeneous population of lymphoma cells without a prominent reactive infiltrate. These data do not permit us to conclude that IL-13 plays a defining role in the pathogenesis of subsets of T-cell lymphomas. In some tumors, IL-13 expression may merely reflect the retention of gene expression patterns of the cell of origin.
In conclusion, we have shown that IL-13 and IL-13Rα1 are frequently expressed in HRS cells of HL. IL-13 is important for normal B-cell physiology, inducing survival and proliferation of these cells.29 Because most cases of classical HL are derived from B cells,27,28 IL-13 may also function as a growth factor for these tumors. The possible role of IL-13 in autocrine growth of HRS cells and paracrine stimulation of the reactive infiltrate suggests that IL-13 signaling is a potential pharmacological target in the treatment of patients with HL. Inhibition of IL-13 signaling for therapeutic purposes may not be seriously detrimental to the immune system of patients with HL. Although the biologic activities of IL-13 and IL-4 overlap, IL-4 is not a cytokine associated with HL.4 Mice deficient for IL-13 (but wild type for IL-4) are still able to develop a Th2 cytokine and Ig response to parasitic infection, although clearance of the pathogen is less efficient.30 Thus, medical intervention to block IL-13–mediated autocrine stimulation of HRS cells should not compromise an IL-4–mediated Th2 response, allowing the patient some measure of immune protection during antitumor treatment.
Acknowledgments
We thank J. Ho for excellent technical support; H. G. Drexler, B. Dörken, and V. Diehl for HL-derived and BL-derived cell lines; M. Bray for valuable critical comments; N. Le for statistical analysis; and M. Saunders for scientific editing.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Tak W. Mak, Ontario Cancer Institute/Amgen Institute, 620 University Ave, Suite 706, Toronto, Ontario, Canada M5G 2C1; e-mail: tmak@amgen.com.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal