Abstract
Fanconi anemia (FA) is an autosomal recessive cancer susceptibility syndrome with 8 complementation groups. Four of the FA genes have been cloned, and at least 3 of the encoded proteins, FANCA, FANCC, and FANCG/XRCC9, interact in a multisubunit protein complex. The FANCG protein binds directly to the amino terminal nuclear localization sequence (NLS) of FANCA, suggesting that FANCG plays a role in regulating FANCA nuclear accumulation. In the current study the functional consequences of FANCG/FANCA binding were examined. Correction of an FA-G cell line with the FANCG complementary DNA (cDNA) resulted in FANCA/FANCG binding, prolongation of the cellular half-life of FANCA, and an increase in the nuclear accumulation of the FA protein complex. Similar results were obtained upon correction of an FA-A cell line, with a reciprocal increase in the half-life of FANCG. Patient-derived mutant forms of FANCA, containing an intact NLS sequence but point mutations in the carboxy-terminal leucine zipper region, bound FANCG in the cytoplasm. The mutant forms failed to translocate to the nucleus of transduced cells, thereby suggesting a model of coordinated binding and nuclear translocation. These results demonstrate that the FANCA/FANCG interaction is required to maintain the cellular levels of both proteins. Moreover, at least one function of FANCG and FANCA is to regulate the nuclear accumulation of the FA protein complex. Failure to accumulate the nuclear FA protein complex results in the characteristic spectrum of clinical and cellular abnormalities observed in FA.
Introduction
Fanconi anemia (FA) is an autosomal recessive cancer susceptibility syndrome characterized by multiple congenital anomalies, progressive bone marrow failure, and cellular sensitivity to DNA cross-linking agents.1,2 Based on somatic cell fusion studies, FA comprises 8 distinct complementation groups.3Four of the FA genes, including the FANCA,FANCG, FANCC, and FANCF genes, have been cloned.4-8 The FANCG gene was found to be identical to human XRCC9, a gene proposed to be involved in cell cycle regulation or post-replication repair.9 Two additional genes, FANCD10 andFANCE,11 have been mapped. The 4 cloned FA proteins have little or no homology to each other or to other proteins in the database, and little is known regarding their cellular function. However, based on the similar clinical and cellular phenotypes observed among the 8 FA complementation groups, the FA genes appear to participate in a common cellular pathway.
Cells from FA patients display a broad range of abnormalities12,13 including hypersensitivity to DNA cross-linking agents and oxygen radicals,14,15 spontaneous chromosome breakage,16 cell cycle abnormalities manifested by a prolongation in the G2 phase of the cell cycle,17,18and increased cellular sensitivity to the apoptotic effects of tumor necrosis factor (TNF)-α and interferon (IFN)-γ.19,20Most recently, FA cells have been shown to have a defect in the fidelity of the DNA double-strand break repair, which suggests a primary defect in a pathway regulating DNA repair.21 22Whether these phenotypic features are shared by all FA complementation groups or only by a subset remains unclear. Moreover, many of the abnormalities described for FA cells could be epiphenomena and may not relate directly to the primary cellular defect.
Increasing evidence demonstrates that the FA proteins cooperate in a novel cellular pathway.23 FANCA, FANCG, and FANCC bind and interact in a protein complex, which is observed in the cytoplasm and the nucleus of normal cells.24-26 Interestingly, this protein complex is not observed in FA cells derived from other FA complementation groups including groups B, E, F, and H.27These results suggest that the products of other FA genes, such as the recently cloned FANCF gene,8 may regulate the formation of the complex, perhaps by serving as adaptor proteins or enzymes regulating protein complex assembly. The FA-D complementation group is distinct from other FA groups.27 In FA-D cells the FA protein complex assembles normally, which suggests that the FANCD protein product may function downstream or independently of the complex.
Patient-derived mutant forms of FA proteins have been instructive in delineating functional domains of these proteins and in defining the biochemical features of the FA pathway. Point mutations in the carboxy-terminal of FANCC, such as the patient-derived FANCC-L554P mutation,5 block complex formation.28Similarly, point mutations in the carboxy-terminal leucine zipper region of FANCA, such as the histidine (His) 1110 to proline (Pro) mutation,29 give rise to a FANCA protein that is defective in FANCC binding, phosphorylation, and nuclear localization. Point mutations thereby help to define many of the normal biochemical events in the FA pathway.27
Recent studies suggest that the FANCA and FANCG proteins participate in a direct interaction.24,30,31 The amino terminal NLS of FANCA, containing amino acids 1-36, forms a direct binding interaction with FANCG.31,32 In contrast, the interaction of FANCC with the FA protein complex appears to be indirect and requires additional post-translational modifications or adaptor proteins.27
In the current study we examined the functional consequences of FANCG/FANCA binding. Interestingly, we found that the FANCG and FANCA proteins stabilize each other and promote the nuclear accumulation of the FA protein complex. Moreover, the binding of FANCA and FANCG initially occurs in the cytoplasm of normal cells prior to the regulated transport of the complex to the nucleus. Accumulation of this nuclear complex is required for the maintenance of chromosome stability.
Materials and methods
Cell culture
Epstein-Barr virus (EBV)–immortalized lymphoblasts were maintained in Roswell Park Memorial Institute (RPMI) media supplemented with 15% heat-inactivated fetal calf serum (FCS) and grown in a humidified 5% carbon dioxide (CO2)–containing atmosphere at 37°C. We used the FA-G lymphoblast line, EUFA316 (Dr Hans Joenje, Free University, Amsterdam, The Netherlands). The SV40-transformed FA-A fibroblasts, GM6914, expressing various mutant forms of the FANCA polypeptide, have been previously described.33
Retroviral infection of FA cell lines
The indicated pMMP constructs were transfected by lipofection into 293 producer cells (human embryonic kidney cells) expressing the VSV-G envelope protein.34 35 Retroviral supernatants were collected on day 5 following lipofection, and they contained 4.6 × 106 infectious U/mL, as estimated by Southern blot analysis of infected National Institutes of Health (NIH)-3T3 cells (data not shown).
FA lymphoblasts were infected with the various pMMP supernatants for 4 hours in the presence of 8 μg/mL polybrene (Sigma Chemical Co, St Louis, MO). Infected cells were washed free of viral supernatant and resuspended in growth media. After 48 hours the cells were transferred to media containing 1 μg/mL puromycin. The dead cells were removed over a Ficoll cushion after 5 days, and the surviving cells were grown under continuous selection in puromycin.
Analysis of FANCA levels by flow cytometry
The indicated FA lymphoblasts were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 minutes at room temperature and permeabilized for 10 minutes using 0.3% Triton X-100 in PBS. After washing, the cells were resuspended in blocking buffer (3% bovine serum albumin [BSA] and 0.1% NP-40 in PBS) and incubated for 1 hour prior to the addition of anti-FANCA or anti-FANCG affinity-purified rabbit antisera at a 1:100 dilution. After either 2 hours at room temperature or 15 hours at 4°C, the cells were washed 3 times with PBS and incubated for an additional hour with a fluorescein isothiocyanate (FITC)–conjugated goat antirabbit antibody (Jackson Immunoresearch Laboratories, West Grove, PA) diluted 1:400 in blocking buffer. The cells were rinsed again with PBS and resuspended in PBS with 2% BSA, and cellular fluorescence was measured using a fluorescence-activated cell sorter (FACScan; Becton Dickinson, San Jose, CA). Data analysis was subsequently performed with the software program CellQuest (Becton Dickinson).
Immunoprecipitation and pulse chase experiments
Whole cell extracts were prepared in lysis buffer comprising 50 mmol/L Tris HCl (tris[hydroxymethyl] aminomethane–hydrochloride) (pH 7.4), 150 mmol/L sodium chloride (NaCl), and 1% (vol/vol) Triton X-100. The buffer was supplemented with protease inhibitors (1 μg/mL leupeptin and pepstatin, 2 μg/mL aprotinin, and 1 mmol/L phenylmethylsulfonylfluoride) and phosphatase inhibitors (1 mmol/L sodium orthovanadate and 10 mmol/L sodium fluoride). Immunoprecipitation and Western blotting were performed as previously described24 using affinity-purified rabbit antisera raised against the C-termini of FANCA, FANCC, or FANCG.24 28 For pulse-chase experiments the cells were metabolically labeled with sulfur 35 (35S)–methionine for 30 minutes, washed, and chased in cold (unlabeled) medium for the indicated time periods. The cells were then lysed in lysis buffer, and the labeled proteins were immunoprecipitated with anti-FANCA or anti-FANCG affinity-purified antisera and resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) followed by autoradiography.
Cell fractionation
Both lymphoblasts and fibroblasts were fractionated into cytoplasmic and nuclear proteins by hypotonic swelling. This was followed by Dounce homogenization, essentially as previously described.36
Generation of human FANCA complementary DNA mutant constructs
Wild type and mutant FANCA complementary DNA (cDNA) was subcloned into the retroviral expression vector pMMP34 as previously described.33 Mutations in the FANCA cDNA were introduced by polymerase chain reaction (PCR) with Pfu polymerase (Stratagene, La Jolla, CA), and cDNA inserts were verified by DNA sequencing.
Immunofluorescence microscopy
Immunofluorescence microscopy of human fibroblasts was performed as previously described.33
Results
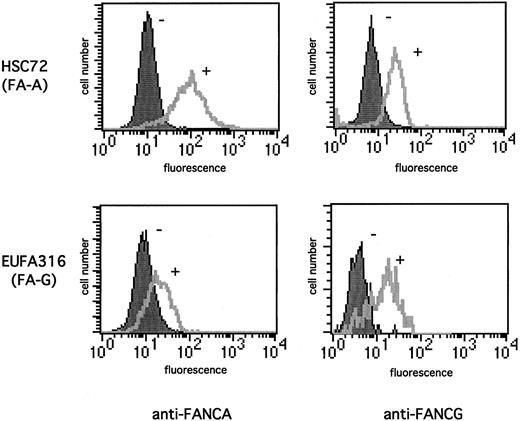
FANCA/FANCG binding regulates the stability of the FA protein complex. We initially demonstrated that FANCG increases FANCA protein levels by using a flow cytometric approach (Figure1) which allows analysis of the cellular contents of a given protein without the need for cell lysis. FA-A lymphoblasts (HSC72) failed to express the FANCA protein28and showed little staining with an anti-FANCA antisera (Figure 1A, upper panel). Functional complementation of these cells with the FANCA cDNA resulted in the correction of MMC sensitivity24 and an increase in the anti-FANCA fluorescence signal (Figure 1A). The FA-G lymphoblast EUFA 316, which failed to express the FANCG protein, also showed little anti-FANCA staining (Figure 1B). However, correction of the MMC sensitivity of these cells by transduction of the FANCG cDNA resulted in a significant increase in the FANCA protein levels (Figure1B). Moreover, functional complementation of the FA-A cells (HSC72) also lead to a significant increase in the anti-FANCG fluorescence signal (Figure 1C), which suggests that the positive effect on the expression level was reciprocal between the 2 FA proteins.
Functional complementation of FA cells increases the expression of FANCA and FANCG proteins.
The indicated lymphoblast lines were stained with 1:100 dilution anti-FANCA antiserum (A-B) or anti-FANCG antiserum (C-D). The cells analyzed were FA-A (HSC72) lymphoblasts, which were uncorrected (−) or corrected (+) with the FANCA cDNA (A, C), or FA-G (EUFA316) lymphoblasts, which were uncorrected (−) or corrected (+) with the FANCG cDNA (B, D). The secondary antibody was FITC-conjugated antirabbit antibody. Cellular fluorescence was measured by FACScan.
Functional complementation of FA cells increases the expression of FANCA and FANCG proteins.
The indicated lymphoblast lines were stained with 1:100 dilution anti-FANCA antiserum (A-B) or anti-FANCG antiserum (C-D). The cells analyzed were FA-A (HSC72) lymphoblasts, which were uncorrected (−) or corrected (+) with the FANCA cDNA (A, C), or FA-G (EUFA316) lymphoblasts, which were uncorrected (−) or corrected (+) with the FANCG cDNA (B, D). The secondary antibody was FITC-conjugated antirabbit antibody. Cellular fluorescence was measured by FACScan.
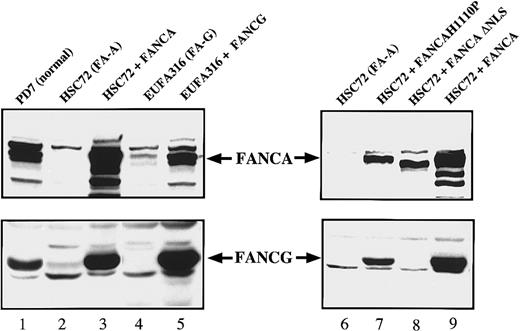
These results were also confirmed by immunoblot analysis (Figure2). Again, correction of FA-A cells with FANCA resulted in a net increase in the cellular FANCG levels (Figure2, lanes 2 and 3); correction of FA-G cells with FANCG resulted in a net increase in the cellular FANCA levels (Figure 2, lanes 4 and 5). We next sought to determine whether the induction in the FANCA and FANCG protein level was either a consequence of the phenotypic correction of the cells or directly related to the FANCA/FANCG interaction. For this purpose, in the FA-A lymphoblasts (HSC72) we expressed 2 previously characterized nonfunctional mutant forms of FANCA24 (Figure 2, lanes 6-9): one that retains the ability to interact with FANCG (FANCA-H1110P) and an amino-terminal truncation mutant lacking the FANCG binding region (FANCA-ΔNLS). Neither mutant conferred MMC resistance to the FA-A cells (data not shown). Interestingly, only FANCA-H1110P was able to increase the FANCG protein levels (Figure 2, compare lanes 7 and 8), which suggests that the FANCA/FANCG protein-protein interaction, and not the resistance to MMC, is critical for this induction.
Expression of FANCA and FANCG protein stabilizes the FA protein complex.
Whole cell extracts were prepared from the indicated lymphoblast lines and analyzed by immunoblotting for expression of the FANCA and FANCG proteins.
Expression of FANCA and FANCG protein stabilizes the FA protein complex.
Whole cell extracts were prepared from the indicated lymphoblast lines and analyzed by immunoblotting for expression of the FANCA and FANCG proteins.
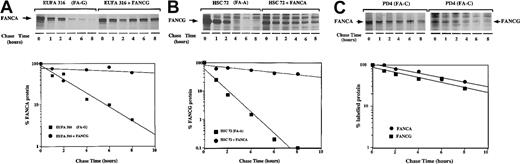
Regulation of the expression level of the FA protein complex could result from one or more mechanisms including transcriptional up-regulation, translational up-regulation, or decreased turnover of a messenger RNA (mRNA) or protein. To directly test the effect of FANCG on FANCA protein turnover, we performed pulse-chase experiments (Figure3). The patient-derived FA-G cell line, EUFA316, was sensitive to mitomycin C and failed to express the FANCG protein.24,37 In these cells the newly synthesized FANCA protein was unstable, with a protein half-life of 1 hour (Figure 3A). In contrast, in EUFA316 cells stably transfected and functionally corrected with FANCG, the half-life of FANCA was more than 10 hours, which suggests that the FANCG protein directly stabilizes the FANCA protein. By a similar analysis, the FANCA protein was shown to complement FA-A cells and to promote the stability of the FANCG protein, as evidenced by an 8- to 10-fold increase in its half-life (Figure 3B). Once again, to discriminate between functional complementation and a more direct effect of the 2 FA proteins on each other, we examined the stability of FANCA and FANCG in a patient-derived MMC-sensitive FA-C cell line (PD4). These cells have no endogenous FANCC protein but have normal FANCA/FANCG binding (Figure3C).24 Consistent with our previous observations, the half-lives of both proteins in these MMC-sensitive cells were comparable to those obtained in the corrected FA-A and FA-G cells. Taken together, our results suggest that the FANCA/FANCG interaction determines the stability of the 2 FA proteins and is thus essential to maintain the cellular levels of the FA protein complex.
FANCG binding prolongs the cellular half-life of the FANCA protein.
(A) EUFA316(FA-G) cells or EUFA316 cells stably transfected and corrected with FANCG cDNA were metabolically labeled with35S-methionine for 30 minutes, washed, and chased in cold (unlabeled) medium for the indicated time periods. The cells were lysed, and labeled proteins were immunoprecipitated with an affinity-purified anti-FANCA antiserum. The indicated bands (endogenous FANCA protein) were scanned, and densitometric values were plotted to quantify protein stability. Wild type lymphoblasts express comparable levels of endogenous FANCA protein (data not shown). (B) HSC72(FA-A) cells or HSC72 cells stably transfected and corrected with the FANCA cDNA were analyzed. Labeled proteins were immunoprecipitated with an affinity-purified anti-FANCG antiserum. (C) PD-4(FA-C) cells or PD4 cells stably transfected and corrected with the FANCC cDNA were metabolically labeled, and FANCA and FANCG proteins were analyzed.
FANCG binding prolongs the cellular half-life of the FANCA protein.
(A) EUFA316(FA-G) cells or EUFA316 cells stably transfected and corrected with FANCG cDNA were metabolically labeled with35S-methionine for 30 minutes, washed, and chased in cold (unlabeled) medium for the indicated time periods. The cells were lysed, and labeled proteins were immunoprecipitated with an affinity-purified anti-FANCA antiserum. The indicated bands (endogenous FANCA protein) were scanned, and densitometric values were plotted to quantify protein stability. Wild type lymphoblasts express comparable levels of endogenous FANCA protein (data not shown). (B) HSC72(FA-A) cells or HSC72 cells stably transfected and corrected with the FANCA cDNA were analyzed. Labeled proteins were immunoprecipitated with an affinity-purified anti-FANCG antiserum. (C) PD-4(FA-C) cells or PD4 cells stably transfected and corrected with the FANCC cDNA were metabolically labeled, and FANCA and FANCG proteins were analyzed.
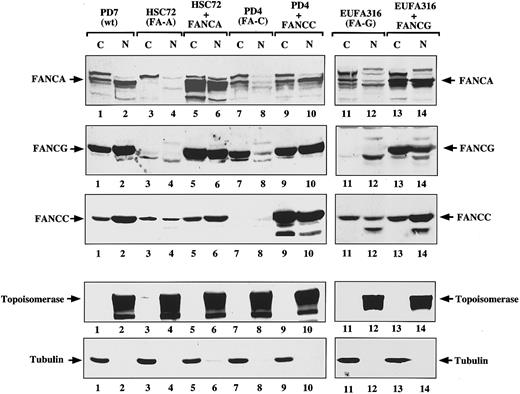
FANCG binding promotes the nuclear accumulation of the FA protein complex. Recent studies have demonstrated that FANCG binds directly to FANCA in the region of the FANCA NLS,31 32 which suggests that FANCG could play a role in modulating the subcellular localization of FANCA. We therefore tested the effect of FANCA/FANCG binding on the nuclear accumulation of FANCA and FANCG (Figure4). Based on cell fractionation studies, FA-A lymphoblasts (HSC72) expressed little FANCG in the nucleus (Figure4, lane 4) compared to normal lymphoblasts (Figure 4, lane 2). Correction of the FA-A lymphoblasts with FANCA resulted in an increase in the total cellular level of FANCG and, more importantly, in the ratio of nuclear to cytoplasmic FANCG protein (Figure 4, compare lanes 3 and 4 to lanes 5 and 6). Similarly, correction of FA-G cells with FANCG lead to an increase in nuclear FANCA levels (Figure 4, compare lanes 12 and 14). Interestingly, in corrected FA-A (Figure 4, lanes 5 and 6) and FA-G cells (Figure 4, lanes 13 and 14), the ratio of nuclear to cytoplasmic FANCC was also higher than in the corresponding mutant cells (Figure 4, lanes 3 and 4 and lanes 11 and 12). This ratio suggests that the FANCA/FANCG interaction may be important for nuclear accumulation of the whole FA protein complex. FA lymphoblasts from other FA complementation groups, such as FA-C (PD-4 cells), also had decreased levels of FANCA and FANCG protein in the nucleus (Figure 4, lane 8) that were restored after complementation with the FANCC cDNA (Figure 4, lane 10). These results demonstrate that formation of the FANCA/FANCG oligomer is necessary but not sufficient for the proper accumulation of the FA proteins in the nucleus. Other FA protein products, such as FANCC, are also required for nuclear accumulation of the FA protein complex. Because we have previously established that the FANCA/FANCG interaction is a prerequisite for FANCC binding, we hypothesize that it is the formation of the multisubunit complex containing FANCA, FANCG, and FANCC which determines the presence of the FA proteins in the nuclear compartment. Whether the effect is on nuclear import/export rates or on protein stability remains to be determined.
FANCG binding promotes the nuclear accumulation of the FANCA protein.
The indicated isogenic pairs of mutant and corrected lymphoblasts were fractionated into cytoplasmic and nuclear components. A control normal lymphoblast line (PD7) was also included. Protein (100 μg) from each sample was electrophoresed; transferred to nitrocellulose; and immunoblotted with anti-FANCA antiserum, anti-FANCG antiserum, or anti-FANCC antiserum (first 3 panels, respectively). To ensure effective fractionation, the same samples were analyzed for topoisomerase (nuclear) and β-tubulin levels (cytoplasmic). Note that the FANCA antiserum reacts nonspecifically with a protein that runs just above the FANCA band (indicated by an arrow in the FANCA panel), and the antiserum is present in all samples including the HSC72 cells, which do not express any FANCA protein.
FANCG binding promotes the nuclear accumulation of the FANCA protein.
The indicated isogenic pairs of mutant and corrected lymphoblasts were fractionated into cytoplasmic and nuclear components. A control normal lymphoblast line (PD7) was also included. Protein (100 μg) from each sample was electrophoresed; transferred to nitrocellulose; and immunoblotted with anti-FANCA antiserum, anti-FANCG antiserum, or anti-FANCC antiserum (first 3 panels, respectively). To ensure effective fractionation, the same samples were analyzed for topoisomerase (nuclear) and β-tubulin levels (cytoplasmic). Note that the FANCA antiserum reacts nonspecifically with a protein that runs just above the FANCA band (indicated by an arrow in the FANCA panel), and the antiserum is present in all samples including the HSC72 cells, which do not express any FANCA protein.
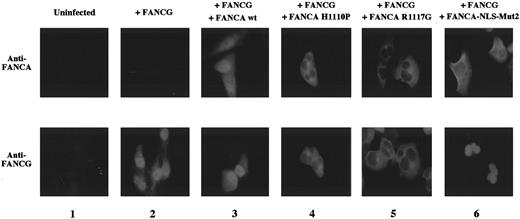
Patient-derived mutant forms of the FANCA protein bind and retain FANCG in the cytoplasm. Previous studies have identified patient-derived missense mutations in the FANCA gene that result in the expression of mutant FANCA proteins.29 38The FANCA-H1110P and FANCA-R1117G mutant proteins, for example, bind FANCG but fail to correct the MMC sensitivity of FA-A cells. To further explore the consequences of the FANCA/FANCG interaction, we analyzed the cellular localization of the FANCA/FANCG protein complex in FA-A fibroblasts transduced with the cDNA encoding these mutant FANCA proteins (Figure 5). The wild type FANCA protein and wild type FANCG protein colocalized to the nucleus, as previously described (Figure 5, column 3). In contrast, the FANCA-H1110P and FANCA-R1117G mutant proteins were localized primarily to the cytoplasm of transduced GM6914 cells (Figure 5, columns 4 and 5). Interestingly, the expression of these mutant FANCA proteins resulted in the retention of the wild type FANCG protein in the cytoplasm.
Patient-derived mutant forms of the FANCA protein bind and retain FANCG in the cytoplasm.
GM6914 fibroblasts (FA-A) were infected with the pMMP-FANCG retrovirus either alone (column 2) or in combination with the pMMP-FANCA wt (column 3), pMMP-FANCA-H1110P (column 4), pMMP-R1117G (column 5), or pMMP-FANCA-NLS-Mut2 (column 6) retrovirus. Pools of infected cells were stained with anti-FANCA (upper panels) or anti-FANCG antiserum (lower panel) and analyzed by immunofluorescence. The GM6914 cells express no FANCA protein (column 1, upper panel) and only low levels of FANCG, which were not detected by our anti-FANCG antibody (column 1, lower panel). The presence of the FANCG protein in the nucleus in panels 4, 5, and 6 may reflect the relative stability of the free (non–FANCA-bound) FANCG protein in fibroblasts compared to the lymphoblasts examined in Figures 3 and 4.
Patient-derived mutant forms of the FANCA protein bind and retain FANCG in the cytoplasm.
GM6914 fibroblasts (FA-A) were infected with the pMMP-FANCG retrovirus either alone (column 2) or in combination with the pMMP-FANCA wt (column 3), pMMP-FANCA-H1110P (column 4), pMMP-R1117G (column 5), or pMMP-FANCA-NLS-Mut2 (column 6) retrovirus. Pools of infected cells were stained with anti-FANCA (upper panels) or anti-FANCG antiserum (lower panel) and analyzed by immunofluorescence. The GM6914 cells express no FANCA protein (column 1, upper panel) and only low levels of FANCG, which were not detected by our anti-FANCG antibody (column 1, lower panel). The presence of the FANCG protein in the nucleus in panels 4, 5, and 6 may reflect the relative stability of the free (non–FANCA-bound) FANCG protein in fibroblasts compared to the lymphoblasts examined in Figures 3 and 4.
We previously described another mutant form of the FANCA protein called FANCA-NLS-Mut2.33 In this protein all of the basic amino acids of the bipartite NLS sequence were mutated. In our previous study, FANCA-NLS Mut2 failed to complement FA-A cells,33to translocate to the nucleus of transduced FA-A cells,33and to bind to FANCG.24 In our new study FANCA-NLS-Mut2 again localized to the cytoplasm (Figure 5, column 6), but FANCG was observed in the nucleus. These results demonstrate that when FANCG is overexpressed, it can translocate to the nucleus independently of FANCA. However, in a normal cell with endogenous levels of FANCA and FANCG, this may not be the preferred pathway. Moreover, given the short half-life of FANCG in the absence of FANCA binding (Figure 3), endogenous FANCG may translocate but fail to accumulate in the nucleus.
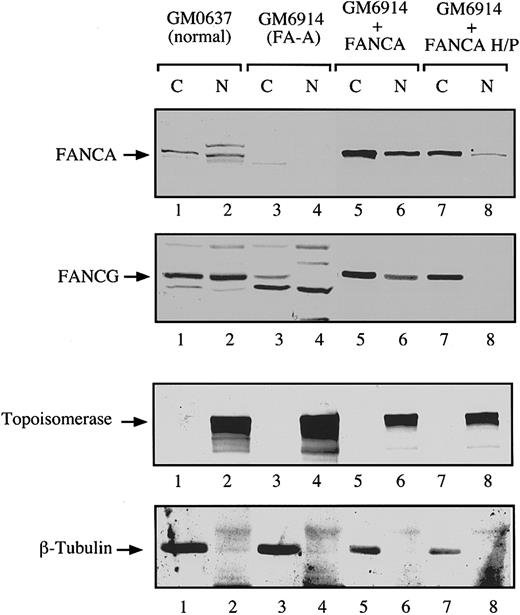
To confirm these results with an endogenous FANCG protein, we performed a series of cellular fractionation experiments (Figure6). In FA-A fibroblasts we observed little or no FANCA or FANCG protein in the cytoplasm and nucleus (Figure 6, lanes 3 and 4). Complementation of these cells with FANCA resulted in the expression of FANCA and FANCG in the cytoplasm and the nucleus (Figure 6, lanes 5 and 6). In contrast, forced expression of FANCA-H1110P resulted in the expression of little or no FANCG in the nucleus, which is consistent with the absence of nuclear FANCA or FANCG immunofluorescence observed in these cells (Figure 5). Similar results were obtained after expression of the FANCA-R1117G (data not shown).
Analysis of the subcellular localization of endogenous FANCG protein by cell fractionation.
The FA-A fibroblasts GM6914 were infected with a retrovirus carrying the wild type FANCA cDNA (lanes 5 and 6) or the mutant FANCA-H1110P cDNA (lanes 7 and 8). The parental (uninfected) and infected GM6914 cells, together with a normal control fibroblast line (GM0637, lanes 1 and 2), were fractionated into cytoplasmic and nuclear components. Samples from all fractions were analyzed by immunoblotting with antisera to FANCA and FANCG. To ensure effective fractionation, the same samples were analyzed for topoisomerase (nuclear) and β-tubulin levels (cytoplasmic).
Analysis of the subcellular localization of endogenous FANCG protein by cell fractionation.
The FA-A fibroblasts GM6914 were infected with a retrovirus carrying the wild type FANCA cDNA (lanes 5 and 6) or the mutant FANCA-H1110P cDNA (lanes 7 and 8). The parental (uninfected) and infected GM6914 cells, together with a normal control fibroblast line (GM0637, lanes 1 and 2), were fractionated into cytoplasmic and nuclear components. Samples from all fractions were analyzed by immunoblotting with antisera to FANCA and FANCG. To ensure effective fractionation, the same samples were analyzed for topoisomerase (nuclear) and β-tubulin levels (cytoplasmic).
Discussion
Several lines of evidence demonstrate that the FA proteins, including FANCA, FANCG, and FANCC, interact in a common cellular pathway and lead to the accumulation of the FA protein complex in the nucleus. Absence of this nuclear protein complex, resulting from biallelic germline mutations of any one of these FA genes, correlates with chromosome instability and a broad array of cellular and clinical abnormalities. The protein products of additional (uncloned) FA genes are required for the assembly and stability of the nuclear FA complex,27 suggesting that many FA genes are involved in the same cellular pathway. While the presence of the FANCC protein product in the nuclear FA complex has been controversial,31 the nuclear complex has recently been confirmed by other investigators.25 26 In the current study we demonstrated that the functional consequence of the FANCG/FANCA interaction is stabilization and increased nuclear accumulation of the FA protein complex.
Formation of the FANCA/FANCG complex results in the stabilization of both proteins. Several mechanisms could account for this effect. First, FANCA/FANCG binding may protect the 2 proteins from proteolysis by masking the proteolytic cleavage sites of the proteins. At present the intracellular proteases responsible for FANCA or FANCG degradation remain unknown. However, we attempted to address this question by using the specific proteosome inhibitor lactacystin. Following drug treatment a significant increase in the level of the p53 protein was observed, but there were no changes detected in the FANCA or FANCG proteins (data not shown). This suggests that the proteosome is not involved in the rapid turnover of these proteins. Second, the FANCA/FANCG interaction may promote translocation of the complex to an intracellular site, such as a nuclear subcompartment, where the 2 proteins are more stable. Third, FANCA/FANCG oligomerization may promote the binding of other FA protein subunits, such as FANCC or FANCF, which further stabilize the multisubunit complex. One or more of these 3 cellular mechanisms may serve to increase the stability of FANCA and FANCG. The systematic analysis of the half-life of the FANCA/FANCG complex in cell lines from other FA complementation groups may help distinguish among these possible mechanisms. In contrast, the cellular level of the FANCC protein is not strongly dependent on the level of FANCA and FANCG in the cell. Instead, FANCC levels may be regulated by GRP94 chaperone binding39 and/or by post-translational modifications.40
Our results demonstrate that the FANCG expression promotes the accumulation of FANCA in the nucleus. Mutant forms of FANCG, which fail to bind FANCA, also fail to promote nuclear accumulation of FANCA (data not shown). Because FANCG has been shown to bind specifically to the NLS region of FANCA,41 it is reasonable to assume that FANCG may play an active role in modulating the nuclear import and/or export processes of the FA protein complex. For instance, FANCG may increase nuclear uptake by acting as an importin protein that recognizes the NLS region of FANCA and allows the interaction with nuclear pore complexes (NPCs)42 or by cooperating with importin-α. However, the interaction of FANCG with proteins involved in nuclear transport, such as importin-α, importin-β, or nucleoporins, has not been reported, and whether FANCG can recognize the NLS region of other nuclear targeted proteins remains unknown. Another interesting possibility is that the effect of FANCA and FANCG on the nuclear accumulation of the FA complex is not due to increased nuclear import (or decreased nuclear export) but rather to the increased stability of the nuclear complex. Regardless of the mechanism, our data indicate that the FANCA/FANCC/FANCG multisubunit complex is important to ensure the efficient accumulation of the FA proteins in the nucleus. In this context the FANCA/FANCG interaction would be required to ensure maintenance of the cellular levels of both proteins and as a precursor step (upstream event) leading to the formation of the multisubunit FA complex containing FANCC and probably some of the other FA proteins.
In transduced fibroblasts, FANCA and FANCG proteins are capable of translocating to the nucleus independently: FANCG translocates to the nucleus of FA-A cells in the absence of functional FANCA protein, and FANCA translocates to the nucleus of FA-G cells in the absence of functional FANCG protein (I.G.-H., unpublished observation, April 2000). Whether this independent nuclear translocation occurs in normal cells and to what extent it occurs, if at all, remain to be determined. However, because FANCA and FANCG depend on each other for their stability (Figure 3), it is unlikely that at normal physiological levels, neither of the 2 proteins accumulate in the nuclear compartment. Our fractionation data would agree with this idea. Moreover, preliminary evidence suggests that the half-life of nuclear FANCA is comparable to that of the cytoplasmic form (unpublished observation). Therefore, FANCA/FANCG binding appears to be a more important determinant of FA complex stability than does subcellular localization.
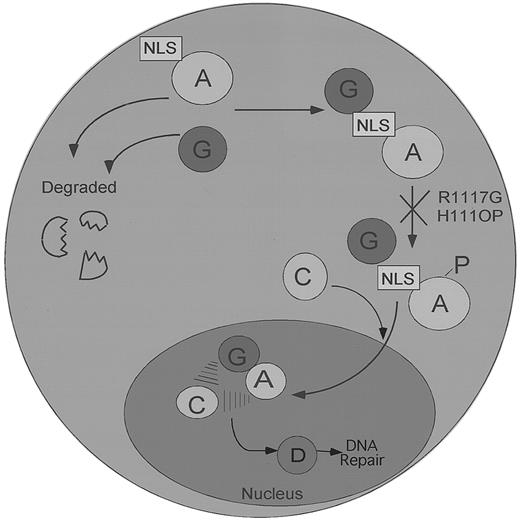
Our data support a model of the FA pathway in which FANCG binds FANCA in the cytoplasm, before their coordinated translocation to the nucleus (Figure 7). In support of this model, some FA patient-derived mutant forms of the FANCA protein (FANCA-H111OP and FANCA-R1117G) bind FANCG in the cytoplasm but are arrested in the pathway, before their translocation and accumulation in the nucleus. Interestingly, these point mutations also ablate the phosphorylation of the FANCA protein, the binding of the FANCC protein, and the accumulation of the stable nuclear complex.29 How the FANCA point mutations H111OP and R1117G block the downstream events in the FA pathway remains unknown, but several mechanisms can be considered. For instance, these point mutations, which are only 7 amino acids apart in the leucine zipper region of FANCA, may block the phosphorylation of FANCA by preventing binding of the relevant FANCA protein kinases (FANCA-PKs). Alternatively, these mutations may prevent the binding of additional protein subunits, perhaps encoded by other FA genes. Regardless of the correct hypothesis, these data demonstrate that several structural features of FANCA, including a functional amino-terminal NLS and an intact carboxy-terminal leucine zipper region, are required for normal nuclear translocation.
A speculative model of the regulated binding and nuclear transport of the FA protein complex.
The FANCA and FANCG proteins bind initially in the cytoplasm of normal cells. Failure to bind results in the rapid degradation of both proteins. FANCG binds directly to the NLS region of the FANCA protein. In contrast, FANCC binding to the FANCA/FANCG complex is indirect and requires FANCA phosphorylation and the products of other FA genes. The FA protein complex subsequently translocates to the cell nucleus, where it executes a nuclear function, such as DNA repair or the segregation of sister chromatids, which helps maintain normal chromosome structure and stability. The FANCD protein may function downstream of the FA protein complex, as indicated. Patient-derived point mutations on the FANCA protein, such as the FANCA-H1110P and the FANCA-R1117G, block FANCA phosphorylation, FANCC binding, and nuclear translocation and retain the FANCG protein in the cytoplasm.
A speculative model of the regulated binding and nuclear transport of the FA protein complex.
The FANCA and FANCG proteins bind initially in the cytoplasm of normal cells. Failure to bind results in the rapid degradation of both proteins. FANCG binds directly to the NLS region of the FANCA protein. In contrast, FANCC binding to the FANCA/FANCG complex is indirect and requires FANCA phosphorylation and the products of other FA genes. The FA protein complex subsequently translocates to the cell nucleus, where it executes a nuclear function, such as DNA repair or the segregation of sister chromatids, which helps maintain normal chromosome structure and stability. The FANCD protein may function downstream of the FA protein complex, as indicated. Patient-derived point mutations on the FANCA protein, such as the FANCA-H1110P and the FANCA-R1117G, block FANCA phosphorylation, FANCC binding, and nuclear translocation and retain the FANCG protein in the cytoplasm.
This model generates new testable hypotheses. For instance, it will be important to determine whether the assembly and transport of the FANCA/FANCG complex is constitutive or regulated by external environmental cues or cell cycle cues. For instance, bifunctional alkylating agents may affect the complex assembly or accumulation. Also, identification of the cytoplasmic kinase of FANCA (FANCA-PK) may elucidate a critical level of regulation of the FA protein complex assembly and function.
Finally, while the accumulation of the FA protein complex appears to be a critical step in the FA pathway, the precise biochemical function of this complex in the nucleus remains unknown. Increasing evidence suggests that FA cells are defective in DNA repair, and the FA complex may play a direct or indirect role in the repair of DNA damage. Also, the FA protein complex assembles normally in FA-D cells, suggesting that the product of the FANCD gene functions downstream or independently of the FA protein complex assembly. Whether the FA protein complex functions by modulating the structure and function of the recently cloned FANCD protein product (Timmers et al, unpublished data) remains unknown.
Acknowledgments
We thank members of the D'Andrea laboratory (Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA) for helpful discussions.
Supported by grants RO1HL52725-04 and PO1HL54785-04 from the National Institutes of Health, Bethesda, MD, and a post-doctoral grant (G.-H.) from the Cancer Research Institute. A.D.D. is a Scholar of the Leukemia Society of America.
Submitted April 12, 2000; accepted June 20, 2000.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Alan D. D'Andrea, Dana-Farber Cancer Institute, Division of Pediatric Oncology, Harvard Medical School, 44 Binney Street, Boston, MA 02115; e-mail: alan_dandrea@dfci.harvard.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal