Abstract
Human granulocytes are characterized by a variety of specific effector functions involved in host defense. Several widely expressed protein kinases have been implicated in the regulation of these effector functions. A polymerase chain reaction–based strategy was used to identify novel granulocyte-specific kinases. A novel protein kinase complementary DNA with an open reading frame of 357 amino acids was identified with homology to calcium-calmodulin–dependent kinase I (CaMKI). This has been termed CaMKI-like kinase (CKLiK). Analysis of CKLiK messenger RNA (mRNA) expression in hematopoietic cells demonstrated an almost exclusive expression in human polymorphonuclear leukocytes (PMN). Up-regulation of CKLiK mRNA occurs during neutrophilic differentiation of CD34+ stem cells. CKLiK kinase activity was dependent on Ca++ and calmodulin as analyzed by in vitro phosphorylation of cyclic adenosine monophosphate responsive element modulator (CREM). Furthermore, CKLiK- transfected cells treated with ionomycin demonstrated an induction of CRE- binding protein (CREB) transcriptional activity compared to control cells. Additionally, CaMK-kinaseα enhanced CKLiK activity. In vivo activation of CKLiK was shown by addition of interleukin (IL)-8 to a myeloid cell line stably expressing CKLiK. Furthermore inducible activation of CKLiK was sufficient to induce extracellular signal-related kinase (ERK) mitogen-activated protein (MAP) kinase activity. These data identify a novel Ca++/calmodulin-dependent PMN- specific kinase that may play a role in Ca++-mediated regulation of human granulocyte functions.
Introduction
Human polymorphonuclear leukocytes (PMN), which include neutrophilic and eosinophilic granulocytes, play an important role in host defense against invading microorganisms.1Recruitment and activation of these cells in vivo occur in a multistep process that involves many different membrane-bound receptors activating an array of diverse intracellular signaling molecules. In short, PMNs in the peripheral blood enter a preactivated state by interacting with cytokines liberated from the inflammatory locus. This is followed by attachment to the endothelium, which is mediated by the interaction with adhesion molecules expressed on the surface of activated endothelial cells. Release of chemokines at the site of inflammation is responsible for the migration of PMNs to this locus. Finally on recognition of the inciting agent by PMNs, phagocytosis, secretion of toxic proteins, and activation of membrane-bound NADPH-oxidase generating reactive oxygen intermediates ensues.2,3 Furthermore, rapid induction of apoptosis in PMNs and subsequent removal of apoptotic cells are important in the rapid resolution of inflammation.4 5 An unfortunate consequence of activation in vivo is tissue damage during acute inflammation and, therefore, the activity of granulocytes is under tight control.
The PMNs express a wide variety of receptors on their plasma membranes, steering the process of priming and activation. On binding of inflammatory mediators, such as formyl peptides, lipopolysaccharides, chemokines, or cytokines, the receptor transmits a signal to the cell interior resulting in the initiation of a cascade of intracellular events. Phosphorylation of effector molecules by kinases is critical for transducing intracellular signals. Thus far several classes of kinases, including (1) serine kinases, such as mitogen activated (MAP) kinases; (2) lipid kinases such as phosphatidylinositol-3 kinase (PI-3K); (3) tyrosine kinases including the src kinases; (4) cyclic adenosine monophosphate (cAMP)-dependent kinases, and (5) Ca++-dependent kinases, are activated in response to inflammatory mediators in human granulocytes.
A role for these widely expressed kinases in neutrophil functions has been intensively studied using pharmacologic inhibitors. A role for MAP kinases, p42ERK1 and p42ERK2, in chemotaxis, respiratory burst, and platelet activating factor (PAF) release is suggested, although the use of a pharmacologic inhibitor for MAPK/ERK kinase (MEK) has resulted in contradictory findings.6,7 A clearer role for PI-3K in neutrophil migration and respiratory burst has been demonstrated by use of PI-3K inhibitors wortmannin and LY294002.7-11 A role for protein kinase C (PKC) has also been postulated in a variety of granulocyte effector functions and protein kinase A (PKA) is suggested to be involved in down-regulating the respiratory burst.12,13Recently a role for src kinases in adhesion-dependent degranulation has also been described.14 Although inhibitory studies support a role for these kinases in regulating neutrophil functions, their activation is not specific for these granulocyte effector functions because they are widely expressed. Furthermore, pharmacologic inhibitors are often limited in their specificity, making the interpretation of data more complex.15
Changes in cytosolic free Ca++ are described during activation of several neutrophil responses, such as degranulation, respiratory burst, and adhesion. An important role for Ca++in these processes has been suggested16,17 and, therefore, Ca++-dependent kinases may well be involved. In addition a role for the Ca++-dependent phosphatase, calcineurin has been shown in the Ca++-dependent recycling of integrins to the front of migrating neutrophils,18 whereas a role for calmodulin and CaMKII is suggested in oxygen production.19
In this report we describe the identification of a novel protein kinase, which we have termed CKLiK (CaMKI-like kinase). This kinase is predominantly expressed in human PMNs and is regulated by Ca++ and calmodulin. Interleukin (IL)-8, which is a potent activator of neutrophil effector functions, induces activation of CKLiK and we show that an inducible active mutant of CKLiK induces ERK MAP kinase activation. These data identify a novel Ca++/calmodulin-dependent protein kinase expressed in PMNs that may play a role in transducing chemokine-induced signals regulating human granulocyte functions.
Materials and methods
Cells, reagents, and antibodies
The COS cells were cultured in Dulbecco modified Eagle medium (Life Technologies, Breda, The Netherlands) supplemented with 8% heat-inactivated fetal calf serum (FCS). CD34+ stem cells were cultured in Iscove modified Dulbecco medium (IMDM) supplemented with 2 mmol/L glutamine, 8% Hyclone, and cytokines. Murine myeloid 32D cells were cultured in RPMI 1640–glutamate supplemented with 8% Hyclone serum (Life Technologies) and mouse IL-3.20 The 32D cells were stably transfected with pBabe-HA_CKLiK by electroporation (0.26 V, 1000 msec, 100 μFD) and selected with puromycin (1 μg/mL; Sigma, Zwijndrecht, The Netherlands) at a concentration of 1 μg/mL. Ionomycin was purchased from Calbiochem (La Lolla, CA). Calmodulin was a kind gift of Dr R. de Groot. Monoclonal antibodies, 12CA5 against HA epitope, were purchased from Boehringer Mannheim Biochemicals (Almere, The Netherlands). Polyclonal anti-VSV tag antibodies were obtained from Medical & Biological Laboratories (MBL, Nagoya, Japan). Human IL-8, trombopoetin (TPO), fms-like tyrosine kinase-3 (FLT-3) ligand, and stem cell factor (SCF) were purchased from PeproTech (Rocky Hill, NJ). Human IL-3 and granulocyte colony-stimulating factor (G-CSF) were purchased from Strathmann Biotech (Hamburg, Germany) and IL-6 was purchased from Roche Molecular Biochemicals (Indianapolis, IN). Phospho-CREB-Ser133 antibodies were obtained from New England Biolabs (Beverly, MA). 4-Hydroxy-tamoxifen and W7 were purchased from Sigma; PD098059 was purchased from Biomol (Plymouth Meeting, PA).
Identification and cloning of granulocyte kinases
For the identification of novel kinases in human granulocytes polymerase chain reaction (PCR) was performed using degenerate primers as previously described.21 Forward primers were based around the consensus amino acid (single-letter symbols) sequence HRDLKPEN, which corresponded to the conserved regions in the serine/threonine protein kinase catalytic domain. Oligonucleotides were also designed against the DXWXXG amino acid motif approximately 65 amino acids downstream and used as reverse primers. PCR was performed on granulocyte complementary DNA (cDNA) with Taq polymerase (Perkin Elmer, Roche Molecular Systems, Branchburg, NJ) using excess of degenerate primers (1 μg) under conditions of 10 mmol/L Tris-Cl pH 8.3, 50 mmol/L KCl, 0.2 mmol/L dNTPs, and 0.8 mmol/L MgCl2. Denaturation, annealing, and extension temperatures of 94°C, 52°C, and 72°C, respectively, were used. PCR products of approximately 200 bp were cloned into pGEM-T vector (Promega, Madison, WI) and sequenced. The identified sequences were screened against the BLAST database (http: //www.ncbi.nlm.nih.gov/BLAST).
Full-length cDNA was obtained by screening a λZAPII eosinophilic library22 with the amplified PCR fragment as a probe. After plaque purification, the pBluescript SK−phagemids containing positive inserts were isolated by in vivo excision using M13K07 helper phages (Promega), and subsequently characterized by restriction mapping and sequencing.
Isolation of human leukocytes
Blood was obtained from healthy volunteers at the donor service of the University Medical Centre (Utrecht, The Netherlands). Granulocytes were isolated from blood treated with 0.4% (wt/vol) trisodium citrate (pH 7.4) as previously described.23Mononuclear cells were removed from granulocytes by centrifugation over isotonic Ficoll from Pharmacia (Uppsula, Sweden). After lysis of the erythrocytes in an isotonic NH4Cl solution, neutrophils were washed and resuspended in incubation buffer (20 mmol/L HEPES, 132 mmol/LM NaCl, 6 mmol/L KCl, 1 mmol/L MgSO4, 1.2 mmol/L KH2PO4, 5 mmol/L glucose, 1 mmol/L CaCl2). Granulocytes were incubated for 30 minutes at 37°C before experiments were performed. Monocytes were further separated from mononuclear lymphocytes by elutriation as described previously.24
Isolation and differentiation of CD34+ stem cells
The CD34+ cells were isolated using the MACS CD34+ progenitor cells isolation kit from Miltenyi Biotech (Auburn, CA). First, mononuclear cells were isolated from human umbilical cord blood cells by density centrifugation over a Ficoll solution. Cells were blocked and additionally incubated with a CD34 antibody in a phosphate buffer containing 2 mmol/L EDTA and 0.5% bovine serum albumin (BSA) (buffer M). Magnetic beads recognizing the CD34 antibody were added and the suspension was applied to an MS+ separation column placed in the magnetic cell separator MiniMACS. CD34− cells were allowed to pass through the column by 3 wash steps with buffer M. CD34+ cells were eluted with buffer M by removal of the column from the separator. To obtain a pure CD34+ cell population the magnetic separation step was repeated on a second column. After final elution stem cells were counted and cultured. Before differentiation, CD34+stem cells were proliferated for 1 week in the presence of TPO (10 ng/mL), SCF (50 ng/mL), FLT-3 ligand (50 ng/mL), and IL-6 (2 ng/mL). At day 0 differentiation was started by addition of SCF, FLT-3 ligand, IL-3 (10 ng/mL), and G-CSF (30 ng/mL). At days 3, 7, 10, 14, and 17, IL-3 and G-CSF were added and at day 21 only G-CSF was added to the cells. At each point cells were counted, diluted to 0.5 × 106 cells/mL and, if possible, samples were taken for mRNA isolation.
RNase protection
Total mRNA from human monocytes, lymphocytes, granulocytes, and hematopoietic cell lines HL60, U937, and THP1 were isolated. In short, 108 cells were lysed in 2 mL GIT-C solution (6 mol/L guanidine thiocyanate, 25 mmol/L sodium citrate, 0.5% N′-lauroyl-sacosine, 100 mmol/L β-mercaptoethanol), and RNAs were further isolated by phenol extraction and ethanol precipitation.32P-UTP–labeled antisense RNA transcript, corresponding to the original PCR fragment coding the catalytic domain of CKLiK, was generated using the Riboprobe in vitro transcription system (Promega) and used as an RNA probe. As internal control a 90-bp antisense RNA probe of GAPDH was used. Total RNA samples (10 μg) were lyophilized and resuspended in 2 μL diethyl pyrocarbonate (DEPC) water. Hybridization was performed with 105 cpm of each antisense RNA probe in 25 μL 80% formamide, 40 mmol/L PIPES pH 6.4, 400 mmol/L NaCl, and 1 mmol/L EDTA overnight at 45°C. Subsequently probes were incubated for 1 hour in RNAse buffer (10 mmol/L Tris-Cl pH 7.5, 5 mmol/L EDTA, 300 mmol/L NaCl supplemented with 0.15 μL/mL T1 RNase) at 37°C to degrade unhybridized RNA. The reaction was stopped by addition of 10 μL proteinase K (5 mg/mL) and 10% sodium dodecyl sulfate (SDS). Hybridized (double-stranded) RNA was purified by phenol extraction and ethanol precipitation. The remaining pellet was resuspended in 2 μL DEPC water and 2 μL RNA loading buffer (80% formamide, 10 mmol/L EDTA, 1 mg/mL xylene cyanol, 1 mg/mL bromphenol blue). Samples were heated for 5 minutes at 95°C and analyzed by polyacrylamide gel electrophoresis (PAGE).
DNA constructs
Epitope-tagged CKLiK was generated by PCR using the oligonucleotides forward XhoI; 5′-CCGCTCGAGTATGGCCCGGGAGAACGGC-3′ and reverse KpnI; 5′-CCGGTACCCAAGTAG-CTGACATTACAGG-5′) and ligated byXhoI/KpnI digest into pMT-HA. HA_CKLiK-309 and HA_CKLiK-296 were also generated by PCR using reverse primers introducing a stop codon at amino acid 310 or 297 309:5′-GCATTTCATGCTTGGCACCATT-3′,296; 5′-GCTCTAGATCA-CTGGGCGCTGACGGACTC-3′). PCR products were cloned into pGEM-T vector and subcloned into pMT2-HA vector. Green fluorescent protein (GFP)-tagged CKLiK was generated by PCR and recloned in frame into pEGFP-C2 vector (Clontech Laboratories, Palo Alto, CA). HA_CKLiK was cloned from pMT-HA into pBabe25 byBamHI/ EcoRI digest. Untagged CKLiK-296 was obtained by PCR and cloned into pSG5.26 VSV-tagged CaMKKα was generated by PCR on rat brain tissue cDNA using the oligonucleotides 5′-CAGTCGACCAGGAATATCCACGGACTGA-3′ and 5′-ATAGCGGCCGCC-GGATGCAGCCTCATCTTC-3′ and cloned into the pMT2-VSV vector. Tamoxifen-inducible CKLiK construct, ER_CKLiK-296, was generated by PCR and cloned into the pCDNA3-ER-N vector. The constructs for HA_PKB and HA_ERK have been previously described.27CREB_GAL4, CREBS133A_GAL4, and GAL4CAT constructs were previously described.28
In vitro kinase assay
The COS cells were transiently transfected with 10 μg of HA-tagged CKLiK-WT, CKLiK-309, or CKLiK-296 using calcium phosphate precipitation, and the medium was refreshed 16 hours later. For the CKLiK kinase assay, 24 hours later cells were stimulated with or without ionomycin for 5 minutes, washed twice with cold phosphate-buffered saline (PBS) and lysed in a buffer containing 1% NP-40, 20 mmol/L Tris-Cl pH 7.5, 150 mmol/L NaCl, 10% glycerol, and 10 mmol/L MgCl2 supplemented with 10 μg/mL aprotinin, 1 mmol/L leupeptin, 1 mmol/L PMSF, 1 mmol/L benzamidine, and 1 mmol/L Na3VO4. The 32D cells stably expressing CKLiK-WT were mIL-3 and serum starved for 4 hours and stimulated with hIL-8 (10−7 mol/L). Cells were washed and lysed in buffer described above. Lysates were precleared for 20 minutes with protein-A beads and immunoprecipitated with 12CA5 antibody. Immunoprecipitates were washed twice in lysis buffer and twice in dilution buffer containing 10 mmol/L Tris-Cl pH 7.4 and 20 mmol/L MgCl2. Kinase assay was performed in the presence of 10 mmol/L Tris-Cl pH7.4, 20 mmol/L MgCl2, 1 mmol/L DTT, 50 μmol/L ATP, 0.1 μL32P-dATP in the presence or absence of 1 mmol/L CaCl2 and 0.5 μg calmodulin, 5 μg CREMβ (33 kd), or mutated CREMτ-S117A (42 kd) as substrate.29 Protein kinase B (PKB) and ERK kinase assay were performed as described previously.27
CAT assay
The COS cells were transiently transfected with 4 μg CKLiK-WT, -309, or -296, together with 2 μg CREB_GAL4 or CREB-S133A_GAL4 fusion expression plasmids and 2 μg GAL4CAT reporter construct using calcium phosphate precipitation. After 16 hours cells were washed twice and medium refreshed. Eight hours later cells were incubated overnight with 1 μmol/L ionomycin. Cells were lysed by repeated freeze-thawing in 100 μL 250 mmol/L Tris-Cl pH7.4 and 25 mmol/L EDTA. Then, 50μL of cellular extract was incubated in a total volume of 100 μL containing 250 mmol/L Tris-Cl 7.4, 2% glycerol, 0.3 mmol/L butyryl coenzyme A, and 0.05 μCi 14C-chloramphenicol for 2 hours at 37°C. Reaction products were extracted using 400 μL xylene/pristane 1:2 and the percentage of acetylated products was determined using liquid scintillation counting. A lacZ reporter was used to correct for transfection efficiency. Data represent at least 3 independent experiments ± SEM.
Detection of GFP fusion-protein localization
Plasmids encoding CKLiK and active CKLiK-296 containing N-terminal enhanced GFP were transiently transfected in COS cells that were grown on coverslips. Thirty-six hours after transfection COS cells were washed with PBS, fixed with 70% ice-cold methanol, and examined by fluorescence microscopy.
Western blotting
After stimulation 106 neutrophils/point were lysed in 1 × Laemmli sample buffer. Protein samples were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Immobilon-P, Millipore, Bedford, MA). The membranes were probed with antiphospho-CREB-Ser133 antibodies (New England Biolabs) and swine antirabbit peroxidase-conjugated antibodies (DAKO, Glostrup, Denmark), following detection by enhanced chemiluminescence (ECL, Amersham-Pharmacia, Biotech, The Netherlands). Protein kinase expression controls were performed by using 12CA5 and rabbit antimouse peroxidase-conjugated antibodies (DAKO) to detect HA_CKLiK, HA_ERK, and HA_PKB. Detection of CaMKK-VSV was performed with anti-VSV antibodies and swine antirabbit peroxidase-conjugated antibodies. Hybridizations were followed by detection with ECL (Amersham).
Results
Cloning and expression of CKLiK
Although several kinases have been implicated in the control of granulocyte effector functions,12,30 most of these proteins are widely expressed. To identify novel granulocyte-specific kinases, degenerate primers against conserved kinase catalytic domains were used as previously described.21 The central core of the catalytic domain consists of subdomains VI and IX and contains 2 well-conserved amino acid triplets APE and DGF.31Degenerate oligonucleotides were designed against subdomains VI and IX and used in a PCR on cDNA of human PMNs. Under stringent conditions PCR products of approximately 200 bp were amplified, cloned, and sequenced. All cloned PCR products contained the 2 conserved amino acid triplets and many were identical to previously identified kinases (data not shown). However, one clone exhibited homology with calcium/calmodulin-dependent kinase I (CaMKI) and subsequently termed CKLiK (CaMKI-likekinase).
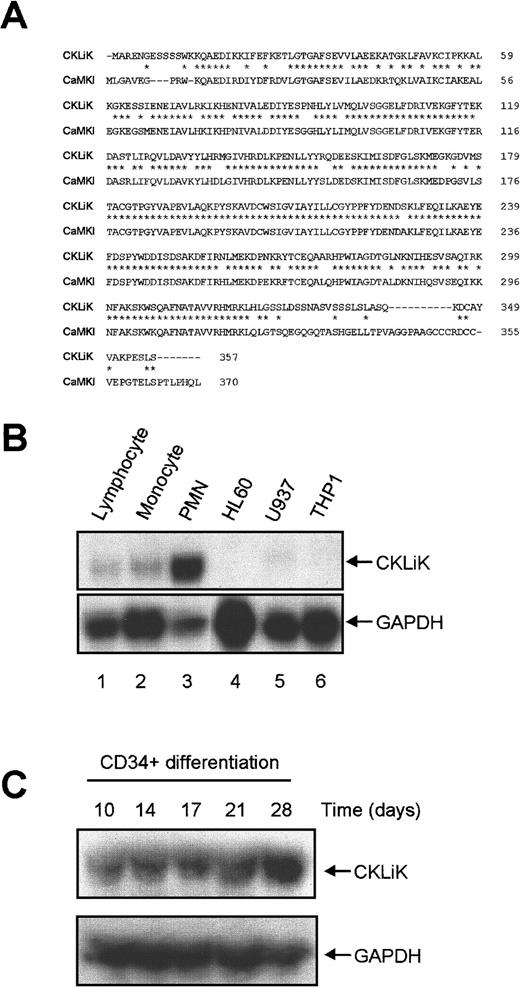
To isolate full-length CKLiK cDNA, a human eosinophil library was screened with the amplified PMN cDNA PCR fragment. Several positive clones were isolated and sequenced, and one cDNA of 1.7 kb contained a 357 amino acid open reading frame encoding a protein of 40 kd as determined by in vitro transcription/translation (data not shown). In Figure 1 the complete cDNA and the translated amino acid sequence is depicted (accession numberAF286366). The context of the ATG codon was in good agreement with eukaryotic translation start-sites.32 As shown in Figure 2A, comparison of the predicted CKLiK protein sequence in BLAST database revealed a 77% homology of CKLiK with CaMKI on the amino acid level. The greatest divergence between the 2 sequences occurs at the N- and C-termini.
CaMKI-like kinase, CKLiK, nucleotide sequence, and open reading frame.
Full-length sequence of the novel kinase was obtained by screening an eosinophil cDNA λZAPII phage library using the initial degenerate PCR product as a probe. Sequencing was performed and the open reading frame was analyzed.
CaMKI-like kinase, CKLiK, nucleotide sequence, and open reading frame.
Full-length sequence of the novel kinase was obtained by screening an eosinophil cDNA λZAPII phage library using the initial degenerate PCR product as a probe. Sequencing was performed and the open reading frame was analyzed.
CKLiK is homologous to CaMKI and highly expressed in human granulocytes.
(A) Alignment of CKLiK with CaMKI. Residues identical to CaMKI are indicated by an asterisk (*) and gaps in the alignment are indicated with a dash (−).The amino acid sequence is indicated on the right. (B) Distribution of CKLiK was determined by RNase protection. The PCR product encoding the catalytic domain of CKLiK was used to generate a radiolabeled RNA probe. RNA isolated from primary hematopoietic cells and cell lines were hybridized for CKLiK expression. A GAPDH probe was used as a control. Lanes 1 to 3 represent primary leukocytes (1, lymphocytes; 2, monocytes; 3, PMNs) and lanes 4 to 6 represent lymphoid and myeloid cell lines as indicated. (C) Expression of CKLiK in cord blood-derived CD34+ stem cells during differentiation toward neutrophils was analyzed by RNase protection as in panel B. Time points indicate the number of days differentiated toward neutrophilic lineage in the presence of G-CSF.
CKLiK is homologous to CaMKI and highly expressed in human granulocytes.
(A) Alignment of CKLiK with CaMKI. Residues identical to CaMKI are indicated by an asterisk (*) and gaps in the alignment are indicated with a dash (−).The amino acid sequence is indicated on the right. (B) Distribution of CKLiK was determined by RNase protection. The PCR product encoding the catalytic domain of CKLiK was used to generate a radiolabeled RNA probe. RNA isolated from primary hematopoietic cells and cell lines were hybridized for CKLiK expression. A GAPDH probe was used as a control. Lanes 1 to 3 represent primary leukocytes (1, lymphocytes; 2, monocytes; 3, PMNs) and lanes 4 to 6 represent lymphoid and myeloid cell lines as indicated. (C) Expression of CKLiK in cord blood-derived CD34+ stem cells during differentiation toward neutrophils was analyzed by RNase protection as in panel B. Time points indicate the number of days differentiated toward neutrophilic lineage in the presence of G-CSF.
To determine the distribution of CKLiK expression we analyzed mRNA levels in primary human leukocytes and hematopoietic cell lines by RNase protection (Figure 2B). The mRNAs from lymphocytes, which include T and B cells, monocytes, and PMNs, were analyzed. Additionally, 3 myeloid cell lines HL60 (promyelocytic leukemia), U937 (histiocytic lymphoma), and THP1 (acute monocytic leukemia) were analyzed for CKLiK expression. As an internal control we used a GAPDH probe (Figure 2B; lower panel). Human PMNs contain high levels of CKLiK (Figure 2B; lane 3), whereas little to no expression was observed in monocytes and lymphocytes (Figure 2B; lanes 1 and 2). In the different lymphoid and myeloid cell lines we could not detect CKLiK (Figure 2B; lanes 4-6). Because CKLiK expression is apparently only detected in mature myeloid cells, we analyzed the expression of CKLiK during differentiation of CD34+ cord blood stem cells toward the neutrophilic lineage. Although after 10 days of differentiation CKLiK mRNA was present, increased mRNA levels are indeed detected during terminal differentiation at day 28 (Figure 2C). Similar results were obtained during the differentiation toward eosinophils in the presence of IL-5 (data not shown).
CKLiK kinase activity is dependent on Ca++/calmodulin and can regulate CREM and CREB
As mentioned, CKLiK has homology with the Ca++/calmodulin-dependent kinase family. The CaM kinases belong to the serine/threonine class of kinases and include CaMKI, CaMKIV, and the upstream CaMKK.33 To investigate whether Ca++ and calmodulin regulated CKLiK we used CRE-binding protein (CREB) or CRE modulator (CREM) as downstream targets. CREB and CREM belong to the basic-leucine-zipper (bZip) class of transcription factors. Serine 133 for CREB and serine 117 for CREM are phosphorylated by several kinases including CaMKI in vitro,28,34resulting in interaction with transcriptional coactivators and induction of transcription.35
CKLiK kinase activity was analyzed by in vitro kinase assay, using CREM as a substrate and by transcriptional reporter assays using CREB_GAL4 fusion constructs.28 CKLiK was immunoprecipitated from transfected COS cell lysates and immunocomplex kinase assays performed (Figure 3A). CREMβ (33 kd) or a mutant CREMτ (42 kd), which has a serine to an alanine substitution on position 117 (CREMτ-S117A), were used as substrates (Figure 3A). Because of the different size of the 2 isoforms, this allows the specific phosphorylation of S-117 to be analyzed within a single assay. The assay was performed in the absence or presence of Ca++and calmodulin. As a positive control for this assay we used recombinant CaMKI, which potently phosphorylated CREMβ. Because of the high level of recombinant CaMKI, some background phosphorylation of the mutated form of CREM (CREMτ-S117A) was detected. A clear phosphorylation of CREMβ but not CREMτ-S117A by CKLiK was observed in the presence of Ca++/calmodulin and this was enhanced by prior ionomycin treatment (Figure 3A). A small activation of CKLiK was also observed by ionomycin treatment in the absence of Ca++/calmodulin during the kinase assay (data not shown). For transcriptional reporter assays, cells were transfected with HA_CKLiK, a GAL4-CAT reporter construct, in combination with a CREB_GAL4 fusion construct or a mutant thereof, in which the phosphorylation site (S-133) is substituted by an alanine (CREB-S133A_GAL4) (Figure 3B). Ionomycin treatment resulted in a 5-fold induction of reporter activity in cells expressing CKLiK with only a modest increase in control cells (Figure 3B; left panel). CKLiK had no effect when using the mutant form of CREB (Figure 3B; right panel). These results demonstrate that CKLiK is regulated by Ca++/calmodulin and that active CKLiK can directly phosphorylate CREM and activate CREB, enhancing its transactivation capacity.
CKLiK activity requires Ca++/Calmodulin and regulates the transcription factors CREM and CREB.
(A) COS cells were transfected with 10 μg HA_CKLiK. Recombinant CaMKI was used as a positive control. Cells were stimulated with or without 1 μmol/L ionomycin as indicated. HA_CKLiK was immunoprecipitated from whole cell lysates and kinase assays were performed in the presence or absence of Ca++/calmodulin as indicated. CREMβ-WT (33 kd) and mutated CREMτ-S117A (42 kd) were used as substrate. Data represent 1 of 4 independent experiments. (B) Cells transfected with GAL4-CAT reporter construct (2 μg) and fusion construct of CREB_GAL4 or CREB-S133A_GAL4 (2 μg) (indicated as CREB-WT and CREB-S133A, respectively), were cotransfected with CKLiK (4 μg) or with control vector as indicated. Stimulation with or without 1 μmol/L ionomycin and CAT reporter assays were performed as described in “Materials and methods.” Data are indicated as counts per minute (cpm) and represent at least 3 independent experiments ± SEM.
CKLiK activity requires Ca++/Calmodulin and regulates the transcription factors CREM and CREB.
(A) COS cells were transfected with 10 μg HA_CKLiK. Recombinant CaMKI was used as a positive control. Cells were stimulated with or without 1 μmol/L ionomycin as indicated. HA_CKLiK was immunoprecipitated from whole cell lysates and kinase assays were performed in the presence or absence of Ca++/calmodulin as indicated. CREMβ-WT (33 kd) and mutated CREMτ-S117A (42 kd) were used as substrate. Data represent 1 of 4 independent experiments. (B) Cells transfected with GAL4-CAT reporter construct (2 μg) and fusion construct of CREB_GAL4 or CREB-S133A_GAL4 (2 μg) (indicated as CREB-WT and CREB-S133A, respectively), were cotransfected with CKLiK (4 μg) or with control vector as indicated. Stimulation with or without 1 μmol/L ionomycin and CAT reporter assays were performed as described in “Materials and methods.” Data are indicated as counts per minute (cpm) and represent at least 3 independent experiments ± SEM.
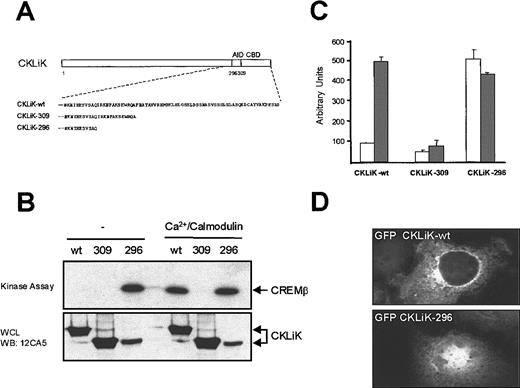
C-terminal truncation of CKLiK results in a constitutively active mutant
Sequence alignment with CaMKI and the results described above suggests that CKLiK contains a calmodulin-binding domain regulating its activity. To investigate this hypothesis we generated 2 truncation mutants (Figure 4A). Truncation of the predicted calmodulin-binding domain (CKLiK-309) should result in an inactive kinase. Indeed mutant CKLiK-309, lacking residues 310 to 357, remains inactive even in the presence of Ca++/calmodulin, unable to phosphorylate CREM (Figure 4B) or to activate CREB-mediated transcription after ionomycin stimulation (Figure 4C). However, if CKLiK also contains an autoinhibitory domain similar to CaMKI, then removal of this domain (CKLiK-296) should generate a constitutively active kinase. As was predicted, truncation of residues 297 to 357 generated a constitutively active CKLiK. CREM was phosphorylated by CKLiK-296 in the absence of Ca++/calmodulin (Figure 4B) and CKLiK-296 greatly enhanced CREB-mediated transcription, which was not further enhanced by ionomycin addition (Figure 4C).
A constitutively active CKLiK mutant activates CREB and CREM independently of Ca++ /calmodulin.
(A) Schematic representation of the predicted calmodulin-binding domain (CBD) and the autoinhibitory domain (AID) of CKLiK. Truncation mutants of these 2 domains are indicated. In CKLiK-309 the predicted CBD and in CKLiK-296 the predicted CBD and AID were deleted. (B) In vitro kinase assays. COS cells were transfected with 10 μg HA_CKLiK-WT (wt), HA_CKLiK-309 (309), or HA_CKLiK-296 (296) as indicated. Immunoprecipitations were performed and CREM phosphorylation was detected in the presence or absence of Ca++/calmodulin as indicated. Expression of the different forms of CKLiK was detected by Western blotting of cell lysates with 12CA5 antibody. (C) Reporter assays. Cells were transfected with GAL4-CAT reporter construct, CREB-GAL4 (2 μg) and cotransfected with CKLiK-WT or mutated CKLiK-309 or CKLiK-296 (4 μg) as indicated. Eighteen hours before harvesting, cells were stimulated with (░) or without (■) 1 μmol/L ionomycin and reporter assays were performed. Data represent at least 3 independent experiments ± SEM and were corrected for transfection efficiency. (D) Active CKLiK is localized in the nucleus. Cells were grown on coverslips and transfected with CKLiK and active CKLiK-296 containing N-terminal enhanced green fluorescent protein (eGFP). Thirty-six hours after transfection cells were examined by fluorescence microscopy for CKLiK localization.
A constitutively active CKLiK mutant activates CREB and CREM independently of Ca++ /calmodulin.
(A) Schematic representation of the predicted calmodulin-binding domain (CBD) and the autoinhibitory domain (AID) of CKLiK. Truncation mutants of these 2 domains are indicated. In CKLiK-309 the predicted CBD and in CKLiK-296 the predicted CBD and AID were deleted. (B) In vitro kinase assays. COS cells were transfected with 10 μg HA_CKLiK-WT (wt), HA_CKLiK-309 (309), or HA_CKLiK-296 (296) as indicated. Immunoprecipitations were performed and CREM phosphorylation was detected in the presence or absence of Ca++/calmodulin as indicated. Expression of the different forms of CKLiK was detected by Western blotting of cell lysates with 12CA5 antibody. (C) Reporter assays. Cells were transfected with GAL4-CAT reporter construct, CREB-GAL4 (2 μg) and cotransfected with CKLiK-WT or mutated CKLiK-309 or CKLiK-296 (4 μg) as indicated. Eighteen hours before harvesting, cells were stimulated with (░) or without (■) 1 μmol/L ionomycin and reporter assays were performed. Data represent at least 3 independent experiments ± SEM and were corrected for transfection efficiency. (D) Active CKLiK is localized in the nucleus. Cells were grown on coverslips and transfected with CKLiK and active CKLiK-296 containing N-terminal enhanced green fluorescent protein (eGFP). Thirty-six hours after transfection cells were examined by fluorescence microscopy for CKLiK localization.
Because we demonstrated that activated CKLiK could phosphorylate CREM and activate CREB-mediated transcription, we were interested in determining whether CKLiK was located in the cytoplasm or nucleus. We generated GFP containing fusion constructs of CKLiK-WT and the active CKLiK-296. COS cells were grown on coverslips and transfected with CKLiK and CKLiK-296 containing N-terminal GFP fusion. Localization was examined by fluorescence microscopy (Figure 4D). GFP-CKLiK was clearly located in the cytoplasm, whereas the active form of CKLiK exhibited a strong nuclear fluorescence. Because the active form of CKLiK is detected in the nucleus, it suggests that CKLiK may regulate transcription through nuclear localization.
Ca++/calmodulin kinase kinaseα enhances CKLiK activity
Although the binding of Ca++/calmodulin is known to activate CaM kinases, an additional activation of CaMKI and CAMKIV by CAMK kinase (CAMKK) has previously been described.36-38CaMKK phosphorylates CAMKI and CaMKIV in the activation loop at position T-177 and T-196/T-200 respectively, stabilizing and thereby enhancing kinase activity. CKLiK contains a threonine at position 180 in alignment with T-177 of CaMKI and T-200 of CAMKIV. To investigate the effect of CAMKKα on the CKLiK activity we performed kinase and reporter assays. Cotransfection of CaMKKα with CKLiK resulted in an enhanced CREM phosphorylation (Figure 5A; right panel). In the absence of Ca++/calmodulin in the kinase assay, CaMKKα enhanced CKLiK-induced CREM phosphorylation (Figure 5A; left panel). Similar results were found using reporter assays (Figure 5B). Cotransfection of CKLiK with CaMKKα resulted in an elevated reporter activity both in untreated (Figure 5B; left panel) and ionomycin-treated cells (Figure 5B; right panel). CaMKKα alone had no effect on CAT activity, suggesting that CaMKKα cannot itself activate CREB directly. Interestingly CaMKKα can enhance CKLiK activity in the absence of Ca++ in the in vitro kinase assay. Indeed there are indications for CaMKKα-induced CaMK activity in the absence of Ca++ and calmodulin33,.39However, Ca++/calmodulin is present in the transfected cells, and phosphorylation of CKLiK by CaMKKα may lead to stabilization of the CKLiK/Ca++/calmodulin complex, resulting in enhanced kinase activity.
CaMKKα enhances CKLiK activity.
(A) HA_CKLiK (8 μg) was transfected in COS cells with or without CaMKKα_VSV (2 μg). Before harvesting, cells were stimulated with or without 1 μmol/L ionomycin as indicated. HA_CKLiK was immunoprecipitated and kinase assays were performed using CREM as substrate in the presence or absence of Ca++/calmodulin. Equal protein expression was demonstrated by Western blotting. For CKLiK antibody 12CA5 (middle panel) and for CaMKKα antibody anti-VSV (bottom panel) is used. (B) CREB_GAL4 and CKLiK (4 μg) were cotransfected without or with CaMKKα (2 μg) as indicated. CaMKKα was transfected without CKLiK as a negative control. Eighteen hours before harvesting, cells were stimulated with or without 1μmol/L ionomycin and reporter assays were performed. Data represent at least 3 independent experiments ± SEM and are corrected for transfection efficiency.
CaMKKα enhances CKLiK activity.
(A) HA_CKLiK (8 μg) was transfected in COS cells with or without CaMKKα_VSV (2 μg). Before harvesting, cells were stimulated with or without 1 μmol/L ionomycin as indicated. HA_CKLiK was immunoprecipitated and kinase assays were performed using CREM as substrate in the presence or absence of Ca++/calmodulin. Equal protein expression was demonstrated by Western blotting. For CKLiK antibody 12CA5 (middle panel) and for CaMKKα antibody anti-VSV (bottom panel) is used. (B) CREB_GAL4 and CKLiK (4 μg) were cotransfected without or with CaMKKα (2 μg) as indicated. CaMKKα was transfected without CKLiK as a negative control. Eighteen hours before harvesting, cells were stimulated with or without 1μmol/L ionomycin and reporter assays were performed. Data represent at least 3 independent experiments ± SEM and are corrected for transfection efficiency.
Activation of CKLiK by IL-8 in bone marrow–derived myeloid precursor cells
Because agonist stimulation of G protein–coupled receptors results in changes of [Ca++]i, these receptors are potential activators of CKLiK. The IL-8 receptor is highly expressed on human neutrophils and IL-8 can activate neutrophil effector functions, such as chemotaxis. To investigate whether IL-8 can activate CKLiK, we used a 32D model system. The 32D cells are IL-3–dependent myeloid precursor cells derived from bone marrow.40 Because these cells express endogenous IL-8R41 we generated stable 32D cell lines expressing HA_CKLiK and used these cells to measure IL-8–induced CKLiK activity. IL-8 was indeed able to induce an increase in intracellular Ca++ in these cells (Figure6A). IL-8 stimulation of HA_CKLiK-expressing 32D cells resulted in a rapid transient increased CKLiK activation (Figure 6B), whereas in Ca++-depleted cells, IL-8–induced CKLiK activity was completely abolished (data not shown). CKLiK activation parallels the rise in intracellular Ca++ induced by IL-8 (Figure 6A). In addition, W7, an antagonist of calmodulin, completely inhibited the IL-8–induced CKLiK activity (Figure 6C). CKLiK activity was also inhibited by W7 in the ionomycin-treated cells, indicating that calmodulin is indeed critical for the CKLiK kinase activity stimulated by agents, which induce a Ca++ influx.
IL-8 stimulation of bone marrow–derived myeloid precursor cells activates CKLiK.
(A) Ca++ response of 32D cells to hIL-8 (10−7mol/L). Cells were incubated with INDO-am for 45 minutes and washed twice. [Ca++]i concentrations were measured by dual excitation at a wavelength of 340 nm and detected at 390 nm using a Hitachi F4500 fluorescence spectrophotometer. Digitonin was added for 100% [Ca++]i as indicated. (B) IL-8 induces CREB phosphorylation in 32D cells stably expressing wild-type HA_CKLiK. Cells were starved for 4 hours and stimulated with IL-8 (10−7 mol/L) for indicated time periods. HA-tagged CKLiK was immunoprecipitated and kinase assays performed in the absence of Ca++/calmodulin using CREM as a substrate. Ionomycin-treated and untransfected 32D cells were used as positive and negative controls, respectively. Data represent 1 of at least 3 independent experiments. (C) W7 inhibits CKLiK kinase activity. HA_CKLiK stable 32D cells were starved and treated for 15 minutes with or without the calmodulin inhibitor W7 prior to IL-8 (10−mol/L) stimulation. Immunocomplex kinase assays were performed as in (B).
IL-8 stimulation of bone marrow–derived myeloid precursor cells activates CKLiK.
(A) Ca++ response of 32D cells to hIL-8 (10−7mol/L). Cells were incubated with INDO-am for 45 minutes and washed twice. [Ca++]i concentrations were measured by dual excitation at a wavelength of 340 nm and detected at 390 nm using a Hitachi F4500 fluorescence spectrophotometer. Digitonin was added for 100% [Ca++]i as indicated. (B) IL-8 induces CREB phosphorylation in 32D cells stably expressing wild-type HA_CKLiK. Cells were starved for 4 hours and stimulated with IL-8 (10−7 mol/L) for indicated time periods. HA-tagged CKLiK was immunoprecipitated and kinase assays performed in the absence of Ca++/calmodulin using CREM as a substrate. Ionomycin-treated and untransfected 32D cells were used as positive and negative controls, respectively. Data represent 1 of at least 3 independent experiments. (C) W7 inhibits CKLiK kinase activity. HA_CKLiK stable 32D cells were starved and treated for 15 minutes with or without the calmodulin inhibitor W7 prior to IL-8 (10−mol/L) stimulation. Immunocomplex kinase assays were performed as in (B).
Activation of downstream signaling pathways by CKLiK
To identify potential downstream targets of CKLiK, we analyzed the effect of CKLiK on the activation of the ERK1 MAP kinase and PKB, an effector of PI-3K. The regulation of ERKs and PKB by [Ca++]i has been demonstrated in several systems.42,43 Cells were transfected with HA-tagged ERK or PKB and cotransfected with constitutively active CKLiK (CKLiK-296) or CaMKKα and immunocomplex kinase assays were performed. Stimulation with 20% FCS for 10 minutes was used as a positive control. Cotransfection of CKLiK-296 had no effect on PKB activity, whereas cotransfection of CaMKKα potently activated PKB similar to levels induced by 20% FCS (Figure 7A; upper panel). This is in agreement with the direct PKB phosphorylation by CaMKK described by Yano and colleagues.42 Interestingly, however, cotransfection of constitutively active CKLiK-296 resulted in an increase in ERK1 activation (Figure 7A; lower panel). To avoid the possibility of autocrine effects due to overexpression of active CKLiK, we constructed a tamoxifen-inducible active CKLiK (ER_CKLiK-296). When transfected into cells, ER_CKLiK-296 is actively repressed by binding of heat-shock proteins. On addition of the estrogen derivative, 4-hydroxy-tamoxifen (4-OHT), heat-shock proteins dissociate and CKLiK-296 is directly activated.44 This system has been successfully used for other protein kinases.45 Cells were transfected with ER_CKLiK-296 and HA epitope–tagged ERK1 or PKB. Before harvesting, cells were stimulated with 4-OHT and ERK/PKB kinase activity was measured. After 2 minutes of treatment an increased ERK activation was detected, whereas no PKB activation occurred (Figure7B). The CKLiK-induced ERK activity could be blocked by the MEK inhibitor PD098059 (Figure 7C), suggesting that CKLiK activates signaling molecules upstream in the MAPK pathway. Thus, these results suggest a role for CKLiK in regulation of ERK MAPK kinases at the level of MEK or higher.
CKLiK activates ERK1 MAP kinase but not PKB.
(A) HA_PKB or HA_ERK1 (2 μg) was transfected in COS cells with or without CaMKKα (8 μg) or CKLiK-296 (8 μg) as indicated. Twenty hours before harvesting cells were serum starved. Unstimulated and 20% FCS-treated cells were used as controls. HA_PKB or HA_ERK were immune precipitated and kinase activity was measured using histone 2B (H2B) or myelin basic protein (MBP) as a substrates. Data represent 1 of at least 4 independent experiments. (B) HA_ERK1 or HA_PKB (2 μg) was cotransfected with ER_CKLiK-296 (8 μg) in COS cells. Before harvesting, cells were stimulated with 4-OHT for 2 or 10 minutes. Immunoprecipitations and kinase assays were performed as in panel A using H2B or MBP as a substrate. Equal expression of HA_PKB and HA_ERK were analyzed by Western blotting of whole cell lysates with 12CA5 antibody. (C) PD098059 inhibits the CKLiK-induced ERK activity. COS cells were transfected with HA_ERK1 (2 μg) and ER_CKLiK-296 (8 μg). Cells were treated with or without the MEK inhibitor PD098059 (50 μmol/L) for 15 minutes before stimulation with 4-OHT for indicated time periods. Cells were lysed and immunocomplex kinase assays were performed as previously described. Equal expression of HA_ERK1 was analyzed by Western blotting.
CKLiK activates ERK1 MAP kinase but not PKB.
(A) HA_PKB or HA_ERK1 (2 μg) was transfected in COS cells with or without CaMKKα (8 μg) or CKLiK-296 (8 μg) as indicated. Twenty hours before harvesting cells were serum starved. Unstimulated and 20% FCS-treated cells were used as controls. HA_PKB or HA_ERK were immune precipitated and kinase activity was measured using histone 2B (H2B) or myelin basic protein (MBP) as a substrates. Data represent 1 of at least 4 independent experiments. (B) HA_ERK1 or HA_PKB (2 μg) was cotransfected with ER_CKLiK-296 (8 μg) in COS cells. Before harvesting, cells were stimulated with 4-OHT for 2 or 10 minutes. Immunoprecipitations and kinase assays were performed as in panel A using H2B or MBP as a substrate. Equal expression of HA_PKB and HA_ERK were analyzed by Western blotting of whole cell lysates with 12CA5 antibody. (C) PD098059 inhibits the CKLiK-induced ERK activity. COS cells were transfected with HA_ERK1 (2 μg) and ER_CKLiK-296 (8 μg). Cells were treated with or without the MEK inhibitor PD098059 (50 μmol/L) for 15 minutes before stimulation with 4-OHT for indicated time periods. Cells were lysed and immunocomplex kinase assays were performed as previously described. Equal expression of HA_ERK1 was analyzed by Western blotting.
Discussion
We have described the identification and characterization of a novel protein serine kinase that is highly expressed in human PMNs. These cells, which include neutrophils and eosinophils, are characterized by a variety of specific effector functions that regulate host defense. Although several protein kinases have been previously postulated to regulate some of these effector functions through pharmacologic inhibition studies, these proteins are ubiquitously expressed and thus more likely to be involved in general cellular processes. Furthermore, the use of pharmacologic inhibitors is often complicated by nonspecific effects15 46
By using degenerate PCR primers against conserved catalytic kinase domains in PCR on cDNA templates from human granulocytes, we have identified a novel protein kinase with an open reading frame of 357 amino acids (Figure 1). This novel kinase shows homology with calcium/calmodulin-dependent kinase I (CaMKI) and therefore we have termed this kinase CaMKI-like kinase (CKLiK). CaMKI belongs to the family of Ca++-and calmodulin-dependent kinases, which includes CaMK I, II, and IV as well as myosin light chain kinase, phosphorylase kinase, and elongation factor 2 kinase.47,48CaMKI has a wide tissue distribution and can phosphorylate a number of substrates in vitro, including the synaptic vesicle-associated proteins synapsin 1 and synapsin 249 and CREB.28Interestingly, analysis of CKLiK mRNA in hematopoietic cells revealed high and almost exclusive expression levels in PMNs, suggesting a role for CKLiK in human granulocytes (Figure 2). Up-regulation of CKLiK mRNA was also observed in cord blood CD34+ stem cells differentiated toward neutrophils, which may indicate the importance of this kinase in terminally differentiated myeloid cells. In granulocytes, changes in [Ca++]i have been associated with multiple functions, including degranulation, phagosome-lysosome fusion, regulation of cytoskeletal binding proteins, and transcriptional control.16 17 These are thus potential processes whereby CKLiK may play a role.
Based on crystal structure and mutational analysis of CamKI, a model has been proposed for CaMKs regulation.50 51 When Ca++ levels rise within the cytosol, calmodulin binds Ca++ and is capable of interacting with calmodulin-binding proteins such as CaMK. Inactive CaMKs are in a folded configuration whereby the N-terminus acts as a pseudosubstrate by interacting with an autoinhibitory domain. On Ca++ recruitment, calmodulin binds to the calmodulin-binding domain located on the C-terminal of CaMK, which then results in unfolding and autophosphorylation of CaMK. It appears that CKLiK is also similarly regulated because truncation of the predicted calmodulin-binding domain or the autoinhibitory domain resulted in an inactive and constitutively active CKLiK, respectively (Figure 4).
Additionally we show that CKLiK can directly activate 2 members of the CREB/ATF family of transcription factors, CREM and CREB, by phosphorylation of serine residues 117 and 133, respectively (Figure3). Although CKLiK is located in the cytoplasm, the constitutively active mutant was clearly translocated to the nucleus (Figure 4D). Possibly this enhanced nuclear translocation uncouples CKLiK from normal regulation resulting in enhanced transcriptional activation. For CaMKIV it has been suggested that it can phosphorylate CREB in vivo because it shows nuclear localization.52 Although a role for CREB in myeloid cells has been proposed in mediating cytokine or chemokine effector functions,53-56 it remains to be determined if CREB and CREM are physiologic substrates for CKLiK in human granulocytes. Indeed we were able to demonstrate CREB phosphorylation by IL-8 in human PMNs (data not shown).
Cotransfection of CaMK kinase (CaMKKα) resulted in an enhanced CKLiK activity measured by in vitro phosphorylation of CREM and in vivo CREB-mediated transcription activation. Enhancement of CKLiK activity by CaMKK is possible via phosphorylation of the threonine residue on position 180, because this is in consensus with threonine residues 177 and 196/200 of CaMKI and CaMKIV, respectively. Interestingly CaMKKα induced CKLiK activity in the absence of Ca++ in the in vitro kinase assay (Figure 5). There is indication of the generation of Ca++-independent CaMKIV activity by CaMKKα; however, this is not the case for CaMKI.33,39 What the mechanism is for generating this enhanced CKLiK activity is unclear, although this may be irrelevant in vivo. It may be due to stabilization of the CKLiK/Ca++/calmodulin complex allowing, for example, a prolonged kinase activity similar to the regulation of CaMKII,57 which might be needed for regulation of transcription.
The IL-8–induced CKLiK activation was rapid and corresponded to the IL-8–induced rise in [Ca++]i (Figure 6). The IL-8 receptor is highly expressed on human neutrophils and could be a possible upstream physiologic receptor for CKLiK. IL-8 is known to induce chemotaxis, and recently a role for intracellular Ca++ in IL-8–induced migration has been described indicating a possible role of CKLiK in this process.58,59 A role for Ca++ and calmodulin-dependent kinases in apoptosis has also been recently demonstrated because CaMKK was found to directly activate PKB and thereby rescue apoptosis.42 Although we found no activation of PKB by CKLiK, we demonstrated an elevated ERK activity by inducing CKLiK activation, which could be blocked by the MEK inhibitor PD098059 (Figure 7). As previously alluded to, pharmacologic inhibitory studies have suggested that ERK MAP kinases possibly play a role in chemotaxis or respiratory burst and PAF release.6,7 60 The mechanism by which CKLiK can activate ERKs however remains to be resolved, although it presumably occurs upstream of ERK.
In conclusion, the approach of degenerate PCR resulted in a successful isolation of a novel PMN protein kinase. We have demonstrated that this kinase is highly and predominantly expressed in human PMNs and terminally differentiated myeloid stem cells and that Ca++/calmodulin regulates its activity. IL-8, which is a potent activator of neutrophil effector functions, is capable of activating CKLiK in bone marrow-derived myeloid cells. Furthermore, a role for CKLiK is suggested in regulation of ERK MAP kinases because induced CKLiK activity was sufficient to activate ERK1. Therefore, the restricted expression of CKLiK in granulocytes can allow the transduction of granulocyte-specific signals induced by a general signal transduction event: the rise in [Ca++]i, initiated by inflammatory mediators.
Acknowledgments
We would like to thank Rolf de Groot for supplying the CREMτ, CREMβ-S117A, and CaMKI proteins and the GAL4 reporter constructs, CREB_GAL4 and CREB-S133A_GAL4 fusion constructs. We would also like to thank Sigrid Oomen and Ivo Touw (Department of Hematology, Erasmus University, Rotterdam) for technical assistance and advice in the generation of 32D stable cell lines.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Paul J. Coffer, Department of Pulmonary Diseases, G03550, University Medical Centre Utrecht, Heidelberglaan 100, 3584 CX Utrecht, The Netherlands; e-mail:P.Coffer@hli.azu.nl.




![Fig. 6. IL-8 stimulation of bone marrow–derived myeloid precursor cells activates CKLiK. / (A) Ca++ response of 32D cells to hIL-8 (10−7mol/L). Cells were incubated with INDO-am for 45 minutes and washed twice. [Ca++]i concentrations were measured by dual excitation at a wavelength of 340 nm and detected at 390 nm using a Hitachi F4500 fluorescence spectrophotometer. Digitonin was added for 100% [Ca++]i as indicated. (B) IL-8 induces CREB phosphorylation in 32D cells stably expressing wild-type HA_CKLiK. Cells were starved for 4 hours and stimulated with IL-8 (10−7 mol/L) for indicated time periods. HA-tagged CKLiK was immunoprecipitated and kinase assays performed in the absence of Ca++/calmodulin using CREM as a substrate. Ionomycin-treated and untransfected 32D cells were used as positive and negative controls, respectively. Data represent 1 of at least 3 independent experiments. (C) W7 inhibits CKLiK kinase activity. HA_CKLiK stable 32D cells were starved and treated for 15 minutes with or without the calmodulin inhibitor W7 prior to IL-8 (10−mol/L) stimulation. Immunocomplex kinase assays were performed as in (B).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/96/9/10.1182_blood.v96.9.3215/5/m_h82100292006.jpeg?Expires=1769173021&Signature=xY7z5DB~KCgp5~Dmz8tX2MEoLoNpoYAVVOQEzUKGREdYLu-~ikMQb2mQEzZzpYREzkyKeK0qLoCViAstpJbqOF6mDWBKmRb3gxHyYmtjU2PqW4phb2DWslPkkJvnPA~uIQLxSLslMDvO7LnpJL9xzUi03ZvJIPEyLCocVJ~zN0-BmjmR82UxXphJi858CiCmSNcWRRBzNfLlROVhJhvzt3hGhgX2rRsoEfz1GEZm9jZNoXqU2w8Cqg9px4Tg54VcR03Tog574zFfVSHKVn1BYw5J--36iUxDdCPz7nsly02xBmGpmIHRr~ByB64clF4A7gScdBZlG1zlVPjgOa20zQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal