Abstract
The transcription of insulin-like growth factor 2 (IGF-2) is affected by genomic imprinting, a multistep process through which the parental origin of a gene influences its transcription. The maternal copy of IGF-2 is silenced in most human tissues, but in the choroid plexus and the adult liver both alleles of IGF-2 are expressed. This study shows that though in peripheral blood mononuclear cells IGF-2shows paternal allele-specific expression, in total bone marrow both alleles are transcribed. This modulation of imprinting is not attributable to use of the P1 promoter, because transcription from the P3 promoter occurred from both alleles. These results suggest that transcriptional recognition of the IGF-2 imprint can be modulated during hematopoiesis and may facilitate the development of in vitro model systems to study the transcriptional recognition of a genomic imprint.
Introduction
Genomic imprinting is a complex multistep process that results in parent-of-origin–specific inactivation of alleles of some genes. The components of the imprinting process include initiation of the imprint in the gamete, postfertilization modification, maintenance during mitosis, and transcriptional recognition. Although it is clear that cytosine methylation is involved in mammalian imprinting, little is known about the structure of an imprint and how it is recognized.
Insulin-like growth factor 2 (IGF-2) is an imprinted growth factor that has a major role in fetal development. In most tissues the paternally derived copy of IGF-2 is expressed, whereas the maternal copy is silent.1 Exceptions to this have been observed in the choroid plexus and leptomeninges of rodents and humans, the rat brain, the adult human liver, the ciliary anlage of the human embryonic retina, and human chondrocytes.2-6Biallelic expression of IGF-2 in the human liver is restricted to late fetuses and adults and occurs as a result of biallelic transcription from the P1 promoter that is normally not imprinted.6 Similarly, the biallelic expression observed in chondrocytes appears to be derived from the P1 promoter. However, not all occurrences of biallelic IGF-2 expression are attributable to use of the P1 promoter. For example, biallelic expression of IGF-2 in the human choroid plexus occurs predominantly from the P3 promoter.6 When IGF-2is expressed from both alleles, it is unknown whether it occurs as a result of erasure of the imprint or because the imprint is not recognized by the transcriptional complex.
We have detected biallelic expression of IGF-2 in human bone marrow, whereas in the peripheral blood, IGF-2 is exclusively expressed from the paternal allele. These results suggest that recognition of the imprint of a gene can be modulated during development.
Materials and methods
Bone marrow and blood samples
Paired bone marrow and peripheral blood samples were taken from 3 healthy volunteers. Peripheral blood samples were taken from 41 healthy volunteers. Diagnostic bone marrow aspirate samples in excess of diagnostic requirements were obtained. Bone marrow samples used for imprinting studies are listed in Table1. Collection and study of patient specimens was approved by The Otago Area Health Board Ethics Committee.
Description of bone marrow samples on which imprinting studies were performed
| Number . | Diagnosis . | Malignant cells (%) . | Sex/age (y) . | IGF-2 . | H19 . | SNRPN . | IGF-2R . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA . | RNA . | DNA . | RNA . | DNA . | RNA . | DNA . | RNA . | ||||
| X12 | ALL in remission | < 5 | F38 | AB | ab | ||||||
| X22 | Normal | 0 | F73 | AB | ab | ||||||
| X26 | MGUS | 3 | M73 | AB | ab | ||||||
| X166 | Normal | 0 | M28 | AB | ab | AB | b | BC | bc | ||
| X175 | NHL staging | Involved | F45 | AB | ab | ||||||
| X177 | Normal | 0 | F30 | AB | ab | AB | a | AB | a | NI | |
| X178 | Normal | 0 | M35 | AB | ab | NI | NI | ||||
| X188 | T-ALL relapse | 28 | M4 | AB | ab | ||||||
| X196 | MDS:RA | NA | M59 | AB | ab | ||||||
| X221 | Megaloblastic | 0 | F80 | AB | ab | AB | b | AB | a | NI | |
| X279 | ALL | > 80 | F4 | AB | a | ||||||
| X289 | NHL staging | 0 | M45 | AB | ab | NI | |||||
| X293 | MPD | NA | M60 | AB | ab | NI | NI | ||||
| X296 | MDS:CMML | NA | M74 | AB | ab | AB | a | BC | bc | ||
| X369 | Neuroblastoma | Involved | M28 | AB | b | ||||||
| X374 | Normal | 0 | F74 | ni | AB | b | AB | a | AB | ab | |
| X381 | ITP | 0 | M88 | AB | ab | AB | a | NI | AC | ac | |
| X388 | Normal | 0 | F28 | AB | ab | AB | b | NI | NI | ||
| X391 | Iron deficiency | 0 | M60 | ni | AB | a | |||||
| X392 | MDS:RA | NA | M68 | ni | AB | a | |||||
| X393 | Rhabdomyoblastoma | ∼2 | M14 | AB | ab | ||||||
| X396 | MGUS | < 5 | M65 | AB | ab | NI | |||||
| X419 | Normal | 0 | F10 | AB | ab | AB | a | NI | BC | bc | |
| X610 | NHL-low grade | Involved | M70 | AB | b | ||||||
| Number . | Diagnosis . | Malignant cells (%) . | Sex/age (y) . | IGF-2 . | H19 . | SNRPN . | IGF-2R . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA . | RNA . | DNA . | RNA . | DNA . | RNA . | DNA . | RNA . | ||||
| X12 | ALL in remission | < 5 | F38 | AB | ab | ||||||
| X22 | Normal | 0 | F73 | AB | ab | ||||||
| X26 | MGUS | 3 | M73 | AB | ab | ||||||
| X166 | Normal | 0 | M28 | AB | ab | AB | b | BC | bc | ||
| X175 | NHL staging | Involved | F45 | AB | ab | ||||||
| X177 | Normal | 0 | F30 | AB | ab | AB | a | AB | a | NI | |
| X178 | Normal | 0 | M35 | AB | ab | NI | NI | ||||
| X188 | T-ALL relapse | 28 | M4 | AB | ab | ||||||
| X196 | MDS:RA | NA | M59 | AB | ab | ||||||
| X221 | Megaloblastic | 0 | F80 | AB | ab | AB | b | AB | a | NI | |
| X279 | ALL | > 80 | F4 | AB | a | ||||||
| X289 | NHL staging | 0 | M45 | AB | ab | NI | |||||
| X293 | MPD | NA | M60 | AB | ab | NI | NI | ||||
| X296 | MDS:CMML | NA | M74 | AB | ab | AB | a | BC | bc | ||
| X369 | Neuroblastoma | Involved | M28 | AB | b | ||||||
| X374 | Normal | 0 | F74 | ni | AB | b | AB | a | AB | ab | |
| X381 | ITP | 0 | M88 | AB | ab | AB | a | NI | AC | ac | |
| X388 | Normal | 0 | F28 | AB | ab | AB | b | NI | NI | ||
| X391 | Iron deficiency | 0 | M60 | ni | AB | a | |||||
| X392 | MDS:RA | NA | M68 | ni | AB | a | |||||
| X393 | Rhabdomyoblastoma | ∼2 | M14 | AB | ab | ||||||
| X396 | MGUS | < 5 | M65 | AB | ab | NI | |||||
| X419 | Normal | 0 | F10 | AB | ab | AB | a | NI | BC | bc | |
| X610 | NHL-low grade | Involved | M70 | AB | b | ||||||
Columns labeled DNA refer to the genotype at a polymorphic site; columns labeled RNA indicate which alleles were expressed.
ALL indicates acute lymphoblastic leukemia; MGUS, monoclonal gammopathy of undetermined significance; NHL, non-Hodgkin lymphoma; MDS:RA, myelodysplasia:refractory anemia; MPD, myeloproliferative disease; ITP, immune thrombocytopenia; NA, not available.
RNA extraction
RNA was extracted from sodium heparin anticoagulated low-density peripheral blood or bone marrow cells, after centrifugation over a Ficoll-Hypaque layer, using the acid guanidinium thiocyanate method.7
IGF-2 imprinting
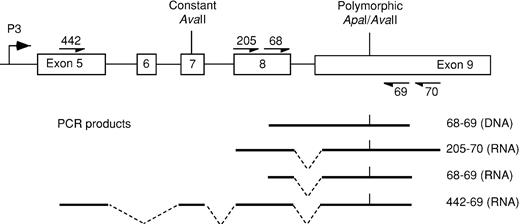
For the assessment of allelic expression of IGF-2, the exon 9 ApaI/AvaII polymorphism was used.8,9 Heterozygous specimens were identified afterApaI digestion of DNA-derived polymerase chain reaction (PCR) products using primers 68 and 69 (see Figure1). Among 186 unrelated persons, the relative frequencies of allele A (not digested by ApaI) and allele B were 0.34 and 0.66. For comparison, published allele frequencies of the allele A are 0.25, 0.38, 0.46, and 0.47.8-10 Bone marrow or peripheral blood RNA was reverse transcribed to cDNA using random hexamers and mMLV. The cDNA was amplified by PCR using primers that spanned intron 8 to eliminate potential misinterpretation arising from contaminating DNA. For bone marrow, cDNA was amplified using primers 68 and 69 (see above), with an annealing temperature of 57°C. For peripheral blood we used nested PCR in which the cDNA was preamplified for 35 cycles using primers 205 and 70 with an annealing temperature of 56°C. Amplified products were digested with ApaI or with AvaII to yield polymorphic fragments of 988 and 891 bp (Figure 1). ApaI andAvaII digest opposite alleles at the polymorphic site; by using each separately, apparent biallelic expression as a result of partial digestion can be excluded. PCR product contamination was consistently excluded by the use of negative controls, in which water was substituted for RNA.
Diagram of exons 5 to 9 of IGF-2showing primers encompassing theApaI/AvaII polymorphic site and the resultant PCR products.
Exons 7 to 9 are used by all 4 promoters, whereas exon 5 is specific to the P3-derived transcript.
Diagram of exons 5 to 9 of IGF-2showing primers encompassing theApaI/AvaII polymorphic site and the resultant PCR products.
Exons 7 to 9 are used by all 4 promoters, whereas exon 5 is specific to the P3-derived transcript.
IGF-2 P3-specific imprinting
Exons 5 to 9 of IGF-2 were amplified from random hexamer-generated cDNA from 4 heterozygous bone marrows using primers 422 and 69. P3-specific products (1372 bp) were obtained and digested with ApaI or AvaII to generate polymorphic fragments of 1372 or 1277 bp and 1084 or 989 bp, respectively.
SNRPN imprinting
Exons 2 and 3 were amplified using primers 654 and 653 that spanned intron 2 and the BstUI polymorphism in the 5′UTR of exon 2 (the first coding exon).11 A 217-bp RNA-derived product was amplified, whereas genomic DNA gave a product approximately 925 bp larger. In the presence of the BstUI polymorphic site, 62 bp are cleaved from the PCR product.
H19 imprinting
The RsaI restriction enzyme site polymorphism in exon 5 was used to determine the imprinting status of H19 in total bone marrow.12 cDNA was amplified using primers that spanned intron 4 (H19 exon 4 and H19 exon 5). Amplification of DNA produced a 704-bp product that was cleaved to 530 bp if the RsaI site was present, whereas cDNA produced a 624-bp product that was cleaved to 450 bp.
IGF-2 receptor imprinting
Primers spanning the dinucleotide repeat polymorphism and 1 of the 4-bp deletion/insertions in the 3′UTR of IGF-2 receptor (IGF-2R) were used to assess imprinting in total bone marrow, as previously described.13 14 cDNA was synthesized from DNAase-treated RNA. The absence of contaminating DNA-derived product was confirmed for each sample with the use of mock cDNA synthesized without reverse transcriptase. The products were visualized by autoradiography after electrophoresis in a 6% denaturing polyacrylamide gel.
Primer sequences
Primer sequences were as follows: IGF-2 68, TCC TGG AGA CGT ACT GTG CTA; IGF-2 69, GGG TTG TTG CTA TTT TCG GAT;IGF-2 205, TTG AGG AGT GCT GTT TCC GC; IGF-2 70, GGT CGT GCC AAT TAC ATT TCA; IGF-2 422, CCC GCT CTG CCC CGT CGC ACA TTC; SNRPN 654, AAC CAG GCT CCA TCT ACT CTT TG;SNRPN 653, TCT TGC AGG ATA CAT CTC ATT CTA; H19exon 4, CGG ACA CAA AAC CCT CTA GCT TGG AAA; H19 exon 5, GCG TAA TGG AAT GCT TGA AGG CTG CTC.
Results
IGF-2 is biallelically expressed in bone marrow
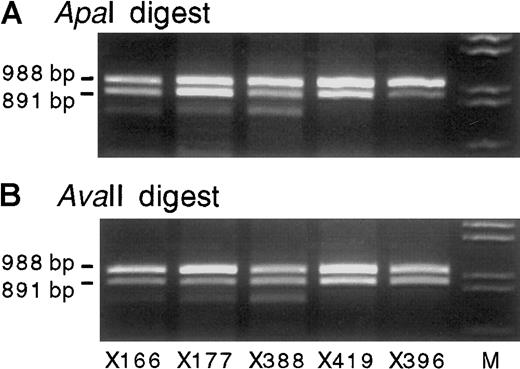
Twenty-four bone marrow samples were informative at theIGF-2 exon 9 ApaI/AvaII polymorphism. In 19 of these samples (from 16 patients and 3 healthy volunteers),IGF-2 transcripts were detected by reverse transcription (RT)-PCR, using primers 68 and 69 as outlined in Figure 1. In 18 of these cases, biallelic expression of IGF-2 was demonstrated by the presence of 2 alleles after digestion with ApaI, whereas in one case the expression of IGF-2 was monoallelic (a case of acute lymphoblastic leukemia) (Table 1). Digestion withAvaII, performed in 8 of these cases, confirmed the presence of 2 alleles (Figure 2). AvaII cleaved the reciprocal allele to that digested by ApaI and excluded the possibility that the apparent biallelic expression was attributable to partial digestion by ApaI. Nine of the biallelically expressing bone marrow samples were from persons with normal or benign biopsies, and the remaining 9 were from patients with diagnostic abnormalities as listed in Table 1.
Exon 8 and 9 IGF-2 RT-PCR products (primers 68 and 69) from 5 representative heterozygous bone marrow RNA samples digested with ApaI orAvaII.
All show biallelic transcription of IGF-2. The use of bothApaI (A) and AvaII (B) excludes misinterpretation from partial enzyme digestion, whereas the product size excludes DNA contamination (Figure 1). Lane M shows λDNA digested withHindIII and EcoRI. All gels shown contain 2% agarose gel with ethidium bromide.
Exon 8 and 9 IGF-2 RT-PCR products (primers 68 and 69) from 5 representative heterozygous bone marrow RNA samples digested with ApaI orAvaII.
All show biallelic transcription of IGF-2. The use of bothApaI (A) and AvaII (B) excludes misinterpretation from partial enzyme digestion, whereas the product size excludes DNA contamination (Figure 1). Lane M shows λDNA digested withHindIII and EcoRI. All gels shown contain 2% agarose gel with ethidium bromide.
IGF-2 is biallelically expressed from the P3 promoter in bone marrow
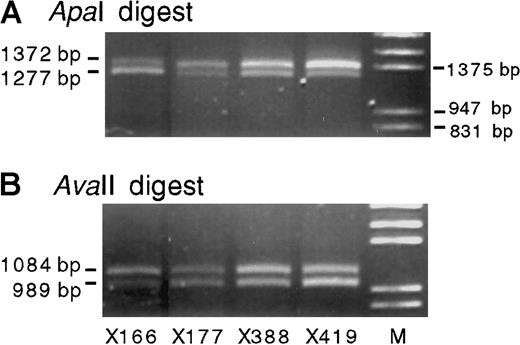
Absence of imprinting of IGF-2 expression has been reported in the adult liver and is attributable to the expression from the P1 promoter, which is not usually imprinted. To determine that the biallelic expression of IGF-2 in the bone marrow did not reflect P1 promoter usage, we used RT-PCR to examine the imprinting of P3-specific transcripts. In 4 heterozygous normal bone marrows, P3-specific transcripts from both alleles were detected (Figure3; the samples shown in Figure 3 are the same as those shown in lanes 1-4 of Figure 2). Because the P3-specific PCR primers span 4 introns and 1 spliced-out exon, the contribution of genomic DNA to these PCR products can be excluded.
Promoter 3-specific IGF-2 RT-PCR products (primers 442-69) from 4 heterozygous normal bone marrow RNA samples.
Digestion with either ApaI (A) or AvaII (B) confirms the presence of 2 transcribed alleles.
Promoter 3-specific IGF-2 RT-PCR products (primers 442-69) from 4 heterozygous normal bone marrow RNA samples.
Digestion with either ApaI (A) or AvaII (B) confirms the presence of 2 transcribed alleles.
IGF-2 is monoallelically expressed in peripheral blood
IGF-2 imprinting status was examined in the peripheral blood of 41 healthy persons, none of whom had hematologic disorders. This group included 18 healthy children (10 girls aged 5 to 11, 8 boys aged 1 to 13 years) and 23 normal adults. IGF-2 RT-PCR products were successfully detected in blood samples from all 18 healthy children and from 22 of 23 healthy adults. After enzyme digestion, all amplified samples showed monoallelic expression ofIGF-2 as previously reported.15 For 12 heterozygous children from 6 families, sufficient material was available to determine the parental origin of the expressedIGF-2 allele, and in each case the expressed allele could only have been derived from the child's father (Table2).
Expression of IGF-2 in peripheral blood
| Child . | Paternal genotype . | Maternal genotype . | Child's genotype . | Expressed allele . |
|---|---|---|---|---|
| M1 | AA | BB | AB | a |
| M2 | AB | a | ||
| M3 | AB | a | ||
| T1 | BB | AA | AB | b |
| T2 | AB | b | ||
| T3 | AB | b | ||
| T4 | AB | b | ||
| D1 | AA | BB | AB | a |
| H1 | AB | BB | AB | a |
| W1 | AB | BB | AB | a |
| O1 | BB | AA | AB | b |
| O2 | AB | b |
| Child . | Paternal genotype . | Maternal genotype . | Child's genotype . | Expressed allele . |
|---|---|---|---|---|
| M1 | AA | BB | AB | a |
| M2 | AB | a | ||
| M3 | AB | a | ||
| T1 | BB | AA | AB | b |
| T2 | AB | b | ||
| T3 | AB | b | ||
| T4 | AB | b | ||
| D1 | AA | BB | AB | a |
| H1 | AB | BB | AB | a |
| W1 | AB | BB | AB | a |
| O1 | BB | AA | AB | b |
| O2 | AB | b |
In 12 children from 6 families, the expressed allele ofIGF-2 was paternally derived.
IGF-2 imprinting was also examined in abnormal peripheral blood from some patients. These included 2 patients with leukemia: one had acute lymphoblastic leukemia and the other had a secondary myeloid leukemia (very early myeloid phenotype in a patient with osteomyelosclerosis). The former showed monoallelic expression, whereas the latter showed biallelic expression of IGF-2. As reported,15 biallelic expression of IGF-2 was detected in the peripheral blood of 4 children with somatic overgrowth.
Comparison of bone marrow and blood from the same persons
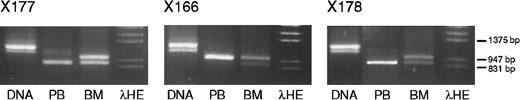
The finding of monoallelic transcription in some samples and biallelic transcription in others raised the question of whether differential allelic transcription was attributable to differences between persons or between tissues. Thus, the imprinting ofIGF-2 in the bone marrow and in the peripheral blood was compared in paired samples from 3 healthy volunteers. In these persons,IGF-2 expression in total bone marrow was biallelic, but that in the peripheral blood was monoallelic, indicating tissue-specific differences in the imprinting. Results from these cases are shown in Figure 4.
IGF-2 PCR and RT-PCR products (primers 68 and 69) from DNA, peripheral blood RNA, and bone marrow RNA from each of 3 healthy volunteers.
Volunteers X177 and X178 express only the short allele in peripheral blood, whereas volunteer X166 expresses the long allele. A small amount of nonspecific PCR product is present in the peripheral blood lane of volunteer X177.
IGF-2 PCR and RT-PCR products (primers 68 and 69) from DNA, peripheral blood RNA, and bone marrow RNA from each of 3 healthy volunteers.
Volunteers X177 and X178 express only the short allele in peripheral blood, whereas volunteer X166 expresses the long allele. A small amount of nonspecific PCR product is present in the peripheral blood lane of volunteer X177.
IGF-2 imprinting in neonatal and cord blood
IGF-2 transcripts were amplified from 3 cord blood and 2 neonatal blood samples informative at the IGF-2 exon 9ApaI/AvaII polymorphism. One of 3 cord blood samples showed partial biallelic expression (Figure5). The presence of both alleles was confirmed by using both ApaI and AvaII digestion of the PCR product. Blood from both neonates (aged 3 and 4 weeks) showed monoallelic expression of IGF-2.
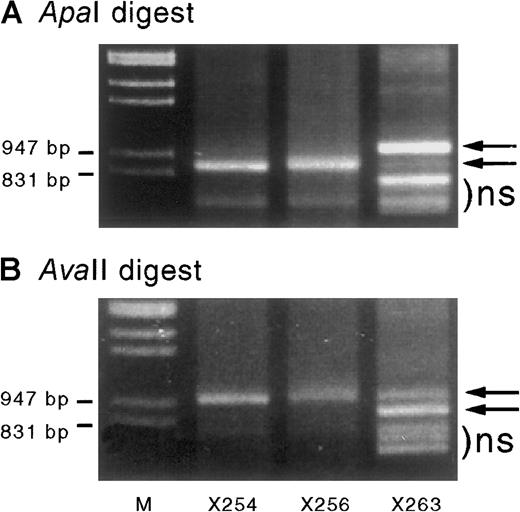
Cord blood–derived IGF-2 RT-PCR products digested with ApaI andAvaII.
When digested with ApaI (A) or AvaII (B) they show monoallelic expression in specimens X254 and X256 but partial biallelic expression in X263 (arrows). Sample X263 also shows some shorter nonspecific (ns) PCR products that migrated in a similar position (below the bands of interest) before enzyme digestion. The ratio of the digested band intensity reflects the effect of heteroduplex formation on the digestion of PCR products. Results are consistent with a starting mix of A and B alleles of approximately 0.8:0.2. Assuming complete heteroduplex formation at the end of PCR cycling, the PCR products consist of a mixture of AA, AB, and BB duplexes in a ratio of a2:2ab:b2—ie, 0.64:0.32:0.04. On incubation with ApaI, only the BB homodimers are digested, resulting in a band ratio of 0.96:0.04 = 24:1 (undigested:digested), whereas incubation with the reciprocal enzyme AvaII products will only digest AA homodimers, resulting in a 0.36:0.64 ratio = 0.56:1. These ratios (24:1 and 0.56:1) are similar to those seen in Figure 5A and B, respectively.
Cord blood–derived IGF-2 RT-PCR products digested with ApaI andAvaII.
When digested with ApaI (A) or AvaII (B) they show monoallelic expression in specimens X254 and X256 but partial biallelic expression in X263 (arrows). Sample X263 also shows some shorter nonspecific (ns) PCR products that migrated in a similar position (below the bands of interest) before enzyme digestion. The ratio of the digested band intensity reflects the effect of heteroduplex formation on the digestion of PCR products. Results are consistent with a starting mix of A and B alleles of approximately 0.8:0.2. Assuming complete heteroduplex formation at the end of PCR cycling, the PCR products consist of a mixture of AA, AB, and BB duplexes in a ratio of a2:2ab:b2—ie, 0.64:0.32:0.04. On incubation with ApaI, only the BB homodimers are digested, resulting in a band ratio of 0.96:0.04 = 24:1 (undigested:digested), whereas incubation with the reciprocal enzyme AvaII products will only digest AA homodimers, resulting in a 0.36:0.64 ratio = 0.56:1. These ratios (24:1 and 0.56:1) are similar to those seen in Figure 5A and B, respectively.
Adherent cells show the highest level of IGF-2expression
Preliminary experiments have shown that the highest levels ofIGF-2 expression (as assessed by semiquantitative RT-PCR) were present in bone marrow cells adhering to plastic tissue culture dishes or nonspecifically binding to paramagnetic beads coated with control antibodies (data not shown). Similarly, among low-density peripheral blood cells, the highest levels of expression were detected in adherent cells, suggesting that monocytes may have the highest expression of IGF-2 in blood and bone marrow.IGF-2 expression could not be detected in the granulocyte fraction of peripheral blood.
SNRPN and H19 showed normal imprinting in bone marrow
To examine whether the biallelic expression ofIGF-2 might reflect generalized changes in imprinting in the bone marrow, we examined the imprinting of SNRPN andH19. Sixteen bone marrow samples were genotyped; 8 were informative at the SNRPN BstUI polymorphism, and 7 were informative at the H19 exon 5 RsaI polymorphism.SNRPN RNA was amplified from the 8 informative bone marrow samples (5 were normal or benign, 1 showed neuroblastoma, 1 showed myelodysplasia, and 1 showed lymphoma; see Table 1). In all cases expression was monoallelic. Examples are shown in Figure6, panel B. Seven bone marrow samples, informative at the H19 exon 5 RsaI polymorphism,12 were used to examine H19imprinting (6 were normal or benign, 1 showed myelodysplasia; see Table1). In all 7 bone marrows, transcripts from only one H19allele were detected (Table 1), indicating that H19imprinting was normal. Resolution of the gel is such that it can be concluded that if relaxation of H19 imprinting did occur, less than 5% of cells were affected.
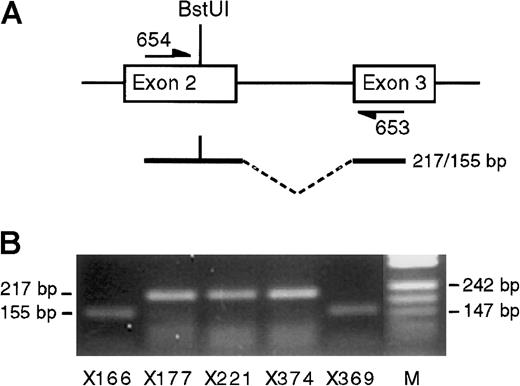
SNRPN transcription.
(A) Diagram of exons 2 and 3 of SNRPN showing polymorphicBstUI site and flanking primers. (B) SNRPN RT-PCR products from 5 heterozygous bone marrow samples after digestion withBstUI. In each case imprinted transcription ofSNRPN is demonstrated. Lane M shows pUC DNA digested with MspI.
SNRPN transcription.
(A) Diagram of exons 2 and 3 of SNRPN showing polymorphicBstUI site and flanking primers. (B) SNRPN RT-PCR products from 5 heterozygous bone marrow samples after digestion withBstUI. In each case imprinted transcription ofSNRPN is demonstrated. Lane M shows pUC DNA digested with MspI.
IGF-2R is not imprinted in bone marrow
To determine whether biallelic expression of IGF-2 in bone marrow might reflect a generalized change in the imprinting of IGF axis genes we examined the imprinting of IGF-2R. Although this gene is imprinted in mice, most investigators have reported that it is not imprinted in humans.3,14,16,17 However, a report of polymorphic imprinting of IGF-2R in some human fetuses18 raises the possibility that the IGF-2Rmight be imprinted in bone marrow to enhance the growth-promoting action of IGF-2. Thirteen bone marrow samples were genotyped for the 3′UTR IGF-2R polymorphism, and 5 informative cases were detected (4 were normal or benign, 1 showed myelodysplasia; see Table1). In all informative bone marrow samples, both alleles ofIGF-2R were detected (Table 1). These results indicate that this gene is not imprinted in most IGF-2R–expressing cells in human bone marrow.
Discussion
We have observed that in bone marrow IGF-2 is expressed biallelically, whereas in the peripheral blood of children and adults,IGF-2 is imprinted. This suggests there is a modulation of the allelic expression of IGF-2 during hematopoietic development. The differences in IGF-2 imprinting between bone marrow and peripheral blood are not attributable to differences between persons but may be associated with the different developmental stages of these tissues. Our observations are in accord with those of a study that reported retention of IGF-2 imprinting in peripheral blood,19 but they differ from those of another study that reported biallelic transcription in most persons aged 6 to 55.20 However, the latter study used a less rigorous RT-PCR methodology in which it is theoretically possible that apparent biallelic expression could have been DNA derived.
In the human liver a similar, but opposite, change in imprinting occurs in that IGF-2 transcription is imprinted in the fetus but not in the adult. However, this apparent modulation of imprinting is attributable to a change in promoter usage, from the normally imprinted P3 promoter to the normally unimprinted P1 promoter. Demonstration of biallelic expression from the P3 promoter in the bone marrow indicates that the apparent “relaxation” of imprinting is not attributable to a shift in promoter usage. In the liver and other tissues, such as the choroid plexus, previous observations of relaxation of IGF-2imprinting could have been attributable to erasure of theIGF-2 imprint rather than disregard of it. In contrast, our observations suggest that the imprint can be present without necessarily resulting in transcriptional silencing—that is, it can be disregarded by the transcriptional process during specific developmental stages of hematopoietic cell maturation. This conclusion is supported by our preliminary observation that peripheral blood mononuclear cells express IGF-2 biallelically within 24 hours of treatment with concanavalin A (data not shown), an observation supported by a previous report of biallelic IGF-2 expression in similarly stimulated mononuclear cells.21
Why is IGF-2 expressed from both alleles in the bone marrow? One possible explanation is that immature bone marrow cells have delayed imprint modification, as seen in the early embryo in which the establishment of genomic imprinting is believed to be dependent on postfertilization modification of the imprint mark. However, we found no evidence for disruption of the imprinting of SNRPN andH19, suggesting that the change in imprinted expression resulted from a process specific to IGF-2. A second explanation suggests that the relaxation of IGF-2 imprinting would provide a mechanism by which effects of the IGF axis are enhanced. A hematopoietic growth-promoting role for the increased local levels of IGF-2 is supported by an extensive literature that suggests IGF-2 can promote erythroid,22-26myeloid,27-29 and lymphoid growth and function.21 Imprinting of IGF-2R in the marrow could further enhance the IGF axis by reducing its function as anIGF-2 “sink.”30 Although most investigators18 have found no evidence for IGF-2Rimprinting in humans, it is imprinted in mice, it maintains some allele-specific methylation in humans, and it has been reported to be imprinted in some persons. Thus we examined whether IGF-2Ris imprinted in human bone marrow but determined that its expression is biallelic.
In the literature there are some reports of biallelic IGF-2expression in hematopoietic tissues. For example, Giannoukakis et al20 have reported biallelic expression of IGF-2in cord blood. We31 have shown the expression of both alleles in one cord blood sample, a tissue known to contain immature hematopoietic cells. Taken together with our current observations of biallelic IGF-2 expression in bone marrow, it appears likely that both alleles of IGF-2 are expressed in tissues that contain immature hematopoietic cells. This is supported by evidence that neoplastic proliferations of immature hematopoietic cells can also have biallelic IGF-2 expression, reflecting the immature phenotype of the affected cells. We have demonstrated this in one case of secondary AML, whereas Wu et al19 reported biallelic transcription of IGF-2 in several cases of AML. Their cases included leukemias with either myeloid or monocytic differentiation, suggesting that both lineages might express IGF-2 during normal hematopoiesis. Similarly, loss of IGF-2 imprinting has been associated with disease progression in chronic myeloid leukemia, an observation that might reflect the increasing proportion of immature cells.32 Given the considerable cellular heterogeneity of bone marrow, it would be informative to examineIGF-2 imprinting in well-defined, lineage-specific developmental subsets of normal bone marrow to confirm that the reported results from chronic myeloid leukemia and acute myeloid leukemia reflect the imprinting state of developmentally comparable normal hematopoietic cells. By defining the lineage and maturational specificity of changes in IGF-2 imprinting, the role of IGF-2 in normal hematopoiesis will be further defined. This could be achieved with the use of appropriate hematopoietic cell lines or purified cell populations. Additionally, given the differences inIGF-2 expression between fetal and adult tissues,33 it is likely that fetal hematopoietic tissues (yolk sac, liver, and bone marrow) will show different patterns ofIGF-2 imprinting than adult bone marrow. The study of a variety of hematopoietic tissues will not only document the role of IGF-2 in hematopoiesis but may also elucidate the mechanisms through which IGF-2 imprinting can be modulated. For example, what is the relation between the processes that establish the imprint in the early embryo and the processes that modulate tissue-specific changes? Does the recently documented chromatin boundary-associated control region upstream of the H19 gene34-36 play a role in tissue-specific imprinting?
Disruption of IGF-2 imprinting is a common event in several malignancies and in some growth disorders. By observing and manipulating the modulation of IGF-2 imprinting with the use of appropriate hematopoietic tissues, the mechanisms through which imprinting is maintained and disrupted will be revealed.
Supported by the Health Research Council of New Zealand, The Cancer Society of New Zealand, and the New Zealand Lottery Grants Board.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Ian M. Morison, Cancer Genetics Laboratory, Department of Biochemistry, University of Otago, PO Box 56, Dunedin, New Zealand; e-mail:ian.morison@stonebow.otago.ac.nz.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal