Abstract
The pathophysiologic role of the Philadelphia chromosome translocation in chronic myelogenous leukemia (CML) has been known for nearly 20 years. However, the most significant morbidity and mortality in CML are caused by progression to blast crisis, about which comparatively little is known at the molecular level. Genomic imprinting is a chromosomal modification leading to parental-origin–specific gene expression in somatic cells. Recently, we and others have described loss of imprinting (LOI) of the insulin-like growth factor-II gene (IGF2), leading to biallelic rather than monoallelic expression in a wide variety of solid tumors. We have now examined the imprinting status of IGF2 in samples from CML patients in stable phase, accelerated phase, and blast crisis. Five of six stable-phase patients showed normal imprinting, but LOI was found in all six cases of advanced disease (three accelerated phase, three blast crisis), which was statistically highly significant (P < .01). Thus, LOI represents a novel type of genetic alteration in CML that appears to be specifically associated with disease progression.
CHRONIC MYELOGENOUS leukemia (CML) is characterized by an expansion of myeloid cells containing the hallmark Philadelphia chromosome translocation.1 This translocation leads to formation of the bcr-abl fusion gene, whose protein product is a highly active tyrosine kinase implicated in the pathogenesis of the disease.2,3 CML has been shown to evolve hematologically from an initial chronic phase to an accelerated phase, finally culminating in an aggressive, fulminant blast crisis.2,3 This evolution is characterized by the appearance of progressively immature myeloid cells as well as the acquisition of additional cytogenetic anomalies.3 The most frequent genetic alterations in progression to blast crisis are deletions and mutations of p53, occurring in 20% to 30% of patients, and amplification of c-myc in 20% of patients.4,5 Other events occurring more rarely include mutations in ras (6%) and rearrangement and deletions of RB (14%) and p16 (15%).6-8
Genomic imprinting is a modification of a gene or the chromosome on which it resides, in the gamete or zygote, leading to differential expression of the two parental alleles of the gene in somatic cells of the offspring. Imprinting usually involves monoallelic expression of either the paternal or maternal allele, depending on the gene. Genomic imprinting is thought to be important in the development of cancer because loss of heterozygosity (LOH) in cancer often involves one of the parental chromosomes preferentially,9 and some imprinted genes are involved directly in the regulation of cell growth, such as insulin-like growth factor-II (IGF2).9 Furthermore, we and others have found loss of imprinting (LOI) in cancer, resulting in loss of parental-origin–specific differential allele expression.10 LOI usually leads to activation of the normally silent maternal copy of IGF2, which is expressed normally only from the paternal allele. LOI can also lead to epigenetic silencing of growth inhibitory genes such as p57KIP2 and H19.11,12 Although LOI was originally observed in embryonal tumors of childhood, including Wilms' tumor, hepatoblastoma, and rhabdomyosarcoma, it has now been found in a wide variety of adult solid tumors, including those of the prostate, breast, liver, and lung.9
Because LOI is one of the most common genetic alterations in solid tumors but has not been examined previously in CML, we sought to determine whether there is abnormal imprinting in CML, and if so whether it is specifically associated with disease progression.
MATERIALS AND METHODS
Screening for informative CML patients by DNA polymerase chain reaction (PCR).
Peripheral blood and bone marrow samples from CML patients were diluted 1:1 with phosphate-buffered saline (PBS) and layered over a discontinuous Percoll (Pharmacia, Uppsala, Sweden) gradient of specific gravity 1.077. The samples were centrifuged at 1,500 rpm for 7 to 10 minutes, and the white cells in the supernatant were washed with PBS and processed for DNA and RNA. For DNA extraction the cells were incubated overnight at 42°C, in a lysis buffer containing 50 mmol/L Tris (pH 8.0), 150 mmol/L NaCl, 25 mmol/L EDTA, 0.5% sodium dodecyl sulfate (SDS), and 0.1 mg/mL Proteinase K. The lysate was extracted with phenol/chloroform, ethanol precipitated, and resuspended in TE buffer. The genomic DNA was analyzed by PCR for anApa I polymorphism in IGF2, as previously described.10 PCR products were electrophoresed on 3%/1% Nuseive/agarose gels.
Imprinting analysis by reverse transcription polymerase chain reaction (RT-PCR).
Samples from heterozygous patients were processed for total RNA using RNAzol B (Tel-Test) following the manufacturer's instructions. To avoid any possible contamination by genomic DNA, the total RNA was digested with DNase I and then extracted with phenol/chloroform. To ensure the absence of genomic DNA contamination, all RT-PCR analysis was performed in duplicate in the presence and absence of reverse transcriptase (AMV-RT; Promega) using primers as described and PCR conditions as follows: 94°C for 3 minutes; 35 cycles of 94°C for 1 minute; 55°C for 1 minute; and 72°C for 1.5 minutes followed by a 10-minute extension at 72°C.12 The samples were electrophoresed on 3%/1% Nusieve/agarose gels.
Analysis of H19 methylation.
DNA extracted from the CML patient samples was screened for a HhaI polymorphism upstream of the H19 gene.13 Heterozygous patients were analyzed for methylation using HpaII. To assess allele-specific methylation, the genomic DNA from informative patients was digested with an excess of HpaII, PCR amplified, and digested with Hha I, as described.13
RESULTS
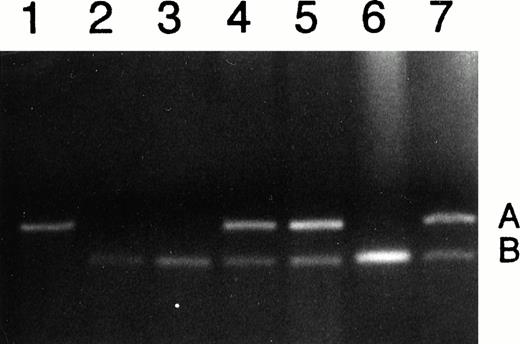
Our primary goal was to determine whether patients with CML maintain normal imprinting or undergo LOI. To analyze genomic imprinting of IGF2, we first identified CML patients who were heterozygous for a transcribed Apa I polymorphism in the IGF2 gene. Of 21 chronic-phase patients 6 were informative (heterozygous); of 7 accelerated-phase patients 3 were informative; and of 12 patients in blast crisis, 3 were informative for the Apa I polymorphism. For example, Fig 1 lane 1 shows a patient homozygous for the A allele (236 bp), and lanes 2, 3, and 6 show patients homozygous for the B allele (173 bp) and thus were not informative. In contrast, lanes 5, 6, and 7 show patients having both A and B alleles and are thus informative for the IGF2 polymorphism.
Analysis of DNA from CML patients for heterozygosity of IGF2. Lane 1 shows patient homozygous for the A allele; lanes 2, 3, and 6 show patients homozygous for the B allele; and lanes 4, 5, and 7 show heterozygotes, with both A and B alleles.
Analysis of DNA from CML patients for heterozygosity of IGF2. Lane 1 shows patient homozygous for the A allele; lanes 2, 3, and 6 show patients homozygous for the B allele; and lanes 4, 5, and 7 show heterozygotes, with both A and B alleles.
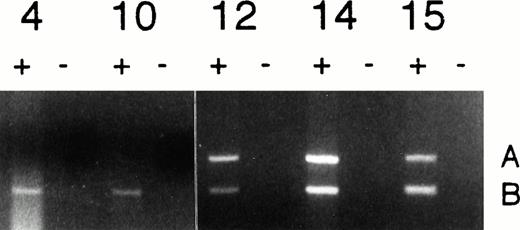
A total of 40 patient samples were tested for the polymorphism, of which 12 informative samples were analyzed for imprinting status by performing RT-PCR for the Apa I polymorphism, as described in Materials and Methods. An example of the analysis of genomic imprinting is shown in Fig 2. Patients 4 and 10, who were in chronic phase, showed expression of only the B allele after RT-PCR, indicating normal monoallelic expression of IGF2 in their leukemic cells. In contrast, patient 12 (accelerated phase) and patients 14 and 15 (both in blast crisis) showed biallelic expression of IGF2 (Fig 2). In 3 samples, LOI was also confirmed using exon connection between exon 8 and exon 9. Of the 6 informative patients in chronic phase, 5 exhibited normal monoallelic IGF2 expression and only one (patient 6) showed biallelic expression of IGF2 gene in his leukemic cells. This patient had an atypical clinical course in that he was unresponsive to therapy, and he died 3 months after his blood sample was obtained because of failure of engraftment after allogeneic bone marrow transplantation.
Analysis of Imprinting of IGF2 in CML. Normal monoallelic expression was seen in patients 4 and 10 (chronic phase), and biallelic expression was observed in patients 12 (accelerated phase), 14, and 15 (blast crisis). All reactions were performed in duplicate in the presence (+) and absence (−) of reverse transcriptase.
Analysis of Imprinting of IGF2 in CML. Normal monoallelic expression was seen in patients 4 and 10 (chronic phase), and biallelic expression was observed in patients 12 (accelerated phase), 14, and 15 (blast crisis). All reactions were performed in duplicate in the presence (+) and absence (−) of reverse transcriptase.
In contrast to most of the stable-phase patients, all three informative patients in accelerated stage and all three informative blast crisis patients exhibited biallelic expression of the IGF2 gene in their leukemic cells (Table 1). The difference between the frequency of LOI in chronic phase (1 of 6) compared with that of accelerated phase (3 of 3) was statistically highly significant (P < .05), as was the difference between the frequency of LOI in chronic phase and that of blast crisis (3 of 3; P < .05). The frequency of LOI in overall advanced-stage disease compared with chronic phase was even more strikingly significant (P < .01). Thus, LOI is a frequent alteration in accelerated phase and blast crisis in CML, and it appears to be specific for disease progression.
Loss of Imprinting in CML
| . | LOI . | Non-LOI . |
|---|---|---|
| Stable phase | 1/6 | 5/6 |
| Accelerated phase | 3/3* | 0/3 |
| Blast crisis | 3/3* | 0/3 |
| Total advanced disease | 6/6-151 | 0/6 |
| . | LOI . | Non-LOI . |
|---|---|---|
| Stable phase | 1/6 | 5/6 |
| Accelerated phase | 3/3* | 0/3 |
| Blast crisis | 3/3* | 0/3 |
| Total advanced disease | 6/6-151 | 0/6 |
*P < .05, compared with stable phase (Fisher's one-tailed exact test).
P < .01.
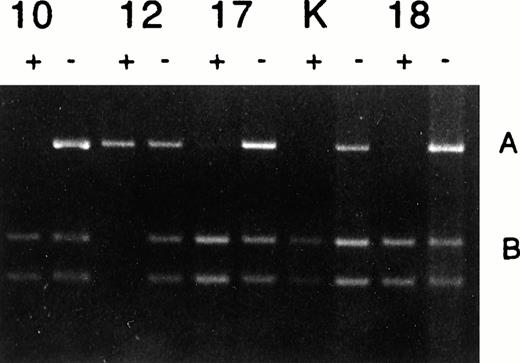
In some solid tumors, such as Wilms' tumor, LOI is associated with abnormal methylation of a CpG island upstream of the neighboring H19 gene, which is normally methylated only on the paternal allele.12 In other patients, LOI of IGF2 can occur independently of methylation changes in the H19 gene.14,15To determine whether LOI in CML was methylation-dependent or independent, we used a PCR-based assay, which we previously described, to detect an Hha I polymorphism within the H19 CpG island and that distinguishes between the two alleles in heterozygous patients.13 Four patients were heterozygous and thus informative for the H19 Hha I polymorphism. As expected, a chronic-phase patient (patient 10) with normal imprinting of IGF2 showed monoallelic methylation of the H19 CpG island. Of the four informative patients, one was in accelerated phase (patient 12) and exhibited LOI of IGF2 (Fig 2). This patient also showed normal methylation of the H19 CpG island, ie, methylation of only one allele (Fig 3). Thus, LOI of IGF2 occurred independent of hypermethylation of the H19 CpG island in this patient. Two additional patients in blast crisis (patients 17 and 18) also showed monoallelic methylation of the H19 CpG island. Thus, it is likely that LOI of IGF2 in CML occurs by a pathway independent of H19 methylation. Interestingly, the blast crisis cell line K562 also displayed a monoallelic pattern of H19 methylation (Fig 3), although it has been reported to show hypermethylation at other loci such as the calcitonin and bcr genes.18-20
Analysis of H19 promoter methylation in CML. Patients 10, 12, 17, 18, and cell line K562 (K) showed normal monoallelic methylation. Samples pretreated (+) or not pretreated (−) withHpaII, PCR amplified and then digested with Hha I, which detects the H19 polymorphism.
Analysis of H19 promoter methylation in CML. Patients 10, 12, 17, 18, and cell line K562 (K) showed normal monoallelic methylation. Samples pretreated (+) or not pretreated (−) withHpaII, PCR amplified and then digested with Hha I, which detects the H19 polymorphism.
DISCUSSION
In summary, we found that six of six patients with advanced CML, three in blast crisis and three in accelerated phase, showed loss of imprinting (LOI) of IGF2, but only one of six patients in chronic phase exhibited LOI. This patient was somewhat unusual in that he was refractory to treatment and died 3 months after the sample was taken. The high frequency of LOI in advanced disease is more frequent than that of other genetic alterations previously observed.4-8
These results are important for two reasons. First, they extend our observation of abnormal imprinting in cancer to this common leukemia in adults. In addition, these results suggest a specific relationship of LOI to disease progression. Recently, it was shown that IGF2 was normally imprinted in blood and in early hematopoietic precursors and that LOI occurs in 50% of acute myelogenous leukemia patients, although relationship to disease progression was not determined.19 We plan to examine LOI prospectively in a larger series of patients to determine whether it will serve as a useful prognostic marker for disease progression in CML. It is intriguing in this regard that the one stable-phase patient with LOI was refractory to therapy. Finally, even though LOI was independent of H19 methylation, alterations in DNA methylation at other loci have been described in CML progression.16-18 Thus, it will be important to examine other imprint-specific sites of DNA methylation, such as within the IGF2 gene itself.14 Regardless of the mechanism of LOI in CML, little is known about the molecular basis of progression to blast crisis, and thus the data reported here provide a new direction for further study.
How might loss of imprinting be functionally related to disease progression in CML? We and others have identified a large (approximately 1 Mb) genomic domain of 11p15 that includes at least eight imprinted genes,9 several of which are predicted to have an effect on cell growth, including the following: IGF29,10; H199,10; p57KIP2, a cyclin-dependent kinase inhibitor11,20; and TSSC3 (the human homologue of mouse apoptosis-related gene TDAG51).21,22 We have also found that loss of imprinting can affect one or more of these genes simultaneously within tumors.9,11 20-22 Our observation of LOI of IGF2 in CML thus leads to the following questions:
Do any of the other genes within this complex show altered imprinting in CML?
What is the consequence of altered imprinting on the expression of these genes in advanced stages of CML? Preliminary analysis does not suggest a significant increase in IGF2 expression in patients with advanced CML (data not shown), although other tumors with LOI also do not show a dramatic increase in IGF2 expression,23 even though LOI is associated with predisposition to malignancy,24 and it appears to be a necessary step in tumor progression.25 Thus, deregulated expression of IGF2, rather than overall expression per se, may be important, or other genes within this cluster may be more important in the pathophysiology of CML.
What is the relationship between LOI and other epigenetic changes in CML, specifically altered DNA methylation? At least six CpG dinucleotide-rich “CpG islands” have been reported within this cluster.10,12-14,26 Because altered DNA methylation at other chromosomal sites is a feature of CML progression,18it will be important to determine whether altered methylation of this imprinted domain also plays a role in CML.
Regardless of the answers to these questions, the data provided here that show that LOI is an important marker for disease progression, and it may serve as an important new prognostic marker for predicting response to therapy. This strong association itself suggests a biologically important role for LOI in CML.
ACKNOWLEDGMENT
We thank Dr Richard Jones for generously providing some of the samples, Leslie Calvert for technical assistance, and Pam Hill for preparing the manuscript.
G.S.R. and H.C. contributed equally to this work.
Supported by Grant Nos. CA49639 and CA65145 from the National Institutes of Health.
Address reprint requests to Andrew P. Feinberg, MD, MPH, Johns Hopkins University School of Medicine, 1064 Ross, 720 Rutland Ave, Baltimore, MD 21205.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal